Abstract
Ligand-independent signals that are produced by the B-cell antigen receptor (BCR) confer an important positive selection checkpoint for immature B cells. Generation of inappropriate signals imposes developmental arrest of immature B cells, though the fate of these cells has not been investigated. Studies have shown that the lack of CD19 results in inappropriate signaling. In immunoglobulin transgenic mice, this inappropriate signaling impairs positive selection and stimulates receptor editing. Here, we studied the extent and significance of receptor editing in CD19-regulated positive selection of normal, nontransgenic B lymphopoiesis, using our bone marrow culture system. We found that the lack of CD19 resulted in elevated tonic signaling and impaired maturation, as revealed by surface marker expression and by functional assays. Immature CD19-/- B cells did not suppress RAG and underwent intensive receptor editing attempts in culture. Finally, in vivo analysis of light-chain isotype expression and Jκ use in CD19-/- mice validated our in vitro observations. Our results suggest that CD19 has an important function in regulating positive selection and maturation of nontransgenic B-cell precursors and that receptor editing is an important salvage mechanism for immature B cells that fail positive selection. (Blood. 2005;105:3247-3254)
Introduction
The developmental transition in B lymphopoiesis from an immature stage to a mature stage is limited by negative and positive selection checkpoints.1-4 Self-antigen binding to surface immunoglobulin M (IgM) imposes developmental arrest and stimulates intensive receptor editing attempts.5,6 This mechanism of negative selection is developmentally regulated, and the competence to undergo receptor editing is lost with maturation.7,8 Impaired maturation and developmental arrest are also imposed in many signaling mutants,9-12 implicating that maturation of non-self B cells is limited by positive selection signals (for reviews, see Niiro and Clark1 and Edry and Melamed2 ). Recently, it has been shown that these signals are generated by the B-cell receptor (BCR) and are ligand independent.13 However, the fate of B cells that fail positive selection because of inappropriate BCR signaling is unknown.
CD19 is a B-cell-restricted signaling molecule that functions as a positive regulator of BCR signaling.14,15 Lack of CD19 results in impaired BCR signaling,16,17 and mice deficient in CD19 have decreased peripheral B-cell numbers that are hyporesponsive to BCR stimuli.18,19 Although negative selection is not impaired in CD19-deficient mice,20 the effect of CD19 in regulating positive selection in B lymphopoiesis is less known. Initial in vivo analysis suggested that bone marrow (BM) development of B lymphocytes is not affected in CD19-/- mice.18,19 However, it was recently shown that the lack of CD19 impairs pre-BCR signaling and blocks positive selection of pro-B to pre-B.21 Using an immunoglobulin transgenic (Ig Tg) mouse model (3-83Tg), we have shown that the lack of CD19 modifies ligand-independent BCR signals22 and blocks BCR-mediated positive selection of immature 3-83Tg B cells.23 As a consequence, these 3-83Tg CD19-/- immature B cells undergo intensive receptor editing.22,23 Despite these findings, it is unclear whether, and how frequently, receptor editing is used in normal, non-Tg immature B cells that fail positive selection. It is also unclear whether CD19 has any regulatory function in positive selection because in vivo BM analysis of CD19-/- mice reveals no apparent defect.18,19 This, in part, may result from the unknown extent of compensatory mechanisms available in the preselected B-cell repertoire and may be attributed to the dynamic nature of B-cell development in a non-Tg system. In the present study, we have used our nonselective BM culture system to probe for the direct effect of CD19 in regulating non-Tg B-cell maturation and stimulation of receptor editing. We report here that in the absence of CD19, immature B cells produce inappropriately elevated ligand-independent signals. These B cells undergo developmental arrest and stimulate intensive receptor editing attempts in culture, which are also evident in vivo. Our results suggest that CD19 has an important function in regulating positive selection and maturation of non-Tg B-cell precursors and that receptor editing is an important salvage mechanism for immature B cells that fail positive selection.
Materials and methods
Experimental mice
Mice used were wild-type (wt) B10.D2nSn/J- or CD19-deficient littermates.19 Mice were housed and bred at the animal facility of the Technion Faculty of Medicine and were used for the experiments at 4 to 8 weeks of age.
Cell culture
BM cultures for B-cell precursors were prepared as previously described.24 Briefly, femoral BM cells, depleted of red blood cells (RBCs), were cultured for 5 days in Iscove modified Dulbecco medium (IMDM) with 50 to 100 U/mL recombinant interleukin-7 (rIL-7). Cells grown in these primary cultures (more than 95% B220+) were used directly for cellular and molecular analysis. In some experiments, cells were washed and recultured in the absence of IL-7 before analysis. Additionally, in some experiments, BM culture cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR) and were injected intravenously into B10D2 mice. Spleen cells of recipient mice were analyzed for the presence of CFSE-labeled cells by fluorescence-activated cell sorter (FACS) 6 hours after transfer.
Flow cytometry and cell sorting
Single-cell suspensions from BM cultures or spleen were stained for surface marker expression using fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, and biotin-conjugated antibodies, visualized with streptavidin SPRD (Southern Biotechnology Associates [SBA], Birmingham, AL). Antibodies used for cell staining were B220 RA3-6B2, CD23 clone 2G8, CD62L Mel14, and Igκ (all from SBA); IgM and Igλ (Caltag Laboratories, Burlingame, CA); and IgD JA12.5.29 Data for 3-color analysis were collected on a FACSCalibur and analyzed using CELLQuest software (BD Biosciences, Immunocytometry Systems, Mountain View, CA). For all analyses, forward and side scatter gates were set to include viable cells and to exclude dead cells and debris. In some experiments, cultured BM cells were fractionated to pro- and pre-B cells (B220+IgM-), immature B cells (IgM+IgD-), and transitional B cells (IgM+IgD+) using magnetic MACS microbeads (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Splenic B cells were also purified using MACS microbeads. In some experiments, cell death was determined by TdT-mediated dUTP nick-end labeling (TUNEL) assay (Roche Applied Science, Mannheim, Germany) as described.7
Analysis of tyrosine phosphorylation
Tyrosine phosphorylation in B-cell precursors grown in BM cultures was performed as previously described.22 Cells were lysed, and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA). Phosphorylation of Igα was determined after immunoprecipitation of Igα/Igβ heterocomplexes using anti-Igβ antibody (HM79) directly coupled to Sepharose beads. For total tyrosine phosphorylation and Igα phosphorylation, blots were probed with antiphosphotyrosine (4G10) followed by rabbit anti-Igα antisera. Tyrosine phosphorylation of ERK1/2 was performed using rabbit antiphosphor-extracellular signal-related kinase (ERK)1/2, followed by rabbit anti-ERK1/2 (both from Cell Signaling Technology, Beverly, MA). Visualization of bound antibodies was performed by enhanced chemiluminescence (ECL) reagent. To determine levels of tyrosine phosphorylation of specific signaling intermediaries, densitometry analysis was performed using the UVIdoc Gel documentation system and UVIdoc analysis software (UVItec Cambridge, United Kingdom), and the ratio between the phosphorylated band and the corresponding total band was calculated as described.22
RNA and cDNA analysis
Total RNA was purified using RNA-Bee (Tel-Test, Friendswood, TX). RNA samples were reverse transcribed to cDNA and polymerase chain reaction (PCR) amplified to detect CXCR4, CXCR5, RAG-1, and β-actin. PCR conditions for CXCR4 were 30 seconds at 94°C, 30 seconds at 50°C, and 45 seconds at 72°C for 31 cycles; primer sequences for CXCR4 were: 5′-ggctgtagagcgagtgttgc-3′ and 5′-gtagaggttgacagtgtagat-3′. PCR conditions for CXCR5 were 30 seconds at 94°C, 40 seconds at 58°C, and 40 seconds at 72°C for 36 cycles; primer sequences for CXCR5 were 5′-aaacgaagcggaaactagagcc-3′ and 5′gcccagcttggtcagaagcc-3′. PCR conditions and primer sequences for RAG-1 and β-actin were as described.24 PCR products were fractionated on 1% agarose and quantified using UVIdoc gel documentation system and UVIdoc software (UVItec), as previously described.25 Results are presented as a semiquantitative estimate, where signal intensity of CXCR4 or CXCR5 or RAG-1 products was normalized to that of β-actin.
Analysis of antibodies
B cells grown in BM cultures were washed and cultured for 48 hours at 106 cells/mL in the presence or absence of lipopolysaccharide (LPS) (50 μg/mL). Total IgM, IgG, Igκ, and Igλ in the supernatants were determined by sandwich enzyme-linked immunosorbent assay (ELISA), using specific goat antimouse polyclonal reagents (SBA). Antibody concentrations were calculated using a reference standard curve of purified IgM or IgG antibodies. In some experiments, IgM/κ, IgM/λ, IgG/κ, and IgG/λ in serum samples were determined using the same reagents and conditions. Serum samples were collected from 6- to 8-week-old mice.
ELISPOT
Detection of antibody-forming plasma cells by enzyme-linked immunospot (ELISPOT) assay was performed as previously described.26 In brief, nitrocellulose membranes were placed in 5-mL tissue culture Petri dishes and were coated with a mix of 2.5 μg/mL goat anti-mouse κ and goat anti-mouse λ (SBA). Membranes were then washed (washing solution, 0.1% Tween 20 in Tris-buffered saline [TBS]) and blocked (blocking solution, 10% of 1% fat durable milk in phosphate-buffered saline [PBS]). Cells from BM cultures stimulated with LPS (50 μg/mL) for 48 hours were washed and recultured on top of membranes for 14 to 16 hours. After incubation, membranes were extensively washed and incubated with goat anti-mouse IgM biotinylated antibody. Streptavidin conjugated with horseradish peroxidase was used as a secondary probe, and spot signals were generated by the ECL reaction.
Jκ use analysis
mRNA samples from sorted IgM+ splenic cells were purified, reverse transcribed to cDNA, and PCR amplified using degenerate Vκ framework-3 (FW3) and Cκ primers, as described.26 The resultant Vκ-Cκ products were cloned, and single colonies were sequenced to determine the Jκ in each, using GenBank and IgBlast databases. For this analysis, only productive Vκ-Jκ rearrangements, as determined based on the presence of conserved amino acid residues,27 were considered.
Chemokine-induced migration
Chemokine-induced migration assay was carried out as described.28 In brief, 5 × 105 BM culture cells or splenic cells in 100 μL RPMI 1640 containing 0.25% bovine serum albumin were added to 5-μm pore size, bare-filter Transwell inserts (Costar, Cambridge, MA). Recombinant CXCL12 (SDF1-α) (250 ng/mL) (PeproTech, Rocky Hill, NJ) or recombinant CXCL13 (B lymphocyte chemoattractant [BLC]) (2 mg/mL) (R&D Systems, Minneapolis, MN) were added to the lower chamber. The assay proceeded for 3 hours. FACS was used to count transmigrating cells. In some experiments, the phenotypes of migrating cells were determined by staining for B220 IgM and IgD, followed by FACS analysis.
Statistical analysis
The statistical significance of differences between experimental groups was determined using unpaired Student t test, with differences considered significant at P less than .05.
Results
Lack of CD19 impairs maturation of developing B cells
The lack of CD19 results in impaired signaling of the antigen receptor16,17 and in a developmental block of the pro-B/pre-B-cell transition.29 We used our in vitro culture system to determine whether lack of CD19 impairs late stages of B-cell maturation. To do so, BM cultures from wt and CD19-deficient mice were prepared, and the maturation of B-cell precursors was determined by surface expression of B220, IgM, IgD, CD23, and CD62L (L-selectin). In both cultures, equivalent cell numbers were obtained (not shown), and the cells grown were predominantly B cells (approximately 95% B220+) (Figure 1A). However, in CD19-/- cultures, there was a profound delay in B-cell maturation, as revealed by a significant reduction in cells expressing IgM (approximately 25% relative to approximately 45% in wt cultures), IgD (approximately 5% relative to approximately 25%), CD23 (approximately 2% relative to approximately 11%), CD62L (approximately 1% relative to approximately 9%), and B220hi (1% relative to approximately 18%) (Figure 1). On removal of IL-7 for 24 hours, further differentiation was induced in both cultures, as measured by the increased proportion of IgM+/IgD+ B cells, though it was more profound in wt cultures (Figure 1B). Because removal of IL-7 also results in extensive cell loss,30 we analyzed for differential apoptosis rates between wt and CD19-/- cultures. We found similar apoptosis rates in both cultures (Figure 1C), suggesting that there is no selective or increased death in CD19-/- cultures. Thus, we conclude that lack of CD19 impairs B-cell maturation in vitro.
Lack of CD19 impairs B-cell maturation in BM cultures. (A) Cells grown in BM cultures were stained for the indicated surface markers and analyzed by FACS. Results from 1 of 4 representative experiments are shown. Numbers in graphs indicate cell percentages within the region. (B) Cells grown in BM cultures (□, wild type [wt];▵, CD19-/-) were washed and recultured in the absence of IL-7 for 12 to 24 hours. Cells were then stained for B220 IgM and IgD and analyzed by FACS. The induction of differentiation was quantified by calculating the relative proportion of IgM+/IgD+ cells in the total IgM+ fraction. Results presented as mean ± SE of 4 different experiments. (C) Apoptosis rates in wt and CD19-/- BM cultures were determined 12 to 24 hours after IL-7 withdrawal by TUNEL. Symbols indicate the same cell types as in panel B. Results presented as mean ± SE of 4 different experiments.
Lack of CD19 impairs B-cell maturation in BM cultures. (A) Cells grown in BM cultures were stained for the indicated surface markers and analyzed by FACS. Results from 1 of 4 representative experiments are shown. Numbers in graphs indicate cell percentages within the region. (B) Cells grown in BM cultures (□, wild type [wt];▵, CD19-/-) were washed and recultured in the absence of IL-7 for 12 to 24 hours. Cells were then stained for B220 IgM and IgD and analyzed by FACS. The induction of differentiation was quantified by calculating the relative proportion of IgM+/IgD+ cells in the total IgM+ fraction. Results presented as mean ± SE of 4 different experiments. (C) Apoptosis rates in wt and CD19-/- BM cultures were determined 12 to 24 hours after IL-7 withdrawal by TUNEL. Symbols indicate the same cell types as in panel B. Results presented as mean ± SE of 4 different experiments.
Chemokine responsiveness of wt and CD19-/- B-cell precursors
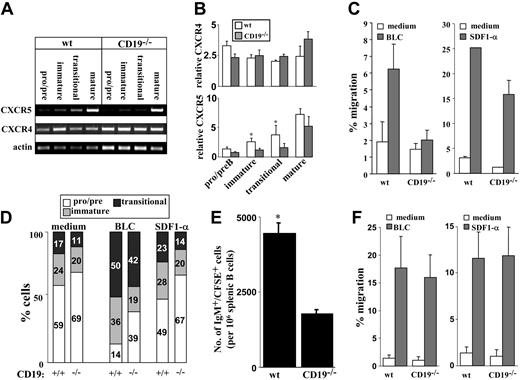
The ability of B cells to leave the BM and migrate to the periphery is coordinated through their chemotactic responses, which are intimately correlated with their developmental stage. Hence, we tested responsiveness to SDF1-α and BLC, which are central cytokines in B lymphopoiesis,31 at the levels of chemokine receptor expression and chemokine-induced migration. Cultured wt and CD19-/- BM B cells were sorted as pro-/pre- (IgM-), immature (IgM+IgD-), and transitional (IgM+IgD+). Mature B cells were isolated from spleen. Expression of CXCR4 (receptor for SDF1-α) and CXCR5 (receptor for BLC) were determined for each sorted population using reverse transcription-PCR (RT-PCR). Results in Figure 2A-B show that in wt and CD19-/- cells, CXCR4 mRNA is detected in all stages of B-cell development, with no significant change. We found that CXCR5 mRNA is increased with maturation but is expressed at lower levels in CD19-/- immature and transitional (P < .05). Thus, although CXCR4 is expressed through-out B-cell development, CXCR5 is developmentally regulated. To further test for chemokine responsiveness, migration assays were performed using transwell inserts. We found that responsiveness of CD19-/- precursors to SDF1-α was slightly reduced, whereas migration of these cells to BLC was severely impaired (Figure 2C). Further phenotypic analysis of CD19-/- and wt cells migrating to SDF1-α revealed no significant advantage to any of the B-cell populations (Figure 2D). In contrast, migration responsiveness to BLC is correlated with the CXCR5 expression and is significantly enhanced in transitional cells. Analysis of wt cultures revealed that most of the BLC-migrating cells are transitional B cells, as reflected by the 2- to 3-fold increase in their relative proportions (from 17% in the culture to 50%; Figure 2D). It should be noted that the small population of CD19-/- precursors that responded to BLC was composed primarily of transitional cells (Figure 2D).
Chemokine receptor expression and chemokine responsiveness of wt and CD19-/- B-cell precursors. Wild-type and CD19-/- B-cell precursors were grown in BM cultures. (A) B-cell precursors were fractionated into pro-/pre- (IgM-), immature (IgM+IgD-), and transitional (IgM+IgD+) populations. Mature B cells were isolated from spleen. Cells were lysed rapidly, and the expression of CXCR4 and CXCR5 was determined for each sorted population by RT-PCR. Results shown are representative of 5 different experiments. Actin blot is shown as an internal control gene expression. (B) Quantification of CXCR4 and CXCR5 expression. Results are presented as a semiquantitative estimate, where signal intensity of CXCR4 or CXCR5 products was normalized to that of β-actin. □ indicates wt; ▦, CD19-/-. Results of 5 experiments are presented as mean ± SE. Asterisk indicates values that are statistically different (P < .05). (C) Total cell migration of wt and CD19-/- B-cell precursors. Cells grown in BM cultures were tested for migration capabilities to BLC (2 mg/mL, left, ▦) or to SDF1-α (250 ng/mL, right, ▦) using transwell inserts. □ indicates medium. Migrating cells were counted by FACS, and the percentage of migration was calculated as: no. migrating cells/no. loaded cells × 100. Results are expressed as mean ± SE of 3 different experiments. (D) Relative proportion of pro-/pre-, immature, and transitional populations in migrating cells. Cells migrating to medium (spontaneous), BLC, or SDF1-α were stained for B220 IgM and IgD to determine the relative proportions of pro-/pre- (□), immature (▦), and transitional cells (▪) by FACS analysis. Results are shown in bars as percentage of cells of each population. Results shown are representative of 3 different experiments. (E) Migration of IgM+ precursors to the spleen. CD19-deficient and wt B-cell precursors grown in BM cultures were labeled with CFSE and injected intravenously into recipient B10D2 mice (total injected cells were adjusted to include 5 × 106 IgM+ cells/mouse). Six hours after transfer, spleens of reconstituted mice were removed, and spleen cells were stained for B220 and IgM. Cells were analyzed by FACS to quantify the number of CFSE-labeled IgM+ cells. Results are presented as the number of CFSE-labeled IgM+ cells/106 spleen cells and are the mean ± SE of 3 different mice. Asterisk indicates values that are statistically different (P < .05). (F) Chemokine-induced migration of splenic B cells. Spleen cells from wt and CD19-/- mice were tested for migration response to SDF1-α (right, ▦) and to BLC (left, ▦) using a transwell system. □ indicates medium. Migrating cells were stained for B220 and IgM to determine the relative proportion of B cells. Results are presented in a migration index and are calculated as: no. migrating B cells/no. loaded B cells × 100. Results are expressed as mean ± SE of 3 different experiments.
Chemokine receptor expression and chemokine responsiveness of wt and CD19-/- B-cell precursors. Wild-type and CD19-/- B-cell precursors were grown in BM cultures. (A) B-cell precursors were fractionated into pro-/pre- (IgM-), immature (IgM+IgD-), and transitional (IgM+IgD+) populations. Mature B cells were isolated from spleen. Cells were lysed rapidly, and the expression of CXCR4 and CXCR5 was determined for each sorted population by RT-PCR. Results shown are representative of 5 different experiments. Actin blot is shown as an internal control gene expression. (B) Quantification of CXCR4 and CXCR5 expression. Results are presented as a semiquantitative estimate, where signal intensity of CXCR4 or CXCR5 products was normalized to that of β-actin. □ indicates wt; ▦, CD19-/-. Results of 5 experiments are presented as mean ± SE. Asterisk indicates values that are statistically different (P < .05). (C) Total cell migration of wt and CD19-/- B-cell precursors. Cells grown in BM cultures were tested for migration capabilities to BLC (2 mg/mL, left, ▦) or to SDF1-α (250 ng/mL, right, ▦) using transwell inserts. □ indicates medium. Migrating cells were counted by FACS, and the percentage of migration was calculated as: no. migrating cells/no. loaded cells × 100. Results are expressed as mean ± SE of 3 different experiments. (D) Relative proportion of pro-/pre-, immature, and transitional populations in migrating cells. Cells migrating to medium (spontaneous), BLC, or SDF1-α were stained for B220 IgM and IgD to determine the relative proportions of pro-/pre- (□), immature (▦), and transitional cells (▪) by FACS analysis. Results are shown in bars as percentage of cells of each population. Results shown are representative of 3 different experiments. (E) Migration of IgM+ precursors to the spleen. CD19-deficient and wt B-cell precursors grown in BM cultures were labeled with CFSE and injected intravenously into recipient B10D2 mice (total injected cells were adjusted to include 5 × 106 IgM+ cells/mouse). Six hours after transfer, spleens of reconstituted mice were removed, and spleen cells were stained for B220 and IgM. Cells were analyzed by FACS to quantify the number of CFSE-labeled IgM+ cells. Results are presented as the number of CFSE-labeled IgM+ cells/106 spleen cells and are the mean ± SE of 3 different mice. Asterisk indicates values that are statistically different (P < .05). (F) Chemokine-induced migration of splenic B cells. Spleen cells from wt and CD19-/- mice were tested for migration response to SDF1-α (right, ▦) and to BLC (left, ▦) using a transwell system. □ indicates medium. Migrating cells were stained for B220 and IgM to determine the relative proportion of B cells. Results are presented in a migration index and are calculated as: no. migrating B cells/no. loaded B cells × 100. Results are expressed as mean ± SE of 3 different experiments.
To determine whether CD19-deficient cells also have impaired migration in vivo, we studied the migration of IgM+ precursors to the spleen by adoptive transfer experiments. B-cell precursors derived from wt or CD19-/- BM cultures were labeled with CFSE and injected intravenously into recipients. Migration of CFSE-labeled IgM+ cells to the spleen was determined and quantified by FACS. Figure 2E shows that the migration and accumulation of IgM+ precursors deficient in CD19 are reduced by 2-fold compared with those of the control. Thus, CD19-/- precursors have impaired migration in vitro and in vivo. However, despite this impaired response, CD19-/- mice have B cells in spleen, though in a reduced number. This suggests that the lack of CD19 can be compensated in vivo through an attempt to promote B-cell maturation and migration from the BM to the spleen.23 To confirm this, we analyzed the chemokine-induced migration of splenic B cells deficient in CD19 compared with wt splenic B cells. Results in Figure 2F show that CD19-/- splenic B cells respond to SDF1-α and to BLC as do wt B cells. Thus, cells that compensated for CD19 deficiency and migrated to the spleen also acquired the necessary chemokine responsiveness.
Responsiveness to LPS stimulation of wt and CD19-/- B-cell precursors
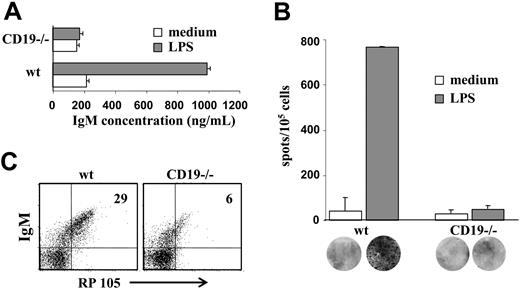
Immature B cells have been shown to proliferate and to secrete antibodies in response to LPS stimulation.32 To determine whether LPS responsiveness is altered in CD19-/- B-cell precursors, wt and CD19-/- BM culture cells were stimulated with LPS for 48 hours. Supernatants were collected, and IgM levels were determined by ELISA. The results in Figure 3A show that CD19-/- cells produced 5-fold less IgM (approximately 200 ng/mL compared with approximately 1000 ng/mL in wt). To determine whether the reduced amounts of IgM obtained in CD19-/- cultures reflected a low frequency of antibody-producing plasma cells or whether they resulted from low secretion capabilities, plasma cells were visualized and quantified by ELISPOT (Figure 3B). The results clearly show a significant (approximately 15-fold) reduction of plasma cell frequencies in CD19-/- cultures after LPS stimulation (approximately 50 clones/105 cells compared with approximately 750 clones/105 cells in wt). This decrease far exceeds what could be attributed to the fewer IgM+ cells in CD19-/- cultures relative to wt (less than 2-fold; Figure 1), suggesting that LPS responsiveness is severely impaired in CD19-/- cultures. Because RP105 is the main receptor for LPS in B lymphocytes, we measured RP105 expression in wt and CD19-/- B-cell precursors. The results in Figure 3C show that RP105 expression was significantly reduced in IgM+ cells derived from CD19-/- cultures (approximately 5% relative to approximately 25% in wt). We concluded that reduced RP105 expression in CD19-/- cultures contributed to the impaired responsiveness to LPS.
LPS responsiveness of wt and CD19-/- B-cell precursors. Wild-type and CD19-/- BM culture cells were stimulated with 50 μg/mL LPS for 48 hours. (A) Supernatants of stimulated cells were collected and assayed for IgM by ELISA. IgM concentrations were determined using a reference IgM standard curve and are expressed as nanograms per milliliter. ▦ indicates LPS; □, medium. Results are expressed as mean ± SE of 3 different experiments. (B) Visualization and quantification of antibody-producing plasma cells in the stimulated cultures. Cultured cells that were stimulated with LPS for 48 hours were then transferred to filters for analysis of plasma cells by an ELISPOT assay. Frequencies of IgM-producing plasma cells were calculated and expressed as number of IgM-producing cells per 105 precursor cells. ▦ indicates LPS; □, medium. Results are expressed as mean ± SE of 3 different experiments. Representative filters are shown for each bar. (C) Expression of RP105 in early B-cell precursors. Cultured cells were stained for IgM and RP105 and analyzed by FACS. Plots shown are representative of 3 experiments. Actin blot is shown as an internal control gene expression.
LPS responsiveness of wt and CD19-/- B-cell precursors. Wild-type and CD19-/- BM culture cells were stimulated with 50 μg/mL LPS for 48 hours. (A) Supernatants of stimulated cells were collected and assayed for IgM by ELISA. IgM concentrations were determined using a reference IgM standard curve and are expressed as nanograms per milliliter. ▦ indicates LPS; □, medium. Results are expressed as mean ± SE of 3 different experiments. (B) Visualization and quantification of antibody-producing plasma cells in the stimulated cultures. Cultured cells that were stimulated with LPS for 48 hours were then transferred to filters for analysis of plasma cells by an ELISPOT assay. Frequencies of IgM-producing plasma cells were calculated and expressed as number of IgM-producing cells per 105 precursor cells. ▦ indicates LPS; □, medium. Results are expressed as mean ± SE of 3 different experiments. Representative filters are shown for each bar. (C) Expression of RP105 in early B-cell precursors. Cultured cells were stained for IgM and RP105 and analyzed by FACS. Plots shown are representative of 3 experiments. Actin blot is shown as an internal control gene expression.
RAG-1 expression is maintained in immature CD19-/- B cells
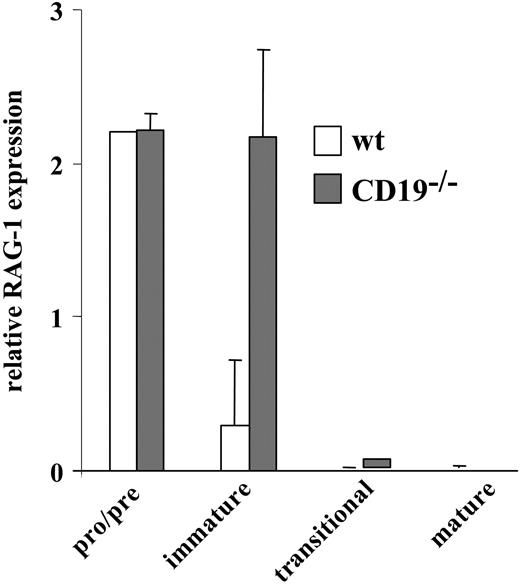
We have previously shown that inappropriate ligand-independent signals block positive selection and stimulate receptor editing.22 To determine the extent of receptor editing in the non-Tg BM culture from CD19-/- and wt mice, we measured RAG-1 expression levels in sorted pro-/pre- (IgM-), immature (IgM+/IgD-), transitional (IgM+/IgD+), and mature B cells (the latter were obtained from spleen) by RT-PCR (Figure 4). As expected, high levels of RAG-1 were detected in pro-/pre-B cells with no apparent difference between CD19-/- and wt cells because of the intensive cell attempts to rearrange and express an IgM receptor. The extent of receptor editing is reflected in levels of RAG-1 expression in the immature compartment. The suppression of RAG-1 in wt immature B cells is a consequence of IgM expression and allelic exclusion. In contrast, despite IgM expression, CD19-/- immature B cells maintained a high level of RAG-1 expression, implicating intensive receptor editing. As expected, transitional and mature B cells from CD19-/- and wt mice had essentially no RAG-1. Thus, the expression of IgM in immature CD19-/- B cells does not terminate RAG-1 expression, and these cells undergo intensive receptor editing.
Rag expression in sorted wt and CD19-/- B-cell precursors. Wild-type (□) and CD19-/- B-cell precursors (▦) grown in BM cultures were sorted to pro-/pre-, immature, and transitional fractions, as described in “Materials and methods.” Mature cells were purified from spleen. Levels of RAG-1 expression in each sorted population were determined by RT-PCR. Results shown are mean ± SE of 3 different experiments.
Rag expression in sorted wt and CD19-/- B-cell precursors. Wild-type (□) and CD19-/- B-cell precursors (▦) grown in BM cultures were sorted to pro-/pre-, immature, and transitional fractions, as described in “Materials and methods.” Mature cells were purified from spleen. Levels of RAG-1 expression in each sorted population were determined by RT-PCR. Results shown are mean ± SE of 3 different experiments.
Increased receptor editing alters the lambda-kappa ratio in CD19-/- mice and in BM cultures
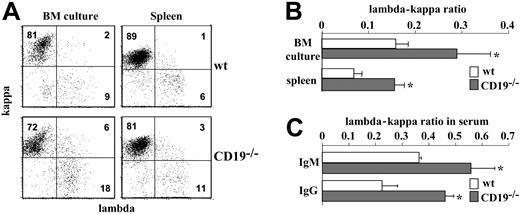
The lambda-kappa ratio serves as an efficient indicator for receptor editing in non-Tg B cells. Ongoing rearrangements lead to an increased production of lambda-expressing cells and alternate the lambda-kappa ratio. To test whether RAG-1 expression in immature CD19-/- B cells also stimulates ongoing rearrangements, we measured the lambda-kappa ratio by surface staining. The results in Figure 5A-B show a significantly increased number of IgM+/lambda+ cells in CD19-/- cultures. The lambda-kappa ratio obtained in CD19-/- cultures was 1:3 to 1:4 compared with 1:9 to 1:10 in wt cultures. Notably, we found an increased frequency of IgM+/kappa+/lambda+ double producers in CD19-/- cultures, perhaps reflecting the cells' alternating light-chain isotype.
Biased lambda-kappa ratio in CD19-/- mice and in BM culture. (A) Bone marrow culture cells and splenic B cells from wt and CD19-/- mice were stained for IgM, kappa, and lambda and analyzed by FACS. Plot analysis for kappa and lambda expression was performed on gated IgM+ cells. Shown is a representative dot plot of 3 different experiments. (B) Histogram summarizing the lambda-kappa ratio expressed in BM culture and spleen cells of wt (□) and CD19-/- mice (▦). Results expressed are mean ± SE of 3 different experiments. (C) The lambda-kappa ratio in serum antibody. Serum samples from 6 wt (□) and 6 CD19-/- mice (▦) were analyzed by ELISA for levels of IgM/kappa and IgM/lambda or IgG/kappa and IgG/lambda. Antibody concentrations were calculated using as reference IgM or IgG standard curves. Results are expressed as mean ± SE. Asterisk indicates values that are statistically different (P < .05).
Biased lambda-kappa ratio in CD19-/- mice and in BM culture. (A) Bone marrow culture cells and splenic B cells from wt and CD19-/- mice were stained for IgM, kappa, and lambda and analyzed by FACS. Plot analysis for kappa and lambda expression was performed on gated IgM+ cells. Shown is a representative dot plot of 3 different experiments. (B) Histogram summarizing the lambda-kappa ratio expressed in BM culture and spleen cells of wt (□) and CD19-/- mice (▦). Results expressed are mean ± SE of 3 different experiments. (C) The lambda-kappa ratio in serum antibody. Serum samples from 6 wt (□) and 6 CD19-/- mice (▦) were analyzed by ELISA for levels of IgM/kappa and IgM/lambda or IgG/kappa and IgG/lambda. Antibody concentrations were calculated using as reference IgM or IgG standard curves. Results are expressed as mean ± SE. Asterisk indicates values that are statistically different (P < .05).
To test whether alternation in the lambda-kappa ratio also applies in vivo, we measured this ratio in splenic B cells and in serum antibodies. We found a lambda-kappa ratio of 1:7 to 1:8 in splenic CD19-/- B cells compared with a ratio of 1:15 to 1:16 in wt (Figure 5A-B). Serum antibody analysis revealed a 1.5-fold increase in the relative proportion of IgM/lambda and a 2-fold increase in the relative proportion of IgG/lambda (Figure 5C). Thus, the increased lambda light chain production and the biased lambda-kappa ratio obtained in vitro are also evident in vivo.
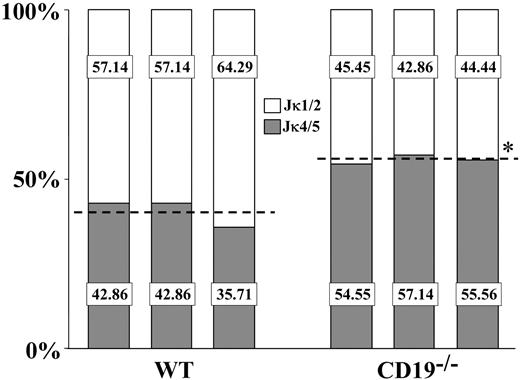
Biased Jκ4-Jκ5 use in CD19-/- splenic B cells
Ongoing rearrangements and receptor editing in non-Tg B cells are also reflected by the increased use of downstream Jκ gene segments for Vκ-Jκ rearrangements. Thus, we next analyzed the Jκ use in splenic B cells from CD19-/- compared with wt mice. For this analysis, mRNA samples from sorted IgM+ cells were subjected to RT-PCR amplification using consensus Vκ FW3 and Cκ oligos. PCR products were cloned and sequenced, and 30 to 50 sequences bearing productive Vκ-Jκ rearrangement were considered for the Jκ analysis. In agreement with the literature, we found that wt splenic B cells used the upstream Jκ1-Jκ2 segments at a higher frequency compared with the downstream Jκ4-Jκ5 segments (approximately 60% compared with approximately 40%, respectively) (Figure 6). In contrast, we found that CD19-/- splenic B cells used the downstream Jκ4-Jκ5 segments at a higher frequency than the upstream Jκ1-Jκ2 segments (approximately 44% of Jκ1-Jκ2 relative to approximately 56% of Jκ4-Jκ5). These results implicate a biased Jκ use toward downstream Jκ in splenic B cells of CD19-/- mice, which might have resulted from increased receptor editing in the BM of these mice.
The Jκ use in splenic wt and CD19-/- B cells. The Jκ use in purified splenic B cells from wt and CD19-/- mice was determined as described in “Materials and methods.” Analysis was performed for 3 wt and 3 CD19-/- mice, and for each mouse 30 to 50 sequences bearing in-frame Vκ-Jκ rearrangements were considered. Results for individual mice are shown. Percentages of rearrangements using Jκ1/2 (□) or Jκ4/5 (▦) for individual mice are presented. For each group, a broken line represents the mean Jκ1/2 and Jκ4/5 use. Asterisk indicates values that are statistically different (P < .05).
The Jκ use in splenic wt and CD19-/- B cells. The Jκ use in purified splenic B cells from wt and CD19-/- mice was determined as described in “Materials and methods.” Analysis was performed for 3 wt and 3 CD19-/- mice, and for each mouse 30 to 50 sequences bearing in-frame Vκ-Jκ rearrangements were considered. Results for individual mice are shown. Percentages of rearrangements using Jκ1/2 (□) or Jκ4/5 (▦) for individual mice are presented. For each group, a broken line represents the mean Jκ1/2 and Jκ4/5 use. Asterisk indicates values that are statistically different (P < .05).
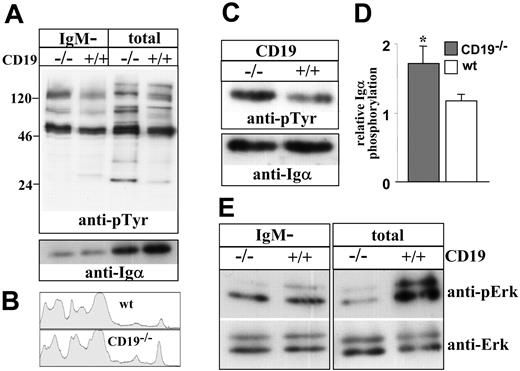
Immature CD19-/- B cells have inappropriate ligand-independent signals
To explain the impaired maturation and the stimulation of receptor editing, we determined whether immature CD19-/- B cells have inappropriate ligand-independent signals. To do so, we measured tonic tyrosine phosphorylation in cell lysates. However, it was impossible to use anti-BCR antibodies to sort the immature B-cell population because BCR ligation rapidly stimulates the phosphorylation of signaling molecules. To overcome this limitation, we measured tonic phosphorylation in total cells and in the IgM-negative (pro-/pre-B) fraction. This provided us with an indirect estimation of the level of tonic signals in the IgM+ immature population. The results in Figure 7A show that tonic tyrosine phosphorylation signals in the IgM-negative fraction are not significantly different in CD19-/- cells compared with the control. In contrast, significantly elevated tonic signals were found in the unsorted CD19-/- cells compared with the control. This finding was observed for total tyrosine phosphorylation (Figure 7A-B) and specifically for Igα phosphorylation assayed by immunoprecipitation (Figure 7C-D). Importantly, the observed differences should still be considered as an underestimation because the relative proportion of IgM+ cells in CD19-/- cultures is 30% to 50% lower compared with wt cultures (Figure 1). In contrast, we found that basal Erk kinase phosphorylation, which has been linked with positive selection and suppression of RAG gene expression in B and T lymphocytes,22,33 was significantly reduced in the unsorted CD19-/- cells compared with wt cells (Figure 7E). No significant difference in basal Erk kinase phosphorylation was found between the sorted IgM- populations. Our interpretation of these results is that the altered tonic signals obtained in the analysis of unsorted cells are primarily contributed by the IgM+ immature fraction. We conclude that immature CD19-/- B cells have inappropriate ligand-independent signals, fail positive selection, and consequentially activate receptor editing.
Ligand-independent tyrosine phosphorylation in wt and CD19-/- B-cell precursors. Wild-type and CD19-/- B-cell precursors were grown in BM cultures. Tonic tyrosine phosphorylation was determined in lysates of total cells or in lysates of sorted IgM- B cells (5 × 106 cell equivalent/lane). (A) Immunoblot analysis of total tyrosine phosphorylation in cell lysates. Membranes were probed with antiphosphotyrosine, stripped, and reprobed with anti-Igα-specific antibody. Blots shown are representative of 4 different experiments. (B) Lane densitometry analysis of tyrosine phosphorylation in lysates of total cells. Tyrosine phosphorylation lanes shown in panel A were subjected to whole-lane densitometry analysis. Plots represent the obtained signal strength on the y-axis compared with its relative location on the x-axis. (C) Igα phosphorylation was determined by immunoprecipitation (10 × 106 cells), as described in “Materials and methods.” Membranes were probed with antiphosphotyrosine, stripped, and reprobed with anti-Igα-specific rabbit antibodies. The blot shown is a representative of 3 different experiments. (D) Relative phosphorylation of Igα was determined in each experiment by densitometry analysis, where values obtained for a phosphorylated band were divided by the values corresponding to the total band. ▦ indicates CD19-/-; □, wt. Summarized results of the 3 experiments are shown in a histogram as mean ± SE (right); asterisk indicates values that are statistically different (P < .05). (E) Erk phosphorylation was determined in cell lysates using specific antibodies to phosphorylated Erk and total Erk. Blots shown are representative of 4 different experiments.
Ligand-independent tyrosine phosphorylation in wt and CD19-/- B-cell precursors. Wild-type and CD19-/- B-cell precursors were grown in BM cultures. Tonic tyrosine phosphorylation was determined in lysates of total cells or in lysates of sorted IgM- B cells (5 × 106 cell equivalent/lane). (A) Immunoblot analysis of total tyrosine phosphorylation in cell lysates. Membranes were probed with antiphosphotyrosine, stripped, and reprobed with anti-Igα-specific antibody. Blots shown are representative of 4 different experiments. (B) Lane densitometry analysis of tyrosine phosphorylation in lysates of total cells. Tyrosine phosphorylation lanes shown in panel A were subjected to whole-lane densitometry analysis. Plots represent the obtained signal strength on the y-axis compared with its relative location on the x-axis. (C) Igα phosphorylation was determined by immunoprecipitation (10 × 106 cells), as described in “Materials and methods.” Membranes were probed with antiphosphotyrosine, stripped, and reprobed with anti-Igα-specific rabbit antibodies. The blot shown is a representative of 3 different experiments. (D) Relative phosphorylation of Igα was determined in each experiment by densitometry analysis, where values obtained for a phosphorylated band were divided by the values corresponding to the total band. ▦ indicates CD19-/-; □, wt. Summarized results of the 3 experiments are shown in a histogram as mean ± SE (right); asterisk indicates values that are statistically different (P < .05). (E) Erk phosphorylation was determined in cell lysates using specific antibodies to phosphorylated Erk and total Erk. Blots shown are representative of 4 different experiments.
Discussion
BCR signaling competence is now recognized as an important positive selection checkpoint that regulates the developmental progression and maturation of B lymphocytes. The current study presents data to show that inappropriate signaling not only blocks positive selection, as has been shown earlier,9-12 it also stimulates intensive secondary light-chain recombination and receptor editing. Thus, receptor editing, which is a main mechanism in negative selection, has an important contribution for the positive selection of immature B cells.
By using an in vitro BM culture system, we show here that the maturation of non-Tg B cells deficient in CD19 is impaired. Our results disagree with earlier in vivo studies showing that BM development in CD19-deficient mice is unaffected.18,19 This discrepancy might have resulted from the fact that, in a non-Tg system, developmental impairment can efficiently be compensated by mechanisms such as receptor editing and selection.34-36 One way to test this is by restricting the repertoire with a transgenic BCR. Indeed, 3-83Tg/CD19-/- B cells have impaired BM development and maturation in vivo.20,23 A major criticism of this approach is the artificial and premature expression of the transgenic BCR, which results in accelerated B-cell development.24 In an alternative approach taken here, we have used a nonselecting in vitro culture system that allows continuous cell sampling for analysis. We report here that inappropriate ligand-independent signals, resulting from CD19 deficiency, impair but do not block the process of B-cell maturation. Thus, the modest CD19-dependent developmental effects in regulating B-cell maturation are significantly detectable in a Tg system in vivo and in a non-Tg BM culture system but may be difficult to assess in non-Tg BM in vivo. Similarly, the impaired pro-B to pre-B transition in CD19-deficient mice becomes apparent only on competitive BM reconstitution or after sublethal irradiation and autoreconstitution.29
In mature B cells, ligand-independent signals determine survival and positive selection and depend on BCR expression and signaling through the Igα/β heterocomplex.13,37,38 Our data suggest that immature CD19-/- B cells have inappropriately elevated levels of tonic signals. This defect blocks positive selection, impairs maturation, and activates receptor editing, as we have previously shown in Ig-Tg B cells deficient in CD19.22,23 Similarly, such tonic signals from the pre-BCR mediate positive selection and pro-B to pre-B transition,39,40 and lack of CD19 impairs this developmental progression.29 CD19 is also required for positive selection and for the generation of marginal zone B cells.41 Thus, CD19 has an important function in regulating ligand-independent signals that are required for positive selection of pro-B and immature B cells.
Earlier studies have shown that a signaling mutation blocks positive selection of immature B cells,1,9-12,42,43 but the consequential stimulation of receptor editing has not been tested. In contrast, the activation of secondary rearrangements in response to inappropriate pre-BCR formation or signaling is well documented.2,44-47 Receptor editing is a developmentally regulated mechanism that has been linked with negative selection. The competence to undergo editing is lost with maturation.7,8 In positive selection, receptor editing may be stimulated in immature B cells expressing a BCR that fails to generate appropriate ligand-independent signals and to promote developmental progression and maturation. In Ig-Tg mice deficient in CD19 or Lyn, an increased number of edited B cells is detected in the periphery.23,48 Receptor editing in this Ig-Tg model is stimulated as a consequence of developmental arrest and failure of positive selection.23 Furthermore, we have recently shown that receptor editing stimulation is directly controlled by modification of the BCR ligand-independent signals.22 In B and T lymphocytes, this tonic, ligand-independent activity determines the physiologic gene expression programs and the induction of RAG genes, and these signals are mediated by Erk kinase.22,33 In agreement with this, we show here that CD19-/- immature B cells undergoing receptor editing have also reduced phosphorylation of Erk kinase. Thus, inappropriate ligand-independent signals in immature CD19-/- B cells block positive selection and stimulate receptor editing.
Lack of positive selection is also reflected in chemokine responsiveness. Studies have shown that SDF-1α is essential for B-cell development in the BM and is less important in the periphery.49 In agreement with this, we found that CXCR4 expression and migration to stromal cell-derived factor 1-alpha (SDF1-α) are not significantly altered in CD19-/- precursors. In contrast, functional responsiveness to BLC has been shown in mature B cells.31,50 Thus, the reduced CXCR5 expression in CD19-/- precursors and their impaired migration to BLC in vitro and to the spleen in vivo may indicate the lack of positive selection. However, despite the impaired development of CD19-/- precursors, some cells are able to compensate for the lack of CD19 and to migrate from the BM to the periphery. This is evident by the detection of a significant number of splenic B cells in CD19-deficient Tg and non-Tg mice.18,19,23 Positive selection of these cells is also reflected by their suppression of RAG23 and by their ability to migrate in response to SDF and to BLC (Figure 2). However, in addition to being hyporesponsive to BCR stimulation, these cells are short-lived.21 This may suggest that tonic signals sufficient for positive selection and migration from the BM are not sufficient for complete maturation and extended survival in the periphery. Such processes also depend on appropriate responsiveness to survival factors such as BAFF and Akt1 or to chemokines.31
Positive selection can also be characterized by the acquisition of LPS responsiveness in immature and mature cells.32 In agreement with this, we show here that RP105 expression is initiated at the immature/transitional stage in wt cultures. Recent studies have shown that CD19 interacts with Lyn and Vav to regulate RP105 signaling on interaction with LPS,51 thereby explaining the reduced LPS responsiveness in mature CD19-deficient B cells.52 In CD19-/- precursors, antibody production in response to LPS, and RP105 expression are substantially reduced, suggesting that unresponsiveness to LPS may result from the combination of impaired maturation and the lack of, or inappropriate, RP105 signaling.
Developmental programs in B lymphopoiesis integrate antigen receptor gene assembly, synthesis and compartmentalization of signaling intermediaries, and acquisition of functional responsiveness to allow progression from 1 stage to another. When one of these programs is aborted or severely impaired, B-cell development is arrested and these cells remain functionally incompetent. Our data suggest that these arrested cells use the receptor editing mechanism to express a new receptor able to signal for positive selection. The detection of receptor editing in positive selection of non-Tg B cells in vivo and in vitro implicates its physiologic relevance. It is yet to be determined whether developmental arrest imposed by other signaling mutations also activates receptor editing.
Prepublished online as Blood First Edition Paper, January 11, 2005; DOI 10.1182/blood-2004-08-3165.
Supported by the National Council for Research and Development, Israel, jointly with the DKFZ Deutsches Krebsforschungszentrum (Heidelberg, Germany) and the Israel Cancer Research Fund-The Rosenwasser Trust.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 1. Lack of CD19 impairs B-cell maturation in BM cultures. (A) Cells grown in BM cultures were stained for the indicated surface markers and analyzed by FACS. Results from 1 of 4 representative experiments are shown. Numbers in graphs indicate cell percentages within the region. (B) Cells grown in BM cultures (□, wild type [wt];▵, CD19-/-) were washed and recultured in the absence of IL-7 for 12 to 24 hours. Cells were then stained for B220 IgM and IgD and analyzed by FACS. The induction of differentiation was quantified by calculating the relative proportion of IgM+/IgD+ cells in the total IgM+ fraction. Results presented as mean ± SE of 4 different experiments. (C) Apoptosis rates in wt and CD19-/- BM cultures were determined 12 to 24 hours after IL-7 withdrawal by TUNEL. Symbols indicate the same cell types as in panel B. Results presented as mean ± SE of 4 different experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/8/10.1182_blood-2004-08-3165/6/m_zh80080576980001.jpeg?Expires=1769081981&Signature=bY63WKsayRl4gY5RzydI46oD5IVW6bPoiwKApJ5I0mas3BTI~qCS1hXyj-gb9ggBAPC8441mC1Wd7xY06pc7IgJXz8Z8GwsYx6p4Pg4AiLZmEldqEXJGb3LenXxfx7y9pm9gBSEhp9psVDhkm6TqHZVTogxaFfR841tsuCEz5Tfb-atD4Mg~JDg0JBjtKM6u9uxYQt2lTZA3VEtAlxRW0-o1IbVyBOQYBYFCbxfkKGSE3zqXMtTEkKKnvbyYu6~LeK36CoV~cecF0Wd~56t-uQ7i0MPXsE-Qi9NT6juq2ofyYWQtyCKceISRCr16nwJj0SpwfOJKXvFu1unxYqLI7A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal