Abstract
Flt3 ligand (FL) enhances hematopoietic cell proliferation and facilitates hematopoietic stem cell mobilization in vivo, while the stromal-derived factor 1α (SDF-1α, CXC ligand 12 [CXCL12])/CXC receptor 4 (CXCR4) axis is critical for their homing and trafficking. We investigated if FL and its receptor, Flt3, functionally interact with CXCL12/CXCR4 to regulate hematopoietic cell migration. FL stimulated chemokinetic activity when used alone, but synergistically enhanced short-term migration of CD34+ cells, Ba/F3 cells expressing human Flt3 (Ba/F3-Flt3), and human RS4;11 acute leukemia cells, induced by CXCL12. Moreover, overexpression of constitutively activated internal tandem duplication (ITD)–Flt3 mutants in Ba/F3 cells dramatically enhanced migration toward CXCL12. In Ba/F3-Flt3 cells, synergistic cell migration to FL plus CXCL12 was associated with enhanced phosphorylation of mitogen-activated protein kinase p42/p44 (MAPKp42/p44), cyclic adenosine monophosphate response element binding protein (CREB), and Akt, and was partially inhibited by pretreatment of cells with selective inhibitors for MAPKp42/p44, protein kinase A (PKA), or phosphatidylinositol 3–kinase (PI3-kinase), implicating these pathways in migration to FL plus CXCL12. In contrast, prolonged exposure of CD34+ or Ba/F3-Flt3 cells to FL down-regulated CXCR4 expression, inhibited CXCL12-mediated phosphorylation of MAPKp42/p44, CREB, and Akt, and impaired migration toward CXCL12. These findings suggest that FL/Flt3 may facilitate hematopoietic cell migration/homing and mobilization by enhancing or inhibiting CXCL12/CXCR4 signaling pathways and that the FL/Flt3 axis participates in trafficking of normal and transformed hematopoietic cells.
Introduction
Homing and mobilization of hematopoietic stem and progenitor cells (HSPCs) are regulated by hematopoietic cytokines,1-3 chemokines,4-6 and adhesion molecules;7,8 however, their mechanisms are poorly understood. Stromal-derived factor 1α (SDF-1α, CXC ligand 12 [CXCL12]) and its receptor CXCR4 play important roles in regulating migration, homing, and mobilization of hematopoietic cells.2,6,9-11 CXCL12 attracts primitive hematopoietic cells expressing CXCR4 to bone marrow,2,11 while disruption of CXCL12/CXCR4 interaction within marrow may facilitate their mobilization to the peripheral circulation.6,12-14 Several studies demonstrate that CXCL12/CXCR4 function is influenced by hematopoietic cytokines. Stem cell factor (SCF) stimulates migration of hematopoietic progenitor cells and enhances migration induced by CXCL12.11,15 Thrombopoietin, granulocyte-macrophage colony-stimulating factor (GM-CSF), and SCF synergistically enhance intracellular signals generated through the CXCL12/CXCR4 axis,16 and treatment of CD34+ cells with SCF plus interleukin-6 (IL-6) enhances their in vivo homing by up-regulating CXCR4 expression.2 These findings suggest that hematopoietic growth factors functionally interact with CXCL12/CXCR4 signaling pathways implicated in cell migration.
Flt3 is a type III tyrosine kinase receptor expressed mainly by primitive hematopoietic cells.17-19 Flt3 ligand (FL) has little direct effect on progenitor cell proliferation but synergizes with other cytokines to enhance proliferation.20,21 FL induces HSPC mobilization when administered in mice and demonstrates synergy when used in combination with granulocyte colony-stimulating factor (G-CSF) or GM-CSF,3,22-24 suggesting that it may enhance HSPC motility. In addition, HSPC mobilization by G-CSF is associated with elevated serum FL, suggesting that FL may be involved in G-CSF–mediated mobilization.25
Flt3 is overexpressed on hematopoietic cells from the majority of patients with acute leukemia,26 and both Flt3 and FL are expressed in most acute myeloid leukemia (AML) cells, suggesting autocrine activation and signaling.26-28 Internal tandem duplication (ITD) mutations of the Flt3 receptor gene that activate the Flt3 tyrosine kinase are found in 25% to 30% of patients with AML and are associated with poor prognosis.27,29,30 Furthermore, their ectopic expression in mice results in myeloproliferative disease with extramedullary hematopoiesis.31,32 AML cells may also have enhanced migratory potential compared with normal cells, since they are often found in extramedullary sites, which is an adverse prognostic factor.33,34 The majority of acute leukemic cells express CXCR4 and migrate in response to CXCL12,9,35,36 suggesting that the CXCL12/CXCR4 axis may be involved in extramedullary disease.
In this report, we investigated whether FL modulates migration of hematopoietic cells and if there are functional associations linking the FL/Flt3 and CXCL12/CXCR4 signaling pathways in migration. Our findings provide an in vitro model for FL-mediated migration of normal and transformed hematopoietic cells and suggest that the FL/Flt3 signaling pathway may regulate hematopoietic cell trafficking through positive and negative effects on the CXCL12/CXCR4 axis.
Materials and methods
Antibodies and cytokines
Phycoerythrin (PE)–conjugated antihuman Flt3 and mouse immunoglobulin G1 (IgG1) were purchased from Immunotech (Westbrook, ME). Fluorescein isothiocyanate (FITC)– or cytochrome (Cy)–conjugated anti-CD34 (BIRMA-K3) and mouse IgG1 were from Dako (Carpinteria, CA). PE-Cy5 antihuman CXCR4 (Clone 12G5) was from eBioscience (San Diego, CA). Biotinylated antimouse CXCR4 (Clone 2B11/CXCR4), biotinylated rat IgG2b, anti-FcγIII/II, FITC antihuman Ki-67 (clone MOPC-21), FITC mouse IgG1 and PE-Cy5 mouse IgG2a were obtained from BD Biosciences (San Diego, CA). Recombinant human (rh) FL was provided by Immunex (Seattle, WA). Recombinant murine (rm) and rhCXCL12 (SDF-1α) and rmIL-3 were purchased from R&D Systems (Minneapolis, MN). Recombinant murine stem cell factor (rmSCF) was a gift from Dr Karl Nocka (UCB Research, Cambridge, MA). Recombinant murine GM-CSF was purchased from BioVision (Palo Alto, CA). The selective pathway inhibitors PD98059 (mitogen-activated protein kinase p42/p44 [MAPKp42/p44]), LY294002 (phosphatidylinositol 3–kinase [PI3-kinase]), H89 (protein kinase A [PKA]), and AG1296 (tyrosine kinase) were purchased from BioMol Research Laboratories (Plymouth Meeting, PA).
Cell lines expressing human Flt3
Full-length wild-type human Flt3 cDNA was a gift from Immunex.18 The empty pLXSN or pLXSN harboring full-length Flt3 was transfected into GP+envAm12 cells, and the virus supernatant was added to GP+E-86 cells.37 IL-3–dependent mouse Ba/F3 cells were incubated with fresh virus supernatant, and G418-resistant Flt3positive cells (Ba/F3-Flt3) were sorted by fluorescence-activated cell sorter (FACS). G418-resistant vector control–transduced Ba/F3 and Ba/F3-Flt3 cells were maintained in RPMI plus 10% heat-inactivated fetal bovine serum (HI-FBS; Hyclone Sterile Systems, Logan, UT) supplemented with 0.1 ng/mL rmIL-3. Ba/F3 cells expressing internal tandem duplication (ITD) Flt3 mutants cloned from patients with AML (W51, W73, and W78) as well as wild-type Flt3 in MSCV vector were kindly provided by Dr D. Gary Gilliland, Harvard Medical School.32 Mouse 32D cells expressing ectopic wild-type Flt3 or ITD-Flt3 were generous gifts from Drs Joachim Schwäble and Hubert Serve, University of Münster.31 Human acute leukemia cell line RS4;11 was maintained in RPMI plus 10% HI-FBS.38
In vitro migration assay
Normal umbilical cord blood (UCB) was obtained with institutional review board approval. Isolation of CD34+ cells39,40 and transwell migration assays were performed as described.11 CD34+ cells (100 000, purity > 95%) were loaded onto the top chamber of transwells (5.0-μm pore size; Corning, Corning, NY) with rhFL and/or rhCXCL12 in the bottom chamber and incubated for 4 to 72 hours at 37°C in 0.5% bovine serum albumin (BSA)/RPMI. Cells completely migrating to the bottom chamber were enumerated by FACS. In some experiments, partially purified CD34+ cells were stained with anti-CXCR4, anti-Flt3, and anti-CD34 antibodies, and Flt3+-CXCR4+-CD34+ cells were isolated by FACS and used in migration assays. The chemokinetic or chemotactic activity of FL on Ba/F3-Flt3 cells was determined by the checkerboard assay.11 The percentage cell migration was calculated by dividing the number of cells migrated to the lower well by the total cell input multiplied by 100.
Actin polymerization assay
Actin polymerization was analyzed as described,11 with minor modifications. Ba/F3-Flt3 cells were resuspended at 1.25 × 106/mL in RPMI-1640/0.5% BSA and 0.2-mL aliquots were stimulated with phosphate-buffered saline (PBS), FL, or CXCL12 for up to 5 minutes. Cells were fixed and stained with FITC-labeled phalloidin (4 × 10–7 M; Sigma Aldrich, St Louis, MO) in 0.5 mg/mL 1-α-lysophosphatidylcholine, 18% formaldehyde in PBS. Cells were incubated for 15 minutes at 37°C, washed, and resuspended in 1% paraformaldehyde. Mean channel fluorescence of phalloidin was analyzed by FACSCalibur (BD Biosciences).
Flow cytometry analysis
Cell surface expression of Flt3 or CXCR4 on Ba/F3-Flt3 or CD34+ cells was analyzed by flow cytometry. Cells were washed with 0.1% BSA/PBS, blocked with 10 μg/mL human IgG or anti-FcγIII/II for 10 minutes, and stained with isotype control or antibodies against CXCR4 or Flt3 for 30 minutes in 0.1% BSA/PBS on ice. Expression of cell surface CXCR4 was compared by normalizing the mean fluorescence intensity (MFI) of CXCR4 with the isotype control. In some experiments, percent change of CXCR4 was determined using the following formula: % change of CXCR4 = 100 × [1 – (MFI of CXCR4 for cells cultured with FL ÷ MFI of isotype for cells cultured with FL) ÷ (MFI of CXCR4 for cells cultured without FL ÷ MFI of isotype for cells cultured without FL)].
Western analysis
Cells (5 million) were lysed in 100 μL sodium dodecyl sulfate (SDS) lysis buffer (20 mM dithiothreitol, 6% SDS, 0.25M Tris [tris(hydroxymethyl)aminomethane, pH 6.8], 10% glycerol, and bromophenol blue), sonicated, and boiled, and the lysates from 1 × 106 cells were subjected to Western analysis. The blot was blocked with 2% nonfat dry milk in PBST (0.05% Tween 20 in PBS) at room temperature for one hour and probed with total or phospho-specific antibodies against MAPKp42/p44 (Thr202/Tyr204), Akt (Ser473), and cyclic adenosine monophosphate response element binding protein, Ser133 (CREB) (Cell Signaling Technology, Beverly, MA) in blocking buffer (1:1000 dilution) overnight at 4°C. The filters were washed 5 times with PBST and incubated with horseradish peroxidase (HRP)–conjugated anti–rabbit or –mouse IgG (Amersham Biosciences, Piscataway, NJ) in blocking buffer (1:2500 dilution) at room temperature for one hour. The blots were washed 5 times with PBST and developed with enhanced chemiluminescence (ECL) reagent (Amersham Biosciences).
Statistics
Data are expressed as mean ± standard error (SEM). Statistical differences between single agents and control were determined using the 2-tailed Student t test in Microsoft Excel (Microsoft, Seattle, WA). Synergy in response to FL plus CXCL12 was compared with FL and CXCL12 used alone by analysis of variance (ANOVA), and the Bonferroni method was used for multiple comparisons. This analysis was performed by the Biostatistics Core Facility of the Indiana University Cancer Center.
Results
FL increases migration of UCB CD34+ cells and Ba/F3-Flt3 cells
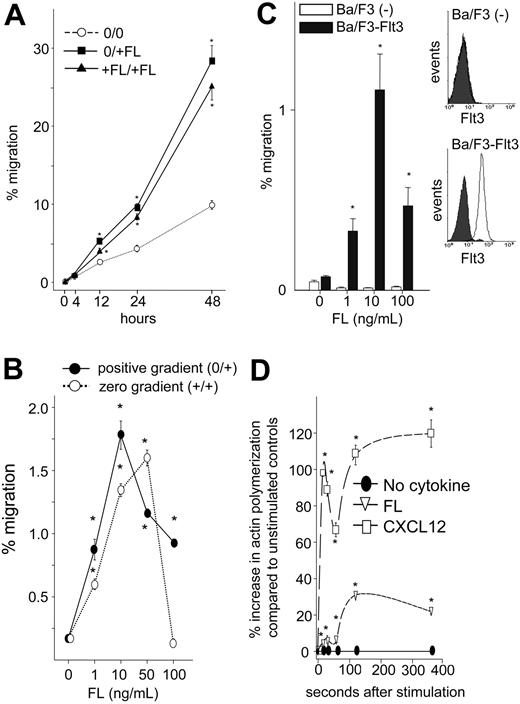
Since several hematopoietic cytokines, notably SCF and IL-3, which, like FL, support proliferation and/or survival of early hematopoietic cells, have been shown to enhance chemotaxis,11,15,41 we examined whether FL affects hematopoietic cell migration/motility. In an in vitro transwell migration assay, both positive (FL in bottom chamber only: 0/+FL) and zero gradients (FL in upper and bottom chamber: +FL/+FL) of rhFL induced migration of UCB CD34+ cells over a 48-hour period (Figure 1A). Despite increased migration to FL at 48 hours (28% ± 2% and 25% ± 2% migration in positive and zero gradients, respectively), FL did not increase total CD34+ cells during this time frame (0.5% ± 0.1% increase in viable cells), suggesting that enhanced migration to FL was not due to an effect on cell proliferation. Similar to CD34+ cells, rhFL induced modest migration of Ba/F3-Flt3 cells both in a positive (0/+) and a zero (+/+) gradient after 4 hours (Figure 1B). Maximum response to FL was observed at 10 ng/mL using a positive gradient and declined with increasing dose. The response was specific to the FL/Flt3 pathway, since no migration was observed in Ba/F3 cells, which do not express Flt3, transduced with empty vector (Figure 1C). Migration of Ba/F3-Flt3 cells to FL was associated with significantly increased actin polymerization (Figure 1D). The maximum migration of Ba/F3-Flt3 cells in a positive (2.2%) and in a negative (1.9%) gradient (Table 1) suggests that FL primarily induces chemokinetic activity.
Migration of UCB CD34+ cells and Ba/F3 cells expressing human wild-type Flt3 (Ba/F3-Flt3 cells) to FL. (A) Freshly isolated CD34+ cells were suspended in 0.5% BSA/RPMI, and migration toward 10 ng/mL rhFL was quantitated by flow cytometry. In replicate cultures, cells were incubated with or without 10 ng/mL FL in 0.5% BSA/RPMI, and cell viability was determined based on forward and side scatter. Percentage migration of CD34+ cells to a positive (0/+FL; ▪) and a zero (+FL/+FL; ▴) gradient along with background migration (0/0; ○) are shown and were calculated based upon migration of CD34+ cells divided by fold change in viable cells incubated with FL. Data are expressed as mean ± SEM of 3 experiments. *P < .05 compared with background migration in the absence of FL. (B) Ba/F3-Flt3 cells were washed and resuspended in RPMI with 0.5% BSA at 5 × 106/mL and 0.1 mL, added to the transwell with either escalating doses of rhFL in the lower chamber (0/+; •) or in both the upper and lower chambers (+/+; ○), and incubated for 4 hours at 37°C. Cells that completely migrated to the bottom chamber were enumerated, and percentage migration was calculated as described in “Materials and methods.” Data represent 1 of the 3 experiments with similar results. *P < .05 compared with background migration in the absence of FL. Error bars indicate mean ± SEM. (C) (Left) Percentage migration of control vector–transduced Ba/F3 cells (□) or Ba/F3-Flt3 cells (▪) in response to a positive gradient of increasing concentration of rhFL. (Right) Flt3 expression was analyzed by flow cytometry. *P < .05 compared with background migration in the absence of FL. Shaded curve indicates isotope staining; open curve, FIT3 staining. Error bars indicate mean ± SEM. (D) Ba/F3-Flt3 cells were extensively washed and incubated with 10 ng/mL rhFL or 100 ng/mL rhCXCL12 for up to 360 seconds, fixed, and stained as described in “Materials and methods.” The percentage increase in mean channel fluorescence of phalloidin-FITC compared with cells incubated without cytokines (•) is shown. □ indicates CXCL12; ▿, FL. Data represent 1 of the 2 experiments with equivalent results. *P < .05 compared with cells without cytokines. Error bars indicate mean ± SEM.
Migration of UCB CD34+ cells and Ba/F3 cells expressing human wild-type Flt3 (Ba/F3-Flt3 cells) to FL. (A) Freshly isolated CD34+ cells were suspended in 0.5% BSA/RPMI, and migration toward 10 ng/mL rhFL was quantitated by flow cytometry. In replicate cultures, cells were incubated with or without 10 ng/mL FL in 0.5% BSA/RPMI, and cell viability was determined based on forward and side scatter. Percentage migration of CD34+ cells to a positive (0/+FL; ▪) and a zero (+FL/+FL; ▴) gradient along with background migration (0/0; ○) are shown and were calculated based upon migration of CD34+ cells divided by fold change in viable cells incubated with FL. Data are expressed as mean ± SEM of 3 experiments. *P < .05 compared with background migration in the absence of FL. (B) Ba/F3-Flt3 cells were washed and resuspended in RPMI with 0.5% BSA at 5 × 106/mL and 0.1 mL, added to the transwell with either escalating doses of rhFL in the lower chamber (0/+; •) or in both the upper and lower chambers (+/+; ○), and incubated for 4 hours at 37°C. Cells that completely migrated to the bottom chamber were enumerated, and percentage migration was calculated as described in “Materials and methods.” Data represent 1 of the 3 experiments with similar results. *P < .05 compared with background migration in the absence of FL. Error bars indicate mean ± SEM. (C) (Left) Percentage migration of control vector–transduced Ba/F3 cells (□) or Ba/F3-Flt3 cells (▪) in response to a positive gradient of increasing concentration of rhFL. (Right) Flt3 expression was analyzed by flow cytometry. *P < .05 compared with background migration in the absence of FL. Shaded curve indicates isotope staining; open curve, FIT3 staining. Error bars indicate mean ± SEM. (D) Ba/F3-Flt3 cells were extensively washed and incubated with 10 ng/mL rhFL or 100 ng/mL rhCXCL12 for up to 360 seconds, fixed, and stained as described in “Materials and methods.” The percentage increase in mean channel fluorescence of phalloidin-FITC compared with cells incubated without cytokines (•) is shown. □ indicates CXCL12; ▿, FL. Data represent 1 of the 2 experiments with equivalent results. *P < .05 compared with cells without cytokines. Error bars indicate mean ± SEM.
Comparison of chemotactic and chemokinetic activities of FL and CXCL12 in Ba/F3-Flt3 cells
. | FL . | CXCL12 . |
|---|---|---|
| MCTA, % | 2.2 | 20.2 |
| MCKA, % | 1.9 | 0.04 |
| CCI | 1.1 | 505 |
| RCKA | 89.9 | 0.2 |
. | FL . | CXCL12 . |
|---|---|---|
| MCTA, % | 2.2 | 20.2 |
| MCKA, % | 1.9 | 0.04 |
| CCI | 1.1 | 505 |
| RCKA | 89.9 | 0.2 |
All calculations were performed based on the checkerboard assay (0-100 ng/mL) as described.11 The maximal response for migration was observed at 10 ng/mL and 100 ng/mL for FL and CXCL12, respectively.
MCTA indicates maximum chemotactic activity (a maximal percentage migration to the lower chamber in the positive gradient of the chemoattractant); MCKA, maximum chemokinetic activity (a maximal percentage migration to the lower chamber in the zero gradient formed by equal concentrations of the chemoattractant in both chambers); CCI, chemotactic-chemokinetic index (calculated by dividing MCTA by MCKA); and RCKA, relative chemokinetic activity (calculated by the following formula: [MCKA/MCTA] × 100).
FL increases migration induced by CXCL12 in CD34+ cells, Ba/F3-Flt3 cells, and human RS4;11 acute leukemia cells
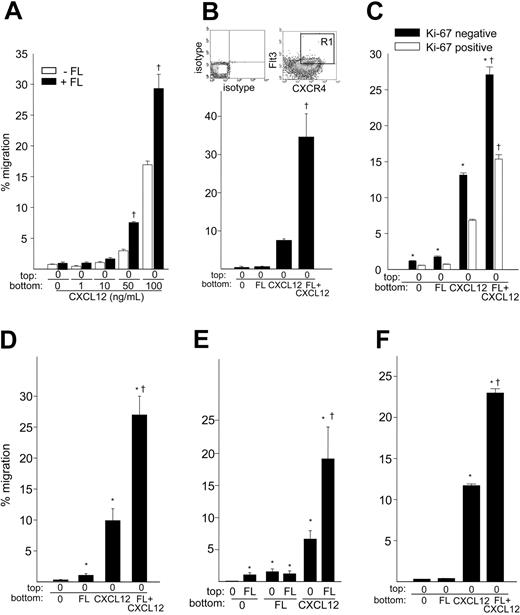
Since the CXCL12/CXCR4 axis regulates migration and homing of HSPCs,2,11 we examined whether FL could enhance migration of CD34+ cells induced by CXCL12. Analysis of CXCR4 and Flt3 expression on freshly isolated UCB CD34+ cells indicated that 36% ± 2% (n = 3) of cells coexpress Flt3 and CXCR4. Transmigration of CD34+ cells toward a positive gradient of 50 or 100 ng/mL CXCL12 for 4 hours was significantly enhanced in the presence of 10 ng/mL FL (Figure 2A). For 50 and 100 ng/mL CXCL12, the presence of 10 ng/mL FL produced significant synergistic migration at both dosage levels (P < .0001). The combination of 10 ng/mL FL plus 50 ng/mL or 100 ng/mL CXCL12 resulted in a 2.6 ± 0.1-fold and 1.7 ± 0.2-fold increase, respectively, compared with CXCL12 alone, although 100 ng/mL CXCL12 resulted in a higher absolute percentage migration than 50 ng/mL CXCL12. FL had no significant effect on CXCR4 expression (1.02 ± 0.2-fold) during this time period; therefore the enhancing effect of FL on CXCL12-mediated migration cannot be explained by alteration in CXCR4 expression. The dramatic synergistic effect of FL on CXCL12-mediated migration was also observed using isolated CXCR4+-Flt3+-CD34+ cells (P < .0001; Figure 2B), likely due to enrichment of double-positive cells that can respond to both FL and CXCL12. However, migration of sorted cells to CXCL12 was lower than unsorted cells (Figure 2A-B), and the enhancing effect of FL on CXCL12-mediated migration of unsorted CD34+ cells was more obvious at a lower dose (50 ng/mL) than a higher dose (100 ng/mL) of CXCL12, suggesting that enhanced migration may also be a consequence of inhibition of CXCR4 by the 12G5 anti-CXCR4 antibody used for cell sorting, which is known to block CXCL12 binding to CXCR4.
Migration of hematopoietic cells in response to the combination of FL and CXCL12. (A) Migration of freshly isolated UCB CD34+ cells to a positive gradient of escalating doses of CXCL12 was analyzed in the absence (□) or presence of 10 ng/mL rhFL (▪) in the bottom chamber for 4 hours. Data are the mean ± SEM of 2 experiments. † denotes synergistic effect of FL plus CXCL12. (B) Migration of CXCR4+, Flt3+, and CD34+ cells to 10 ng/mL FL and/or 100 ng/mL CXCL12. Partially purified UCB CD34+ cells were stained with FITC anti-CD34, PE anti-Flt3, and Cy-chrome anti-CXCR4, and CD34+, CXCR4+, and Flt3+ cells (gate R1) were sorted by FACS. The isotype and CXCR4/Flt3 staining are shown. Cells were subjected to migration for 4 hours. Data represent the mean ± SEM of 2 experiments. † denotes synergistic effect of FL plus CXCL12. (C) Total CD34+ cells were subjected to migration to 10 ng/mL rhFL and/or 100 ng/mL rhCXCL12 for 4 hours. Intracellular Ki-67 expression in CD34+ cells was performed as described.40 Migrated cells were fixed with 1% paraformaldehyde overnight at 4°C and washed with PBS containing 0.25% Triton X-100 and 1% BSA. Cells were blocked with human IgG on ice for 10 minutes and then stained with FITC-conjugated isotype control or anti-Ki67 antibody in the same buffer on ice for 60 minutes. Cells that appeared below isotype staining were defined as Ki-67negative cells. The percentage of Ki-67negative cell migration (▪) was calculated as the number of migrated Ki-67negative cells divided by the number of Ki-67negative CD34+ input cells multiplied by 100. A similar calculation was performed for Ki-67positive cells (□). Data are the mean ± SEM from 4 experiments. *P < .05 compared with Ki-67positive cells by t test; † denotes synergistic effect of FL plus CXCL12. (D) Percentage migration of Ba/F3-Flt3 cells to FL, CXCL12, or the combination of FL and CXCL12. Exponentially growing Ba/F3-Flt3 cells were extensively washed and 5 × 105 cells subjected to in vitro transmigration assay to 10 ng/mL rh FL, 100 ng/mL rmCXCL12 or rhCXCL12, or FL plus CXCL12. Data are shown as mean ± SEM percentage migration for 3 experiments. *P < .05 compared with background migration; † indicates synergistic effect of FL plus CXCL12. (E) Percentage migration of Ba/F3-Flt3 cells toward FL (10 ng/mL) or CXCL12 (100 ng/mL) in the presence or absence of negative gradient of FL (10 ng/mL). Data are shown as mean ± SEM percentage migration for 3 experiments. *P < .05 compared with background migration; † indicates synergistic effect of FL plus CXCL12. (F) Migration of the human RS4;11 cells in response to 10 ng/mL FL and/or 100 ng/mL CXCL12. Cells were extensively washed and evaluated for migration for 4 hours in 0.5% BSA/RPMI. The data represent the mean ± SEM percentage migration for 1 of 2 experiments. *P < .05 compared with background migration; † indicates synergistic effect of FL plus CXCL12.
Migration of hematopoietic cells in response to the combination of FL and CXCL12. (A) Migration of freshly isolated UCB CD34+ cells to a positive gradient of escalating doses of CXCL12 was analyzed in the absence (□) or presence of 10 ng/mL rhFL (▪) in the bottom chamber for 4 hours. Data are the mean ± SEM of 2 experiments. † denotes synergistic effect of FL plus CXCL12. (B) Migration of CXCR4+, Flt3+, and CD34+ cells to 10 ng/mL FL and/or 100 ng/mL CXCL12. Partially purified UCB CD34+ cells were stained with FITC anti-CD34, PE anti-Flt3, and Cy-chrome anti-CXCR4, and CD34+, CXCR4+, and Flt3+ cells (gate R1) were sorted by FACS. The isotype and CXCR4/Flt3 staining are shown. Cells were subjected to migration for 4 hours. Data represent the mean ± SEM of 2 experiments. † denotes synergistic effect of FL plus CXCL12. (C) Total CD34+ cells were subjected to migration to 10 ng/mL rhFL and/or 100 ng/mL rhCXCL12 for 4 hours. Intracellular Ki-67 expression in CD34+ cells was performed as described.40 Migrated cells were fixed with 1% paraformaldehyde overnight at 4°C and washed with PBS containing 0.25% Triton X-100 and 1% BSA. Cells were blocked with human IgG on ice for 10 minutes and then stained with FITC-conjugated isotype control or anti-Ki67 antibody in the same buffer on ice for 60 minutes. Cells that appeared below isotype staining were defined as Ki-67negative cells. The percentage of Ki-67negative cell migration (▪) was calculated as the number of migrated Ki-67negative cells divided by the number of Ki-67negative CD34+ input cells multiplied by 100. A similar calculation was performed for Ki-67positive cells (□). Data are the mean ± SEM from 4 experiments. *P < .05 compared with Ki-67positive cells by t test; † denotes synergistic effect of FL plus CXCL12. (D) Percentage migration of Ba/F3-Flt3 cells to FL, CXCL12, or the combination of FL and CXCL12. Exponentially growing Ba/F3-Flt3 cells were extensively washed and 5 × 105 cells subjected to in vitro transmigration assay to 10 ng/mL rh FL, 100 ng/mL rmCXCL12 or rhCXCL12, or FL plus CXCL12. Data are shown as mean ± SEM percentage migration for 3 experiments. *P < .05 compared with background migration; † indicates synergistic effect of FL plus CXCL12. (E) Percentage migration of Ba/F3-Flt3 cells toward FL (10 ng/mL) or CXCL12 (100 ng/mL) in the presence or absence of negative gradient of FL (10 ng/mL). Data are shown as mean ± SEM percentage migration for 3 experiments. *P < .05 compared with background migration; † indicates synergistic effect of FL plus CXCL12. (F) Migration of the human RS4;11 cells in response to 10 ng/mL FL and/or 100 ng/mL CXCL12. Cells were extensively washed and evaluated for migration for 4 hours in 0.5% BSA/RPMI. The data represent the mean ± SEM percentage migration for 1 of 2 experiments. *P < .05 compared with background migration; † indicates synergistic effect of FL plus CXCL12.
Similar synergistic migration to CXCL12 was observed for Ki-67negative CD34+ cells (P < .0001), that is, quiescent cells,42,43 and for the Ki-67positive population (P < .0001; Figure 2C), although migration to FL and/or CXCL12 was significantly higher in Ki-67negative cells (P < .05). FL also synergistically enhanced CXCL12-induced migration of Ba/F3-Flt3 cells (Figure 2D) after 4 hours of incubation (P < .0001). Although the effect was weak, migration of Ba/F3-Flt3 cells toward CXCL12 in the face of a negative gradient of FL (FL in top chamber and CXCL12 in bottom chamber) was also synergistically increased (P = .01; Figure 2E). Similarly, FL synergistically enhanced migration of human RS4;11 acute leukemia cells, which express wild-type Flt3 and CXCR4 (not shown) to CXCL12 (P < .0001; Figure 2F).
Long-term migration of CD34+ cells induced by FL and/or CXCL12
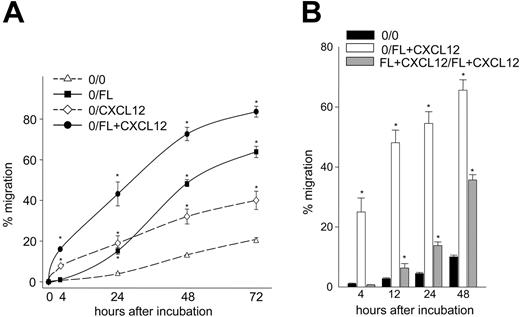
Since the stimulatory effect of FL on CD34+ cell migration was time dependent (Figure 1A), we compared the long-term versus short-term effects of FL on CXCL12-mediated migration. Migration of CD34+ cells to a positive gradient of FL and/or CXCL12 was evaluated for up to 72 hours. Time-dependent increased migration was observed in all groups (Figure 3A) but did not result from an increase in cell proliferation (not shown). At 48 and 72 hours, migration to FL alone became greater than migration to CXCL12 alone and was higher than migration induced by the combination of FL plus CXCL12 at 4 hours. Migration to the combination of FL plus CXCL12 was the highest at all time points. The effect of FL plus CXCL12 was additive beyond 24 hours. We also compared migration of CD34+ cells induced by a zero gradient of FL plus CXCL12 (FL+CXCL12 in upper and lower chambers) with a positive gradient of FL plus CXCL12 (FL+CXCL12 in lower chamber only) (Figure 3B). The zero gradient of FL plus CXCL12 had no effect on migration at 4 hours but significantly induced migration after 12 hours, although the response was lower than the positive gradient of FL plus CXCL12, suggesting that FL plus CXCL12 can induce random cell migration over time.
Long-term migration of CD34+ cells to FL and/or CXCL12. (A) Freshly isolated UCB CD34+ cells were subjected to in vitro migration to a positive gradient of 10 ng/mL FL (▪), 100 ng/mL CXCL12 (⋄), or the combination of FL plus CXCL12 (•). Cell migration was quantitated at 4, 24, 48, and 72 hours. Replicate cultures were incubated with 10 ng/mL rhFL, 100 ng/mL CXCL12, or a combination of FL plus CXCL12 in the chemotaxis medium for 72 hours, and viable cell counts were determined by the forward and side scatter analysis. Percentage of migration was calculated based upon migration of CD34+ cells divided by fold change in viable cells. Data represent the mean ± SEM from 2 experiments. *P < .05 compared with background migration (0/0; ▵). (B) Migration of CD34+ cells in response to positive (0/FL+CXCL12; □) and zero (FL+CXCL12/FL+CXCL12; ▦) gradients of FL plus CXCL12 were determined for 48 hours. Data are the average ± SEM of 3 experiments. *P < .05 compared with background migration (0/0; ▪).
Long-term migration of CD34+ cells to FL and/or CXCL12. (A) Freshly isolated UCB CD34+ cells were subjected to in vitro migration to a positive gradient of 10 ng/mL FL (▪), 100 ng/mL CXCL12 (⋄), or the combination of FL plus CXCL12 (•). Cell migration was quantitated at 4, 24, 48, and 72 hours. Replicate cultures were incubated with 10 ng/mL rhFL, 100 ng/mL CXCL12, or a combination of FL plus CXCL12 in the chemotaxis medium for 72 hours, and viable cell counts were determined by the forward and side scatter analysis. Percentage of migration was calculated based upon migration of CD34+ cells divided by fold change in viable cells. Data represent the mean ± SEM from 2 experiments. *P < .05 compared with background migration (0/0; ▵). (B) Migration of CD34+ cells in response to positive (0/FL+CXCL12; □) and zero (FL+CXCL12/FL+CXCL12; ▦) gradients of FL plus CXCL12 were determined for 48 hours. Data are the average ± SEM of 3 experiments. *P < .05 compared with background migration (0/0; ▪).
MAPKp42/p44, PI3-kinase, and PKA are involved in migration induced by FL plus CXCL12
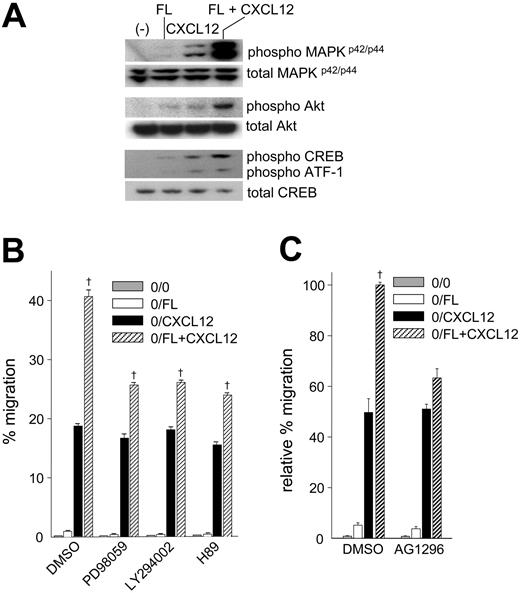
Our results demonstrating synergistically enhanced migration to CXCL12 by FL suggested the possibility of cross talk between FL/Flt3 and CXCL12/CXCR4. Ligand binding to Flt327 or chemokine receptors activates MAPKp42/p44 and PI3-kinase.16,44 In order to investigate pathways involved in synergistic migration to FL plus CXCL12, we examined phosphorylation of MAPKp42/p44, Akt, CREB (cAMP response element binding protein), a downstream target of PKA, and its family member activating transcriptional factor-1 (ATF).45 In Ba/F3-Flt3 cells, FL induced barely detectable MAPKp42/p44, Akt, and CREB phosphorylation, while CXCL12 stimulation resulted in significant phosphorylation (Figure 4A). The combination of FL plus CXCL12 synergistically enhanced phosphorylation of MAPKp42/p44, Akt, and CREB (Figure 4A), consistent with synergistically enhanced migration. To confirm the involvement of these pathways in migration toward FL plus CXCL12, we evaluated migration of Ba/F3-Flt3 cells pretreated with PD98059, LY294002, and H89, selective inhibitors for MAPKp42/p44, PI3-kinase, and PKA, respectively (Figure 4B). Although synergistic migration was not completely blocked and remained statistically significant, pretreatment with PD98059, LY294002, or H89 significantly inhibited migration induced by the combination of FL plus CXCL12 (P < .05). The inhibitors had little effect on migration to CXCL12 alone but reduced migration in response to FL alone (Figure 4B; Table 2), suggesting that suppression of synergistic migration was a consequence of inhibition of the FL/Flt3 signaling pathway rather than the CXCL12/CXCR4 pathway. Consistent with this finding, pretreatment of cells with AG1296, which inhibits Flt3 tyrosine kinase,46,47 blocked synergistic migration to FL plus CXCL12 (73% ± 10% reduction in synergistic response) (Figure 4C).
Phosphorylation of MAPKp42/p44, Akt, CREB, and ATF-1 in Ba/F3-Flt3 cells stimulated with FL and/or CXCL12 and effects of selective pathway inhibitors on migration to FL and/or CXCL12. (A) IL-3–starved cells were incubated with rhFL (10 ng/mL) and/or rhCXCL12 (100 ng/mL) for 5 minutes. Lysates were subjected to Western analysis for phosphorylation of MAPKp42/p44, Akt, CREB, and ATF-1. (B) Cells were preincubated with dimethyl sulfoxide (DMSO), 50 μM PD98059, 50 μM LY294002, or 10 μM H89 for 1 hour, washed twice, and subjected to migration to 10 ng/mL rhFL (□), 100 ng/mL rhCXCL12 (▪), or the combination of FL plus CXCL12 (▨) for 4 hours. ▦ indicates background migration (0/0). Cell viability was evaluated by forward and side scatter analysis following migration. Data represent mean ± SEM of percentage migration in 1 of 6 experiments with similar results. † indicates synergistic effect of FL plus CXCL12; P < .0001. (C) Ba/F3-Flt3 cells were preincubated with 10 μM AG1296 for one hour, washed, and evaluated for migration to 10 ng/mL FL (□), 100 ng/mL CXCL12 (▪), or a combination of FL plus CXCL12 (▨) as described in panel B. ▦ indicates background migration (0/0). Data represent the mean ± SEM relative percentage migration compared with cells pretreated with DMSO and migrating to FL plus CXCL12 (expressed as 100%) from 2 experiments of triplicate counts. † indicates synergistic migration to FL plus CXCL12; P < .0001.
Phosphorylation of MAPKp42/p44, Akt, CREB, and ATF-1 in Ba/F3-Flt3 cells stimulated with FL and/or CXCL12 and effects of selective pathway inhibitors on migration to FL and/or CXCL12. (A) IL-3–starved cells were incubated with rhFL (10 ng/mL) and/or rhCXCL12 (100 ng/mL) for 5 minutes. Lysates were subjected to Western analysis for phosphorylation of MAPKp42/p44, Akt, CREB, and ATF-1. (B) Cells were preincubated with dimethyl sulfoxide (DMSO), 50 μM PD98059, 50 μM LY294002, or 10 μM H89 for 1 hour, washed twice, and subjected to migration to 10 ng/mL rhFL (□), 100 ng/mL rhCXCL12 (▪), or the combination of FL plus CXCL12 (▨) for 4 hours. ▦ indicates background migration (0/0). Cell viability was evaluated by forward and side scatter analysis following migration. Data represent mean ± SEM of percentage migration in 1 of 6 experiments with similar results. † indicates synergistic effect of FL plus CXCL12; P < .0001. (C) Ba/F3-Flt3 cells were preincubated with 10 μM AG1296 for one hour, washed, and evaluated for migration to 10 ng/mL FL (□), 100 ng/mL CXCL12 (▪), or a combination of FL plus CXCL12 (▨) as described in panel B. ▦ indicates background migration (0/0). Data represent the mean ± SEM relative percentage migration compared with cells pretreated with DMSO and migrating to FL plus CXCL12 (expressed as 100%) from 2 experiments of triplicate counts. † indicates synergistic migration to FL plus CXCL12; P < .0001.
Percentage inhibition of Ba/F3-Flt3 cell migration by pretreatment with signaling molecule inhibitors
. | PD98059 . | LY294002 . | H89 . | PD98059 + LY294002 . | PD98059 + H89 . | LY294002 + H89 . |
|---|---|---|---|---|---|---|
| FL | 26 ± 6 | 34 ± 4 | 23 ± 5 | 13 ± 6 | 44 ± 3 | 38 ± 4 |
| CXCL12 | -7 ± 6 | 2 ± 6 | 4 ± 6 | 29 ± 7 | 29 ± 4 | 21 ± 6 |
| FL + CXCL12 | 23 ± 3* | 26 ± 3* | 22 ± 3* | 38 ± 2 | 37 ± 2 | 38 ± 2* |
. | PD98059 . | LY294002 . | H89 . | PD98059 + LY294002 . | PD98059 + H89 . | LY294002 + H89 . |
|---|---|---|---|---|---|---|
| FL | 26 ± 6 | 34 ± 4 | 23 ± 5 | 13 ± 6 | 44 ± 3 | 38 ± 4 |
| CXCL12 | -7 ± 6 | 2 ± 6 | 4 ± 6 | 29 ± 7 | 29 ± 4 | 21 ± 6 |
| FL + CXCL12 | 23 ± 3* | 26 ± 3* | 22 ± 3* | 38 ± 2 | 37 ± 2 | 38 ± 2* |
Ba/F3-Flt3 cells were washed and preincubated with 50 μM PD98059, 50 μM LY294002, 10 μM H89, or combinations of PD98059 and LY294002, PD98059 and H89, or LY294002 and H89 for one hour. Cells were extensively washed and subjected to migration toward 10 ng/mL FL and/or 100 ng/mL CXCL12 for 4 hours. Percentage inhibition of migration by the inhibitors compared with DMSO is shown. Data are the average of 6 experiments.
P < .05 compared with CXCL12 alone.
Treatment of cells with the combinations of PD98059 plus LY294002, PD98059 plus H89, or LY294002 plus H89 resulted in a greater inhibitory effect on migration to CXCL12 or FL plus CXCL12 compared with each inhibitor used alone (Table 2). The inhibitory effects of PD98059 plus LY294002 or PD98059 plus H89 on migration to FL plus CXCL12 were the same as CXCL12 alone, whereas pretreatment with LY294002 plus H89 resulted in greater inhibition of migration to FL plus CXCL12 than migration to CXCL12 alone. Inhibition of migration was not due to compound toxicity since forward and side scatter analysis demonstrated that more than 95% of cells remained viable following treatment with inhibitors and during the migration assay.
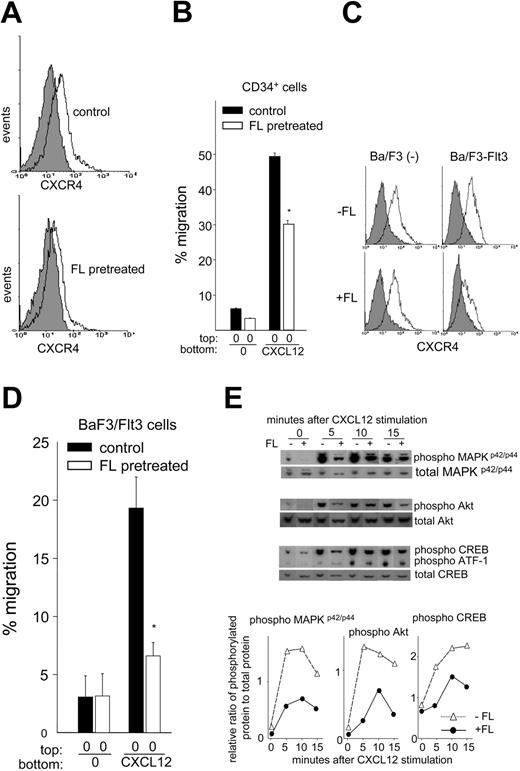
Prolonged incubation with FL down-modulates CXCR4, reduces signaling molecule phosphorylation, and negatively regulates migration induced by CXCL12
Disruption of CXCL12/CXCR4 interaction13,14 or multiday administration of FL to mice induces HSPC mobilization,3,22,23 and elevated serum FL is associated with mobilization induced by a multiday regimen of G-CSF.25 This finding suggests that prolonged exposure of CD34+ cells to FL might antagonize CXCL12/CXCR4 function and facilitate mobilization. Incubation of CD34+ cells with FL for 24 hours or Ba/F3-Flt3 cells for 8 hours had no effect on CXCR4 expression; however, culture of CD34+ cells with FL for 48 hours or Ba/F3-Flt3 cells for 24 hours significantly decreased CXCR4 expression (Figure 5A,C), coincident with significantly reduced migration toward CXCL12 (Figure 5B,D) compared with cells incubated without FL. CD34+ cell CXCR4 expression and migration to CXCL12 were reduced by 31% ± 5% (P < .001, n = 6) and 23% ± 6% (P < .05, n = 3), respectively, while Ba/F3-Flt3 cell CXCR4 expression and migration to CXCL12 were reduced by 52% ± 8% (P < .005, n = 3) and 64% ± 7% (P < .005, n = 3), respectively. FL had no effect on CXCR4 expression (Figure 5C) and migration (not shown) in vector-transduced Ba/F3 cells that do not express Flt3. Moreover, pretreatment of Ba/F3-Flt3 cells with FL for 24 hours inhibited phosphorylation of MAPKp42/p44, Akt, CREB, and ATF-1 induced by CXCL12 (Figure 5E). In control cells, MAPKp42/p44 phosphorylation peaked between 5 and 10 minutes after CXCL12 incubation and remained elevated but started to decline by 15 minutes. In contrast, FL pretreatment resulted in significantly weaker and delayed phosphorylation. CXCL12-induced Akt phosphorylation also peaked at 5 minutes and declined slightly thereafter in control cells, whereas in cells pretreated with FL, Akt phosphorylation reached maximum at 10 minutes and diminished rapidly at 15 minutes. Maximum CREB phosphorylation in response to CXCL12 was observed at 10 minutes and was sustained until 15 minutes in control cells but was weaker and reduced at 15 minutes in FL-pretreated cells. Similarly, ATF-1 phosphorylation was weaker in FL-treated cells than cells without FL treatment at 10 and 15 minutes (Figure 5E). These results suggest that with time, FL can downmodulate CXCR4 expression, inhibit CXCL12/CXCR4 signaling, and negatively regulate migration to CXCL12.
Effects of preculture of UCB CD34+ and Ba/F3-Flt3 cells with FL on migration to CXCL12, CXCR4 expression, and phosphorylation of intracellular signaling molecules. (A) Flow cytometric analysis of expression of cell surface CXCR4 on UCB CD34+ cells cultured with or without 100 ng/mL FL for 48 hours in 20% HI-FBS using antihuman CXCR4 antibody (clone 12G5). The filled and open histograms show isotype and CXCR4 staining, respectively. The top panel shows CXCR4 expression in cells incubated without FL, and the bottom panel shows cells pretreated with FL. The histogram is representative of 6 samples analyzed. (B) Percentage migration of UCB CD34+ cells cultured without (▪) or with 100 ng/mL FL (□) for 48 hours to CXCL12. After culture, cells were washed and subjected to migration assay with 100 ng/mL CXCL12. The panel represents 1 of the 3 experiments with similar results. *P < .001 compared with control cells without FL pretreatment. Data expressed are mean ± SEM. (C) Expression of cell surface CXCR4 on Ba/F3 cells or Ba/F3-Flt3 cells cultured with FL. Exponentially growing Ba/F3 cells or Ba/F3-Flt3 cells were cultured with (+) or without (–) 100 ng/mL rhFL in the presence of IL-3 for 24 hours. CXCR4 expression was analyzed by flow cytometry using antimouse CXCR4 antibody (clone 2B11). The filled and open histograms show isotype and CXCR4 staining, respectively. The data represent 1 of 3 experiments. (D) The graph shows mean ± SEM percent migration of Ba/F3-Flt3 cells to CXCL12 precultured without (▪) or with FL (□) for 24 hours from 3 experiments. *P < .001 compared with control cells without FL pretreatment. (E, top) Phosphorylation of MAPKp42/p44, Akt, CREB, and ATF-1 upon stimulation with 100 ng/mL CXCL12 in Ba/F3-Flt3 cells preincubated with (+) or without (–) FL. Following culture as described in panel C, cells were washed extensively and incubated with 100 ng/mL CXCL12 for up to 15 minutes. Protein phosphorylation relative to total protein was quantified by densitometry and shown in the bottom panel. ▵ indicates –FL; •, +FL.
Effects of preculture of UCB CD34+ and Ba/F3-Flt3 cells with FL on migration to CXCL12, CXCR4 expression, and phosphorylation of intracellular signaling molecules. (A) Flow cytometric analysis of expression of cell surface CXCR4 on UCB CD34+ cells cultured with or without 100 ng/mL FL for 48 hours in 20% HI-FBS using antihuman CXCR4 antibody (clone 12G5). The filled and open histograms show isotype and CXCR4 staining, respectively. The top panel shows CXCR4 expression in cells incubated without FL, and the bottom panel shows cells pretreated with FL. The histogram is representative of 6 samples analyzed. (B) Percentage migration of UCB CD34+ cells cultured without (▪) or with 100 ng/mL FL (□) for 48 hours to CXCL12. After culture, cells were washed and subjected to migration assay with 100 ng/mL CXCL12. The panel represents 1 of the 3 experiments with similar results. *P < .001 compared with control cells without FL pretreatment. Data expressed are mean ± SEM. (C) Expression of cell surface CXCR4 on Ba/F3 cells or Ba/F3-Flt3 cells cultured with FL. Exponentially growing Ba/F3 cells or Ba/F3-Flt3 cells were cultured with (+) or without (–) 100 ng/mL rhFL in the presence of IL-3 for 24 hours. CXCR4 expression was analyzed by flow cytometry using antimouse CXCR4 antibody (clone 2B11). The filled and open histograms show isotype and CXCR4 staining, respectively. The data represent 1 of 3 experiments. (D) The graph shows mean ± SEM percent migration of Ba/F3-Flt3 cells to CXCL12 precultured without (▪) or with FL (□) for 24 hours from 3 experiments. *P < .001 compared with control cells without FL pretreatment. (E, top) Phosphorylation of MAPKp42/p44, Akt, CREB, and ATF-1 upon stimulation with 100 ng/mL CXCL12 in Ba/F3-Flt3 cells preincubated with (+) or without (–) FL. Following culture as described in panel C, cells were washed extensively and incubated with 100 ng/mL CXCL12 for up to 15 minutes. Protein phosphorylation relative to total protein was quantified by densitometry and shown in the bottom panel. ▵ indicates –FL; •, +FL.
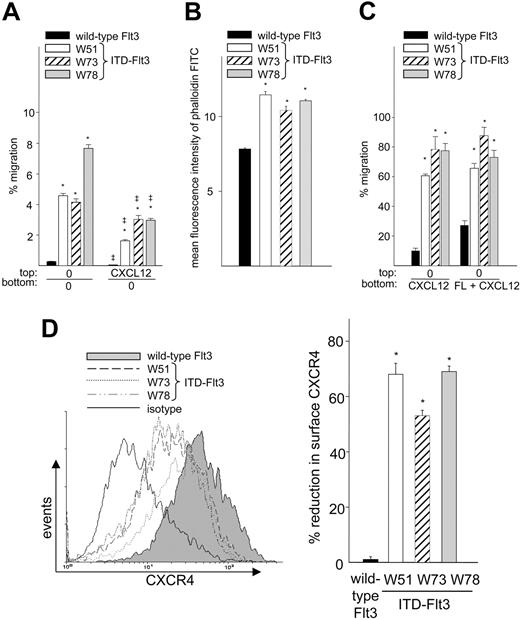
ITD-Flt3 increases migration to CXCL12
Flt3 is constitutively activated by autocrine mechanisms or ITD-Flt3 mutations in the majority of AML cells.26,28,29 Since AML cells can migrate to CXCL12,35 we investigated whether ITD-Flt3 affects CXCL12-induced migration. Forced expression of the ITD-Flt3 mutants W51, W73, and W78 in Ba/F3 cells resulted in respective 18 ± 1-, 17 ± 2-, and 31 ± 4-fold increases in spontaneous migration compared with cells expressing wild-type Flt3 (Figure 6A; 0/0) and were associated with increased baseline actin polymerization (46% ± 3%, 33% ± 3%, and 41% ± 1% increases in W51, W73, and W78, respectively, P < .001, compared with wild-type Flt3) (Figure 6B). In addition, migration of ITD-Flt3 Ba/F3 cells to CXCL12 was significantly increased compared with cells expressing wild-type Flt3 (0/CXCL12; Figure 6C). Identical results were observed in myeloid 32D cells expressing ectopic ITD-Flt3 compared with cells expressing wild-type Flt3 (not shown). No further enhancement in migration to FL alone (not shown) or FL plus CXCL12 (0/FL+CXCL12; Figure 6C) was observed in Ba/F3 cells expressing ITD-Flt3.
Migration, actin polymerization, and CXCR4 expression of Ba/F3 cells expressing wild-type or ITD-Flt3 mutations (W51, W73, and W78). (A) Ba/F3 cells expressing wild-type Flt3 (▪) or ITD-Flt3 (W51, □; W73, ▨; and W78, ▦) were subjected to transwell migration in the presence or absence of a negative gradient of 100 ng/mL CXCL12 with no cytokine in the lower chamber. These mutations in the juxtamembrane domain of Flt3 cloned from the patients with AML induce IL-3–independent growth of Ba/F3 cells and the W51 and W78 ITD-Flt3 induce lethal myeloproliferative disease in a murine bone marrow transplantation assay.32 Data represent mean ± SEM percentage migration for 3 experiments. *P < .001 compared with wild-type Flt3, and ‡P < .001 compared with 0/0 gradient. (B) Baseline actin polymerization of Ba/F3 cells expressing wild-type or ITD-Flt3 expressed as mean fluorescence intensity of phalloidin-FITC. Symbols indicate same information as in panel A. Data are the mean ± SEM of triplicate measurements from 1 of 2 experiments with similar results. *P < .001 compared with wild-type Flt3. (C) Migration of Ba/F3-Flt3 (wild type or ITD-Flt3) to 100 ng/mL rhCXCL12 or 10 ng/mL FL plus 100 ng/mL CXCL12. Symbols indicate same information as in panel A. Data are expressed as mean ± SEM percentage migration for 3 experiments. *P < .0001 compared with wild-type Flt3. (D, left) Expression of cell surface CXCR4 on Ba/F3 cells expressing ITD-Flt3 compared with cells expressing wild-type Flt3 analyzed by FACS. Histogram represents 1 of the 3 experiments with similar results. The open histogram with bold line shows isotype staining of cells expressing wild-type Flt3. The isotype for other cells was equivalent to cells expressing wild-type Flt3 (not shown). The right panel represents mean ± SEM percentage reduction of CXCR4 in cells expressing ITD-Flt3 compared with cells expressing wild-type Flt3 from 3 experiments. Symbols indicate same information as in panel A. *P < .001 compared with wild-type Flt3.
Migration, actin polymerization, and CXCR4 expression of Ba/F3 cells expressing wild-type or ITD-Flt3 mutations (W51, W73, and W78). (A) Ba/F3 cells expressing wild-type Flt3 (▪) or ITD-Flt3 (W51, □; W73, ▨; and W78, ▦) were subjected to transwell migration in the presence or absence of a negative gradient of 100 ng/mL CXCL12 with no cytokine in the lower chamber. These mutations in the juxtamembrane domain of Flt3 cloned from the patients with AML induce IL-3–independent growth of Ba/F3 cells and the W51 and W78 ITD-Flt3 induce lethal myeloproliferative disease in a murine bone marrow transplantation assay.32 Data represent mean ± SEM percentage migration for 3 experiments. *P < .001 compared with wild-type Flt3, and ‡P < .001 compared with 0/0 gradient. (B) Baseline actin polymerization of Ba/F3 cells expressing wild-type or ITD-Flt3 expressed as mean fluorescence intensity of phalloidin-FITC. Symbols indicate same information as in panel A. Data are the mean ± SEM of triplicate measurements from 1 of 2 experiments with similar results. *P < .001 compared with wild-type Flt3. (C) Migration of Ba/F3-Flt3 (wild type or ITD-Flt3) to 100 ng/mL rhCXCL12 or 10 ng/mL FL plus 100 ng/mL CXCL12. Symbols indicate same information as in panel A. Data are expressed as mean ± SEM percentage migration for 3 experiments. *P < .0001 compared with wild-type Flt3. (D, left) Expression of cell surface CXCR4 on Ba/F3 cells expressing ITD-Flt3 compared with cells expressing wild-type Flt3 analyzed by FACS. Histogram represents 1 of the 3 experiments with similar results. The open histogram with bold line shows isotype staining of cells expressing wild-type Flt3. The isotype for other cells was equivalent to cells expressing wild-type Flt3 (not shown). The right panel represents mean ± SEM percentage reduction of CXCR4 in cells expressing ITD-Flt3 compared with cells expressing wild-type Flt3 from 3 experiments. Symbols indicate same information as in panel A. *P < .001 compared with wild-type Flt3.
In a negative gradient of CXCL12 (CXCL12/0; Figure 6A), spontaneous migration of BaF3 cells expressing either wild-type or ITD-Flt3 was repressed; however, migration of ITD-Flt3 cells was significantly higher than cells expressing wild-type Flt3, indicating that ITD-Flt3 facilitates migration even in a negative gradient of CXCL12. Despite enhanced migration to CXCL12, cell surface CXCR4 was down-regulated by 53% to 69% in all ITD-Flt3–expressing Ba/F3 cells (Figure 6D) and by 39% ± 2% in 32D cells expressing ITD-Flt3 compared with cells expressing wild-type Flt3 (P < .001).
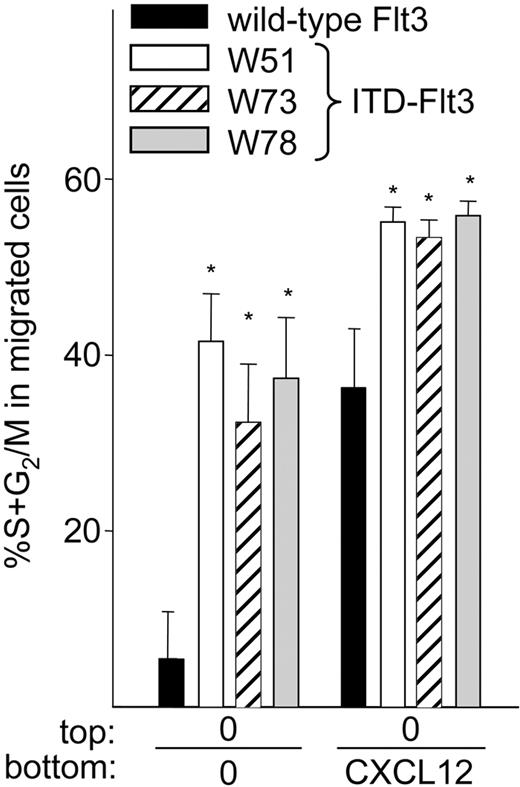
ITD-Flt3 increases migration of cells in S+G2/M phase
It has been suggested that hematopoietic cells that migrate in vitro represent a quiescent cell population.48,49 We evaluated the cell cycle distribution of migrated Ba/F3 cells expressing wild-type or ITD-Flt3 (Figure 7) and found that less than 10% of spontaneously migrating Ba/F3 cells expressing wild-type Flt3 were in S+G2/M phase, while more than 36% to 44% of spontaneously migrating Ba/F3 cells expressing ITD-Flt3 were in S+G2/M phase. Similarly, a significantly higher proportion of Ba/F3 cells expressing ITD-Flt3 and migrating to CXCL12 were in S+G2/M phase compared with cells expressing wild-type Flt3. The proportion of Ba/F3 cells in S+G2/M phase was comparable (50%-60%) between Ba/F3 cells expressing wild-type or ITD-Flt3. Incubation of Ba/F3 cells expressing wild-type or ITD-Flt3 with CXCL12 for 4 hours had no effect on cell cycle distribution (not shown), indicating that the larger S+G2/M fraction of migrating cells expressing ITD-Flt3 did not result from a larger number of S+G2/M input cells.
Cell cycle analysis of migrated wild-type or ITD-Flt3 Ba/F3 cells. Cells that had migrated after 4 hours were collected, fixed overnight in 1% paraformaldehyde, and stained with 1 μg/mL propidium iodide (Sigma Aldrich) in 0.6% nonidet P-40 (NP-40)/PBS with 1 μg/mL RNase (Sigma Aldrich), as described.40 Data were analyzed using a FACScan and ModFIT and Cell Quest software (Becton Dickinson) and expressed as the mean ± SEM of spontaneously or CXCL12-induced migrating cells in S+G2/M phase from 3 experiments. ▪ indicates wild-type Flt3; □, W51; ▨, W73; and ▦, W78. *P < .05 compared with wild-type Flt3.
Cell cycle analysis of migrated wild-type or ITD-Flt3 Ba/F3 cells. Cells that had migrated after 4 hours were collected, fixed overnight in 1% paraformaldehyde, and stained with 1 μg/mL propidium iodide (Sigma Aldrich) in 0.6% nonidet P-40 (NP-40)/PBS with 1 μg/mL RNase (Sigma Aldrich), as described.40 Data were analyzed using a FACScan and ModFIT and Cell Quest software (Becton Dickinson) and expressed as the mean ± SEM of spontaneously or CXCL12-induced migrating cells in S+G2/M phase from 3 experiments. ▪ indicates wild-type Flt3; □, W51; ▨, W73; and ▦, W78. *P < .05 compared with wild-type Flt3.
Discussion
Flt3 ligand and its receptor Flt3 have been implicated in survival, proliferation, and adhesion of HSPCs.27,50,51 We now provide compelling evidence that the FL/Flt3 axis also regulates migration of normal and transformed hematopoietic cells and that this effect is mediated through the CXCL12/CXCR4 axis. The combination of FL and CXCL12 synergistically enhances migration of CD34+ cells, Ba/F3-Flt3, and human acute leukemia RS4;11 cells. Furthermore, ITD-Flt3 derived from AML patients significantly enhanced spontaneous and CXCL12-induced migration when overexpressed in Ba/F3 and 32D cells. In contrast to enhanced migration in short-term assays, prolonged exposure to FL can down-modulate CXCR4 expression and impair migration of CD34+ cells and Ba/F3-Flt3 cells toward CXCL12. These findings indicate that FL can provide both positive and negative effects on CXCL12/CXCR4-mediated migration and suggest that the FL/Flt3 axis participates in the trafficking of normal and transformed hematopoietic cells.
The facts that FL had an effect only on Ba/F3 cells expressing ectopic Flt3 but not control vector–transduced Ba/F3 cells, which do not express Flt3; that the effect of FL on CD34+ cell migration increased with time; and that actin polymerization induced by FL directly paralleled cell migration all support a direct effect of FL on cell migration. The fact that positive or zero gradients of FL resulted in similar effects suggests that the predominant effect of FL is to enhance random cell migration or cell motility (ie, chemokinesis).
Although signaling pathways involved in hematopoietic cell migration are not completely understood, we found that synergistic migration to FL plus CXCL12 was associated with synergistic phosphorylation of MAPKp42/p44, Akt, and CREB, implicating these pathways in hematopoietic cell migration. The effects of inhibitors for MAPKp42/p44, PI3-kinase, or PKA were not selective for synergistic migration and did not completely block synergistic migration. Rather, the inhibitory effects of PD98059, LY294002, or H89 on migration by FL plus CXCL12 were almost identical with reduction in migration to FL alone (Table 2), suggesting that the inhibition of synergistic migration is likely due to antagonizing Flt3 signaling pathways. This is supported by the reduction in synergistic migration by AG1296, which blocks Flt3 activity.46,47 However, the failure of inhibitors for MAPKp42/p44, PI3-kinase, or PKA pathways to inhibit migration to CXCL12 under the conditions tested does not rule out involvement of these pathways in CXCL12-mediated migration. A role for PKA in CD14+ peripheral blood mononuclear cell migration or cytokine-mediated cancer cell migration has been demonstrated,52,53 and PI3-kinase has been implicated in CXCL12-mediated migration.54-56 A role for MAPKp42/p44 in CXCL12-mediated migration appears to be cell-type specific.54,57-59 Phosphorylation of MAPKp42/p44, Akt, and CREB by 100 ng/mL CXCL12 was consistently more than 10 ng/mL FL (Figure 4A), suggesting that the inhibitors may be less effective on CXCL12/CXCR4 than FL/Flt3 at the doses and time frame tested. Indeed, migration to CXCL12 was dramatically inhibited by combinations of inhibitors, compared with any of them used alone, coincident with further reduction in migration to FL plus CXCL12 (Table 2). This suggests that MAPKp42/p44, Akt, or PKA are involved in migration to CXCL12 and that inhibition of these pathways downstream of CXCL12/CXCR4 may also account for reduction of migration to FL plus CXCL12. Overall, these data suggest involvement of MAPKp42/p44, PI3-kinase, or PKA pathways in migration to FL plus CXCL12; however, our data also suggest the presence of pathways other than MAPKp42/p44, PI3-kinase/Akt, or PKA/CREB that regulate synergistic migration, since even the combination of all 3 inhibitors failed to completely inhibit migration to FL plus CXCL12 (not shown).
Our results suggest that positive and negative cross talk between the FL/Flt3 and CXCL12/CXCR4 pathways may differentially regulate HSPC homing and mobilization. The synergistic migration to FL plus CXCL12 observed in primary hematopoietic cells suggests that FL and CXCL12 may cooperatively enhance homing of HSPCs to the bone marrow micronenvironment, which is mediated by short-term activation of key intracellular signaling cascades. In contrast, multiday administration of FL can induce HSPC mobilization in mice.3,22-24 It has been suggested that marrow stroma–derived CXCL12 forms a positive gradient that maintains HSPCs within the marrow microenvironment,1,11 and disruption of CXCL12/CXCR4 interactions has been implicated in stem cell mobilization.6,12-14 Our findings demonstrate that prolonged incubation of CD34+ cells or Ba/F3-Flt3 cells with FL down-modulates migration to CXCL12, blocks CXCR4 signaling, and reduces cell surface CXCR4 expression, and therefore may be one mechanism that facilitates HSPC mobilization in response to FL. Consistent with the negative effect of FL on in vitro migration to CXCL12, preliminary studies indicate that preincubation of mouse bone marrow cells with FL significantly reduces homing of colony-forming unit–granulocyte-macrophage (CFU-GM) to recipient bone marrow in vivo (S.F. and L.M.P., unpublished observation, July 2004), suggesting that long-term exposure to FL can negatively regulate in vivo homing of progenitor cells.
FL increases CD34+ cell migration in a time-dependent fashion in vitro, suggesting that in vivo, the effects of FL on CXCL12-induced migration may also be time dependent. When short-term versus long-term migration of CD34+ cells was compared, the effect of FL alone after 48 hours became greater than the effect of FL plus CXCL12 at 4 hours. However, in contrast to FL, which induced random migration over the long term, FL plus CXCL12 induced directed migration to a positive gradient in short-term (4 hours) assays; although it also stimulated moderate random migration after 12 hours, indicating that the nature of long-term migration by FL and the early effect of FL plus CXCL12 are different. Synergistic migration by FL plus CXCL12 was observed within only 24 hours, suggesting that FL and CXCL12 synergy is an early effect. This may be due to desensitization of the CXCR4 receptor and/or disruption of a CXCL12 gradient. Although our data are limited, it would suggest that an early effect of FL in vivo may be to favor recruitment of cells to bone marrow through synergistic activation of CXCL12/CXCR4 pathways, whereas in the long term, FL or FL plus CXCL12 would increase random cell migration, and eventually, antagonize CXCL12/CXCR4 function, which would contribute to cell peripheralization.
The majority of AML cells or cell lines express Flt3 as well as CXCR49,26-28,35,36 and migrate in response to CXCL12. Our data indicate that RS4;11 acute leukemic cells, which coexpress wild-type Flt3 and CXCR4, demonstrated synergistic migration to FL plus CXCL12. Our results demonstrating that expression of ITD-Flt3 significantly accelerates migration of Ba/F3 and 32D cells to CXCL12 support a positive role of Flt3 in regulating migration to CXCL12, and are in good agreement with superior engraftment of AML cells expressing ITD-Flt3 in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice compared with cells expressing wild-type Flt3.60 This suggests that activation of Flt3 signaling either by ITD-Flt3 or autocrine mechanisms may enhance migration/homing of AML cells to secondary organs that express CXCL12, a notion supported by recent reports demonstrating a positive role of the CXCL12/CXCR4 axis in metastasis of breast and prostate cancers.61-63 Ba/F3 cells expressing ITD-Flt3 demonstrate significantly elevated migration even in the presence of a negative gradient of CXCL12, providing further evidence that Flt3 activation enhances cytokinesis and motility. Enhanced cell motility/migration resulting from activation of Flt3 signaling may allow transformed hematopoietic cells to migrate out of bone marrow despite existence of a positive marrow CXCL12 gradient. In addition, ITD-Flt3 facilitates migration of cells in S+G2/M phase, which may accelerate homing or peripheralization of actively dividing cells.
The percentage of Ba/F3 cells harboring ITD-Flt3 and migrating to CXCL12 was significantly higher than wild-type Flt3–expressing Ba/F3 cells migrating to FL plus CXCL12, suggesting that signals generated by ITD-Flt3 are not identical to those induced by FL and wild-type Flt3. ITD-Flt3–expressing Ba/F3 cells do not directly migrate to FL and show no enhanced migration to FL plus CXCL12, likely because autoactivation of ITD-Flt3 already provides a maximal response. Despite enhanced migration to CXCL12, CXCR4 expression on ITD-Flt3–expressing cells was down-regulated compared with cells expressing wild-type Flt3. This is consistent with down-regulation of CXCR4 in CD34+ cells and Ba/F3-Flt3 cells following prolonged culture with FL and suggests that activation of intracellular signaling pathways is likely responsible for enhanced migration. On the other hand, a recent report suggests a positive correlation between ITD-Flt3 expression and higher CXCR4 expression in AML cells, although it lacks direct evidence that higher CXCR4 levels are a consequence of ITD-Flt3.64
In summary, our data provide evidence that CXCL12-mediated hematopoietic cell migration can be modulated in positive and negative fashion by FL/Flt3, which may be important for trafficking of normal or malignant hematopoietic cells. Manipulation of their interaction could be clinically beneficial for hematopoietic cell transplantation and for treatment of hematopoietic malignancies in which both functional Flt3 and CXCR4 are expressed. Activation of FL/Flt3 and CXCL12/CXCR4 signaling pathways may enhance homing of normal HSPCs to the bone marrow microenvironment. In this regard, enhanced migration of quiescent Ki-67negative CD34+ cells42,43 to CXCL12 by FL suggest that FL may facilitate homing of CD34+ cells with long-term in vivo engraftment ability.65,66 Stimulating the FL/Flt3 axis could potentially facilitate collection of HSPCs in patients and donors undergoing peripheral blood stem cell mobilization for transplantation by attenuating CXCL12/CXCR4 function while blocking interaction of the FL/Flt3 and CXCL12/CXCR4 signaling pathways by Flt3 inhibitors currently used in clinical trails,67,68 and AMD3100, which can block cell migration to CXCL12,69,70 may help to reduce extramedullary leukemia infiltration in patients with AML.
Prepublished online as Blood First Edition Paper, December 23, 2004; DOI 10.1182/blood-2004-04-1440.
Supported by US Public Health Service grants HL69669 and HL79654 (L.M.P.); and DK53674, HL67384, and HL56416 (H.E.B.) from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Chang H. Kim, PhD, Department of Pathobiology, Purdue University for critical reading of the manuscript; Huimin Bian for technical assistance; Susan Rice and Denessa Luckett for cell sorting; and Constantin T. Yiannoutsos, PhD, and Qianqian Zhao, Biostatistics Core Facility, Indiana University Cancer Center for statistical analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal