Abstract
We have identified a homozygous G>A substitution in the donor splice site of intron 6 (IVS6 + 1G>A) of the cytidine monophosphate (CMP)–sialic acid transporter gene of Lec2 cells as the mutation responsible for their asialo phenotype. These cells were used in complementation studies to test the activity of the 2 CMP–sialic acid transporter cDNA alleles of a patient devoid of sialyl-Lex expression on polymorphonuclear cells. No complementation was obtained with either of the 2 patient alleles, whereas full restoration of the sialylated phenotype was obtained in the Lec2 cells transfected with the corresponding human wild-type transcript. The inactivation of one patient allele by a double microdeletion inducing a premature stop codon at position 327 and a splice mutation of the other allele inducing a 130–base pair (bp) deletion and a premature stop codon at position 684 are proposed to be the causal defects of this disease. A 4-base insertion in intron 6 was found in the mother and is proposed to be responsible for the splice mutation. We conclude that this defect is a new type of congenital disorder of glycosylation (CDG) of type IIf affecting the transport of CMP–sialic acid into the Golgi apparatus.
Introduction
Four years ago we described a new syndrome in a 4-month-old boy with macrothrombocytopenia, neutropenia, and complete lack of the sialyl-Lex antigen, or CD15s (NeuAcα2,3Galβ1,4(Fucα1,3)GlcNAc-R), on polymorphonuclear cells (PMNs).1 The patient presented initially with a spontaneous massive bleeding of the posterior chamber of the right eye and cutaneous hemorrhages. The clinical status worsened over a period of 30 months with more severe hemorrhages due to severe thrombocytopenia (2 × 109/L to 6 × 109/L), respiratory distress syndrome, and opportunistic infections. Treatment with transfusions and steroids temporarily ameliorated his condition, and bone marrow transplantation was performed at the age of 34 months. However, complications that included graft-versus-host disease, pulmonary viral infection, and massive pulmonary hemorrhage with refractory respiratory failure led to death at the age of 37 months. Ultrastructural studies showed hypogranular giant platelets. Bone marrow aspirates gave normocellular marrow with megakaryocytic hyperplasia. Abnormalities of the megakaryocyte morphology included numerous small mononuclear or hyposegmented megakaryocytes, vacuolated cells, and abnormal fragmentation of megakaryocyte cytoplasm into large platelet masses.1
Besides the lack of sialyl-Lex antigen on PMNs, no other abnormalities in chemotaxis or generation of reactive oxygen species were detected. The expression of β2-integrin adhesion molecules and of L-selectin was normal. The genes involved in the coagulation cascade (Wiskott-Aldrich syndrome protein gene, von Willebrand factor VIII, glycoprotein (Gp) Ib binding domain, GpV, and GpIX) were normal. The α1,3-fucosyltransferases (FUT4, FUT7) and the α2,3-sialyltransferases (ST3Gal-IV, ST3Gal-VI) involved in the synthesis of sialyl-Lex and the corresponding enzyme activities were also normal.1 At this time, we were unsuccessful in our attempts to delineate the causal molecular defects, but a congenital disorder of glycosylation (CDG) was suspected due to the lack of sialyl-Lex on PMNs.
In a broad sense, CDGs comprise all genetic defects leading to incomplete N-glycans or other altered glycoconjugates including O-glycans and glycosaminoglycans (for review see Jaeken,2 Marquardt and Denecke,3 Dupré et al,4 and Jaeken and Carchon5 ). They comprise 2 main types: (1) type I CDGs are caused by assembly defects in the endoplasmic reticulum (ER) of the dolichylpyrophosphate-linked oligosaccharide N-glycan precursor or its transfer to glycoproteins; (2) type II CDGs were initially described as defects of the Golgi compartment glycosylation machinery resulting in alterations of the N-glycan maturation steps but have recently been extended to many other glycosylation defects.
Among the type II CDGs, the inactivation of the Golgi guanosine diphosphate (GDP)–fucose transporter,6,7 a member of the nucleotide sugar transporter family,8-11 has been named CDG-IIc or leukocyte adhesion deficiency II (LAD II).12 This CDG-IIc is characterized by the absence of all fucosylated glycoconjugates including ABH, Lea, and Leb blood groups; the Lex antigen or CD15; and the sialyl-Lex antigen or CD15s implicated in the leukocyte selectin adhesion system. However, no alteration of the O-fucosylated glycans was observed.13
Since ABH, Lea, and Leb blood groups and Lex antigens as well as all the glycosyltransferases involved in the biosynthesis of sialyl-Lex were normal in our patient, we looked for an eventual deficiency of the cytidine monophosphate (CMP)–sialic acid transporter gene, which encodes for a protein responsible for the translocation of CMP–sialic acid from the cytosol to the Golgi apparatus,14 as the origin of the lack of sialyl-Lex in this patient.
A similar lack of Golgi CMP–sialic acid, secondary to a deficiency of the CMP–sialic acid transporter, occurs in the Lec2 mutant cells derived from the Chinese hamster ovary (CHO) cell line.14-16 In this work, we have characterized the mutation in the gene of the transporter of the CMP–sialic acid leading to its inactivation in the Lec2 cell line, and we have used these cells for complementation studies to test the activity of the 2 cDNA alleles of the CMP–sialic acid transporter expressed in the leukocytes of this patient.
Materials and methods
Cell cultures
Wild-type CHO cell lines and their mutated Lec2 variant were obtained from the American Type Culture Collection (Rockville, MD). Both cell lines were cultured at 37°C in a 5% CO2 atmosphere in an alpha-modified minimum essential medium (α-MEM; Eurobio Laboratories, Les Ulis, France) supplemented with ribosides and deoxyribosides and with 10% decomplemented fetal calf serum (Eurobio Laboratories), 2 mM glutamine, 1% antibiotics solution (penicillin 100 units/mL and streptomycin 100 μg/mL).
Amplification of cDNA alleles of the CMP–sialic acid transporter gene by reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was isolated from a buffy coat containing an enriched fraction of PMNs from the patient1 and from a healthy individual using the trizol reagent (Invitrogen SARL, Cergy-Pontoise, France). Total RNA was also isolated from the CHO and Lec2 cell lines. Contaminating DNA was removed from each RNA preparation by digestion with RNAse free DNAseI, as already described.17 To initiate the first cDNA strand synthesis, 1 μg of total RNA was reverse transcribed at 42°C for 90 minutes, using the oligo-dT-cDNA synthesis primer and 200 units of the superscript II RNAse H–reverse transcriptase from the superscript system kit (Invitrogen SARL).
The cDNA alleles of the CMP–sialic acid transporter gene of the patient and the control individual were amplified using a 2-step PCR with human primers (TRN-1s and TRN-6as; Table 1). The first PCR was carried out with the Klentaq mixture (Clontech-Ozyme, Saint-Quentin en Yvelines, France) and 2 μL of the cDNAtemplates. The second PCR was made with 1 μL of the corresponding first PCR products diluted 1:10. All of the PCRs were performed in 50 μL Klentaq buffer containing 0.2 μM of each primer, 1 unit of Klentaq DNApolymerase, and 0.2 mM deoxynucleoside triphosphate (dNTP) with the touch-down program: initial denaturation at 94°C for 90 seconds, followed by 5 cycles of 94°C for 30 seconds and 72°C for 4 minutes; 5 cycles of 94°C for 30 seconds and 70°C for 4 minutes; and 25 cycles of 94°C for 30 seconds and 68°C for 4 minutes. For the alleles of CHO and Lec2 cell lines, we used the same type of PCR mixture with the corresponding primers (HA-s and HA-as; Table 1) and the corresponding cDNA of CHO and Lec2 cells. In all cases we amplified the corresponding putative CMP–sialic acid transporter cDNA encompassing the entire coding region.
Oligonucleotide primer sequences used for CMP-sialic acid transporter amplifications
Species primers . | Sequences 5′ to 3′ . | cDNA positions* . | Comments . |
|---|---|---|---|
| Human | |||
| TRN-1s | GGCTGTCGGGGAACCATGGCTGC | - 15..8 | cDNA cloning |
| NTR-6as | CCGGAACAAGAGAACAGTTAGTACAG | 1407..1382 | cDNA cloning |
| TRN-2s | GAACCATGGCTGCCCCGAGAGAC | - 5..18 | SfaNI digestion |
| TRN-5as | TCCACATAACCGCACTGATCATGCC | 1144..1120 | SfaNI digestion |
| NTR-3s | CTGGATGCAGCAGTGTACCAGGTG | - 334..-357 | Hybridization probe |
| NTR-4as | GGAAAGGACAATGGCCGCTGCTGC | 848..825 | Hybridization probe |
| INT-6s | GTACAGTATGTCATGTGTCACACAACC | intron 5 | Intron 6 amplification |
| NTR-4as | GGAAAGGACAATGGCCGCTGCTGC | 848..825 | Intron 6 amplification |
| Hamster | |||
| HA-s | CTCTGCACCATGGCTCAGGCGAG | - 9..14 | cDNA cloning |
| HA-as | AATCTCACACCAATGACTCTTTCT | 1230..1180 | cDNA cloning |
| HA-Ex6s | CAGGGATCGTGGTGACGTTAGTTGG | 647..671 | Intron 6 amplification |
| HA-NTRas | AGAAAGAACAATGGCTGCAGCTGC | 849..826 | Intron 6 amplification |
Species primers . | Sequences 5′ to 3′ . | cDNA positions* . | Comments . |
|---|---|---|---|
| Human | |||
| TRN-1s | GGCTGTCGGGGAACCATGGCTGC | - 15..8 | cDNA cloning |
| NTR-6as | CCGGAACAAGAGAACAGTTAGTACAG | 1407..1382 | cDNA cloning |
| TRN-2s | GAACCATGGCTGCCCCGAGAGAC | - 5..18 | SfaNI digestion |
| TRN-5as | TCCACATAACCGCACTGATCATGCC | 1144..1120 | SfaNI digestion |
| NTR-3s | CTGGATGCAGCAGTGTACCAGGTG | - 334..-357 | Hybridization probe |
| NTR-4as | GGAAAGGACAATGGCCGCTGCTGC | 848..825 | Hybridization probe |
| INT-6s | GTACAGTATGTCATGTGTCACACAACC | intron 5 | Intron 6 amplification |
| NTR-4as | GGAAAGGACAATGGCCGCTGCTGC | 848..825 | Intron 6 amplification |
| Hamster | |||
| HA-s | CTCTGCACCATGGCTCAGGCGAG | - 9..14 | cDNA cloning |
| HA-as | AATCTCACACCAATGACTCTTTCT | 1230..1180 | cDNA cloning |
| HA-Ex6s | CAGGGATCGTGGTGACGTTAGTTGG | 647..671 | Intron 6 amplification |
| HA-NTRas | AGAAAGAACAATGGCTGCAGCTGC | 849..826 | Intron 6 amplification |
Numbering considers the A of the starting codon ATG as position 1.
PCR products were purified from agarose gels and subcloned into the TA cloning PCR3.1 vector (Invitrogen SARL) and used to transform TOP10F bacteria. Sixty clones from culture-positive plasmids were selected by PCR using the corresponding primers: NTR-3s and NTR-4as for the human and HA-s and HA-as for the hamster alleles (Table 1). Full-length cDNA isoforms of CMP–sialic acid transporter, containing the inserts in sense orientation (with respect to the PCR3.1-cytomegalovirus promoter) and devoid of PCR errors, were selected after sequencing.
Cloning of the human cDNA alleles into a recombinant adeno-associated virus for gene complementation of Lec2-deficient cells
We gel purified 2 BamHI-XbaI fragments containing the 2 major patient cDNA alleles of the CMP–sialic acid transporter gene and the wild-type sequence amplified from a healthy individual. These 3 sequences were cloned in the multiple cloning site (MCS) of the plasmid adeno-associated virus (pAAV)–MCS vector of the Stratagene AAV-helper-free system (Stratagene, La Jolla, CA) via the BamHI and XbaI sites to generate recombinant AAV expression plasmids. After cloning, the presence of the inserts was verified by PCR and by sequence analysis. The inverted terminal repeat sequences present in the vector provide the cis-acting elements necessary for the adenovirus replication and the packaging of the viral particles. To produce the recombinant viral particle supernatants with titers of 1010 viral particles/mL, we cotransfected into HEK293 cells 10 μg of the 3 recombinant expression plasmids with pHelper (carrying adenovirus-derived genes) and pAAV-RC (carrying AVV2 replication and capside RC genes) vectors, which together supply all the trans-acting factors required for AVV replication and packaging in the HEK293 cells, following the manufacturer's recommendations.
The recombinant AVV2 viral particles were prepared from infected HEK293 cells following the manufacturer's recommendations, and before complementation of the Lec2 cells, 10 μL of virus-containing supernatant was used to confirm the presence of the insert by PCR amplification using the specific primers (NTR-3s and NTR-4as; Table 1) of the CMP–sialic acid transporter cDNA inserts.
About 3 × 105 Lec2 cells were plated in a 6-well plate, and after 24 hours 1 mL of the viral stock supernatant (∼ 1010 viral particles/mL) diluted with 1 mL of complete α-MEM medium was added to each well. The cells were then incubated at 37°C in a 5% CO2 atmosphere for 24 hours. The following day we renewed the viral supernatant for 24 hours under the same conditions. After 48 hours of transduction, we evaluated the rate of complementation by immunofluorescence labeling of the cells with the fluorescein isothiocyanate (FITC)–labeled lectin of Maakia amurensis (MAA) and the tetramethyl rhodamine isothiocyanate (TRITC)–labeled peanut agglutinin (PNA; Vector Laboratories, Burlingame, CA). The MAA lectin reacts mainly with the sialylated structures Neu5Acα(2,3)Galβ(1,4)GlcNAc/Glc or Neu5Acα(2,3)Galβ(1,3)GalNAc, whereas the PNA lectin reacts mainly with the nonsialylated Galβ(1,3)GalNAc precursor structure.
Immunofluorescent detection of α2,3-sialyl-glycans and their precursors on Lec2 cells
The 2 × 105 cells of the native or transduced Lec2 cells were seeded on glass cover slides in 35-mm culture Petri dishes and incubated for 24 hours. Then the cells were washed with phosphate-buffered saline (PBS) and fixed for 15 minutes in 2% paraformaldehyde in PBS, then 50 mM NH4Cl in PBS was added for 20 minutes to neutralize aldehyde residues. Thereafter, cells were permeabilized with 0.075% saponin in PBS with 0.1% bovine serum albumin (BSA) for 15 minutes, followed by 1-hour incubation with the FITC-labeled MAA lectin at a final concentration of 10 μg/mL. After 3 washings with PBS, the cells were incubated with the second lectin labeled with TRITC (PNA at 10 μg/mL). Final washings stopped the reaction, and the coverslips with the labeled cells were mounted on slides with Mowiol 4-88 (Hoechst, Frankfurt, Germany) for observation on a Leica DMR epifluorescence microscope equipped with a PLAN/APO 63×/1.32-06 oil objective lens and an MC PLAN 10×/0.25 ocular (Leica, Heidelberg, Germany). Images were captured using a Leica LEI-750 CE digital camera and Lida Grad software volme 54 (Leica). Images were further processed with Adobe Photoshop 7.0 (Adobe, San Jose, CA).
Genomic DNA amplification of intron 6
A genomic fragment, from Lec2 and CHO cells, of 703 bp containing the entire intron 6 was amplified with primers located in exons 5 and 7 (Table 1) and the AccuPrime Pfx DNA polymerase from Invitrogen SARL, following the manufacturer's recommendations. A similar fragment of 896 bp from the human maternal genomic DNA was amplified with the corresponding human primers (Table 1).
DNA sequencing
All cDNAs and genomic samples of the CMP–sialic acid transporter were cloned into the expression vector PCR3.1 (Eukaryotic TA cloning kit; Invitrogen SARL) and sequenced (Biofidal SARL, Vaulx-en-Velin, France) in both directions with the corresponding primers. Twenty positive plasmids containing the A1 and A2 alleles of the patient CMP–sialic acid transporter gene were sequenced (European Molecular Biology Laboratory [EMBL] accession nos. AJ810302, AJ810303). Seventy clones of the coding region of the CMP–sialic acid transporter gene of the individual heterozygous for the CACT intron 6 insertion were sequenced (EMBL accession nos. AJ851888, AJ851889, and AJ851890). Ten clones for the hamster CHO and Lec2 intron 6 fragment (EMBL accession nos. AJ810300 and AJ810301) and 10 clones of the maternal intron 6 fragment were also sequenced.
Results
Molecular mechanism of the inactivation of the transporter of CMP–sialic acid in the hamster model
The Lec2 mutants exhibit an asialo phenotype at the cell membrane18 and are unable to translocate CMP–sialic acid to the lumen of the Golgi apparatus. Previous studies19,20 have described the cDNA profile of the mutated Lec2 cells, which have a homozygous skipping of exon 6. Since the peptide encoded by this exon contains 2 highly conserved peptide motifs that have been proposed to participate in the recognition of CMP–sialic acid,10 this homozygous lack of exon 6 is probably responsible for the asialo phenotype of the Lec2 cells. The expression of transcripts with skipping of the entire exon 6 suggested the existence of a splice site mutation. We amplified, from genomic DNA of CHO and Lec2 cells, a 703-bp DNA fragment encompassing the entire intron 6 region with the 5′ and 3′ intron/exon boundaries. All the genomic clones of Lec2 had a G>A mutation in the donor splice site of intron 6 (Figure 1; IVS6 + 1G>A21 ). This homozygous substitution of G by A changes the regular splice donor site “GT” to “AT,” which cannot work as a donor splice site. Therefore, this mutation creates a novel splicing event, joining exon 5 directly to exon 7.
Hamster CMP–sialic acid transporter transcripts of CHO and Lec2 cells. The heterozygous exon 2 deletion is present in both CHO and Lec2 cell lines, whereas the homozygous deletion of exon 6 is present only in the mutated Lec2 cell line. This deletion is generated by the G>A substitution in the donor splice site of intron 6 (IVS6 + 1G>A). aa, amino acid.
Hamster CMP–sialic acid transporter transcripts of CHO and Lec2 cells. The heterozygous exon 2 deletion is present in both CHO and Lec2 cell lines, whereas the homozygous deletion of exon 6 is present only in the mutated Lec2 cell line. This deletion is generated by the G>A substitution in the donor splice site of intron 6 (IVS6 + 1G>A). aa, amino acid.
Cloning of the CMP–sialic acid transporter transcripts found in the patient
The human CMP–sialic acid transporter gene encodes for a protein of 337 amino acids.22 The cDNAs were synthesized by RT-PCR from total mRNA extracted from the patient's PMNs. The first PCR with the primers TRN-1s and NTR-6as (Table 1) gave only a faint smear. Two second PCRs were performed, one with the same primers and the other with the nested primers (TRN-2S and TRN-5as; Table 1). Both second PCRs gave 2 major bands corresponding to the wild-type transcript size and a weaker band corresponding to a shorter transcript. These 2 bands amplified with TRN-1s and NTR-6as were subcloned in the PCR3.1 vector. The sequence analysis of the normal size transcript (1410 bp) revealed the nucleotide substitution 147T>C resulting in no change at the amino acid level but generating a new SfaNI restriction site in this allele. In addition, this allele showed 2 microdeletions of G277 and C281 that create another new SfaNI restriction site. These 2 microdeletions generate a frameshift at position 277 and the appearance of a premature stop codon at position 327, reducing the protein by two thirds of its length (A1 allele; Figure 2; EMBL accession no. AJ810302).
Genomic organization of the human normal wild-type and the patient A1 and A2 transcripts of the CMP–sialic acid transporter gene. The A1 allele contains a 147T>C substitution and 2 microdeletions (G277 and C281) that generate a frameshift (▨) and the appearance of a premature stop codon at position 327. The A2 allele has a 130-bp deletion covering two thirds of exon 6 and generating also a frameshift (▨) and a premature stop codon at position 684.
Genomic organization of the human normal wild-type and the patient A1 and A2 transcripts of the CMP–sialic acid transporter gene. The A1 allele contains a 147T>C substitution and 2 microdeletions (G277 and C281) that generate a frameshift (▨) and the appearance of a premature stop codon at position 327. The A2 allele has a 130-bp deletion covering two thirds of exon 6 and generating also a frameshift (▨) and a premature stop codon at position 684.
The sequence of the plasmid with the insert corresponding to the smaller PCR band (1293 bp) revealed a deletion of 130 bp corresponding to a partial skipping of exon 6 that could be due to the use of a cryptic donor splice site within exon 6. This 130-bp deletion results also in a frameshift and the appearance of a premature stop codon at position 684 with a reduction by about one third of the length of the translated protein (A2 allele; Figure 2; EMBL accession no. AJ810303).
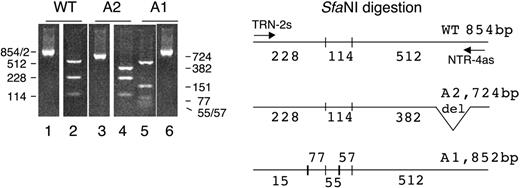
Figure 3 represents the SfaNI restriction profile of the cDNA transcripts observed in the patient compared with a wild-type cDNA profile. The PCR with the nested primers TRN-2s and NTR-4as (Table 1) gives a broad product of 854 bp (Figure 3, lane 1) in the normal wild-type sequence and 2 bands of 724 bp (Figure 3, lane 3) and 852 bp (Figure 3, lane 6) with the 2 patient alleles. After SfaNI digestion of the 3 PCR products, we obtained 3 distinct SfaNI profiles. The wild-type allele has 2 SfaNI restriction sites giving 3 fragments of 228 bp, 114 bp, and 512 bp (Figure 3, lane 2). The patient allele A2 with the 130-bp deletion gave the same 2 smaller fragments of 228 bp and 114 bp plus a fragment of 382 bp instead of the 512-bp fragment, due to the 130-bp deletion in this allele (Figure 3, lane 4). The A1 allele with the 147T>C mutation and the double microdeletion of G and C had 2 new SfaNI restriction sites, giving a restriction profile with 5 fragments of 151 bp, 77 bp, 55 bp, 57 bp, and 512 bp (Figure 3, lane 5).
SfaNI restriction of the normal wild-type 854-bp fragment (WT) and the 2 patient fragments of 724 bp (A2) and 852 bp (A1). The cDNAs were amplified by PCR with the primers TRN-2s and NTR-4as (Table 1). The SfaNI digestion of the wild type gives 3 fragments of 228, 114, and 512 bp in the wild type; 3 fragments of 228, 114, and 382 bp in the patient's A2 allele; and 5 fragments of 151, 77, 55, 57, and 512 bp in the patient's A1 allele. The size reduction from 512 to 382 of the third A2 digestion fragment is due to the lack of the 130 bp corresponding to the partial skipping of exon 6. The 5 digestion fragments of the A1 allele are due to the creation of 2 new SfaNI restriction sites, one in the 228 fragment that is split in 2 fragments of 151 and 77 bp and one in the 112 fragment that is split into 2 fragments of 55 and 57 bp.
SfaNI restriction of the normal wild-type 854-bp fragment (WT) and the 2 patient fragments of 724 bp (A2) and 852 bp (A1). The cDNAs were amplified by PCR with the primers TRN-2s and NTR-4as (Table 1). The SfaNI digestion of the wild type gives 3 fragments of 228, 114, and 512 bp in the wild type; 3 fragments of 228, 114, and 382 bp in the patient's A2 allele; and 5 fragments of 151, 77, 55, 57, and 512 bp in the patient's A1 allele. The size reduction from 512 to 382 of the third A2 digestion fragment is due to the lack of the 130 bp corresponding to the partial skipping of exon 6. The 5 digestion fragments of the A1 allele are due to the creation of 2 new SfaNI restriction sites, one in the 228 fragment that is split in 2 fragments of 151 and 77 bp and one in the 112 fragment that is split into 2 fragments of 55 and 57 bp.
Parental alleles
Both parents had an apparently normal life. The father was dead at the time of the study and no biologic samples were available, but genomic DNA of the mother was available. This maternal DNA had a homozygous insertion of 4 bases (CACT) in intron 6 of the CMP–sialic acid transporter gene, creating a new U2 snRNA site ACTCAAT (Figure 4). This U2 site is in competition with the putative normal U2 snRNA site GAGTGAT23,24 and can induce alterations in the splicing of exon 6. The 2 microdeletions G277 and C281 and the 147T>C substitution of the A1 allele were not found in the maternal cDNA, suggesting a paternal origin.
Sequence of a fragment of the CMP–sialic acid transporter gene of the maternal genomic DNA amplified with the underlined primers (Table 1). This fragment contains the 3′ terminus of intron 5, the exon 6, the intron 6, and the beginning of exon 7 (exons in capital letters and introns in lowercase letters). The sequence of the intron 6 revealed the homozygous insertion of 4 bases CACT (bold capitals) inducing a new snRNA U2 site (ACTcaat, double underlined) in all the clones of the maternal genomic DNA. The original putative snRNA U2 site at 27 base pairs downstream from the acceptor splice site (gagtgat) is also double underlined. The sequence of the eukaryote consensus snRNA U2 site is YNYYRAY.23,24
Sequence of a fragment of the CMP–sialic acid transporter gene of the maternal genomic DNA amplified with the underlined primers (Table 1). This fragment contains the 3′ terminus of intron 5, the exon 6, the intron 6, and the beginning of exon 7 (exons in capital letters and introns in lowercase letters). The sequence of the intron 6 revealed the homozygous insertion of 4 bases CACT (bold capitals) inducing a new snRNA U2 site (ACTcaat, double underlined) in all the clones of the maternal genomic DNA. The original putative snRNA U2 site at 27 base pairs downstream from the acceptor splice site (gagtgat) is also double underlined. The sequence of the eukaryote consensus snRNA U2 site is YNYYRAY.23,24
Transcript variants induced by the CACT insertion in intron 6 of the CMP–sialic acid transporter gene
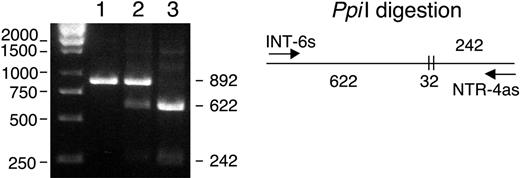
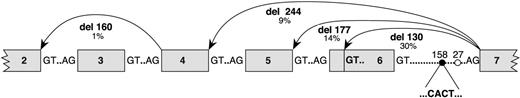
A population of 53 blood donors was screened for the presence of the intron 6 CACT insertion with the restriction enzyme PpiI (MBI Fermentas GMBH; St Leon-Rot, Germany; Figure 5). The cDNA of an individual heterozygous for the CACT insertion, like the patient, was amplified. Two major bands corresponding to the wild-type and the 130-bp deletion, respectively, were detected, but some faint lower bands suggested the presence of smaller transcripts. The expressed transcripts were subcloned in the PCR3.1 vector and 70 clones were sequenced: 33 clones (47%) had a wild-type sequence with no alterations; 20 clones (29%) had the same 130-bp deletion observed in the patient A2 allele corresponding to the partial skipping of exon 6; 10 clones (14%) had a larger deletion of 177 bp corresponding to skipping of the entire exon 6 (EMBL accession no. AJ851888); 6 clones (9%) had a deletion of 244 bp corresponding to skipping of exons 5 and 6 (EMBL accession no. AJ851889); 1 clone (1%) had the 130-bp deletion together with a 160-bp deletion corresponding to skipping of exons 3 and 6 (EMBL accession no. AJ851890; Figure 6).
PpiI restriction pattern of the human intron 6 fragments amplified with primers INT-6s and NTR-4as (Table 1). Wild type (1), heterozygous blood donor (2), and maternal homozygous (3) with the double CACT insertion. A single undigested band of 892 bp is seen with the wild type. The heterozygous individual presents the wild-type 892-bp band and 2 lower bands of 622 and 242 bp from the digested mutated allele. The maternal sequence presents the homozygous mutated profile with the 2 digested bands of 622 and 242 bp. The Ppi I enzyme cuts twice before and after the digestion site, giving 2 large fragments of 622 and 242 bp and a small fragment of 32 bp (not shown).
PpiI restriction pattern of the human intron 6 fragments amplified with primers INT-6s and NTR-4as (Table 1). Wild type (1), heterozygous blood donor (2), and maternal homozygous (3) with the double CACT insertion. A single undigested band of 892 bp is seen with the wild type. The heterozygous individual presents the wild-type 892-bp band and 2 lower bands of 622 and 242 bp from the digested mutated allele. The maternal sequence presents the homozygous mutated profile with the 2 digested bands of 622 and 242 bp. The Ppi I enzyme cuts twice before and after the digestion site, giving 2 large fragments of 622 and 242 bp and a small fragment of 32 bp (not shown).
Splice mutation variants expressed in a heterozygous CACT individual. The insertion CACT detected in the intron 6 of the CMP–sialic acid transporter is located at 158 bp upstream from the splice acceptor site (•). This insertion creates a new U2 snRNA site that is in competition with the putative normal U2 snRNA site (○) located 27 bp upstream from the splice acceptor site. Five different transcript variants were detected among the 70 sequenced clones: wild-type gene (47%), partial skipping of exon 6 (29%), full skipping of exon 6 (14%), full skipping of exons 5 and 6 (9%), and partial skipping of exon 6 plus full skipping of exon 3 (1%).
Splice mutation variants expressed in a heterozygous CACT individual. The insertion CACT detected in the intron 6 of the CMP–sialic acid transporter is located at 158 bp upstream from the splice acceptor site (•). This insertion creates a new U2 snRNA site that is in competition with the putative normal U2 snRNA site (○) located 27 bp upstream from the splice acceptor site. Five different transcript variants were detected among the 70 sequenced clones: wild-type gene (47%), partial skipping of exon 6 (29%), full skipping of exon 6 (14%), full skipping of exons 5 and 6 (9%), and partial skipping of exon 6 plus full skipping of exon 3 (1%).
Complementation of Lec2 with the wild-type human CMP–sialic acid transporter sequence and lack of complementation with either of the 2 patient cDNA alleles
Before proceeding to genetic complementation studies we confirmed the asialo phenotype of the Lec2 cell line by lectin analysis using peanut agglutinin (PNA), which recognizes preferentially the nonsialylated precursor Galβ1,3GalNAc, and the Maackia amurensis agglutinin (MAA), which recognizes α2,3 bound sialic acid on glycoproteins or glycolipids. All the Lec2 cells were stained with PNA, which reveals the presence of the nonsialylated precursor oligosaccharide. No MAA-stained cells were seen, illustrating the absence of α2,3-sialylated structures on the Lec2 mutant cells. The parental CHO cell line gives the opposite profile with 100% of the cells labeled with MAA and none labeled with PNA. This demonstrates that all the detected precursor structures expressed in the wild-type CHO cells are sialylated and recognized by the MAA lectin (Figure 7A).
Fluorescent staining with PNA and MAA lectins of CHO, Lec2, and complemented Lec2 cells with the 2 patient alleles. (A) All cells are MAA– and PNA+ in the Lec2 cell line (expression of nonsialylated precursor). Unlike this, the opposite phenotype is seen with the parental CHO cell line, where all cells are MAA+ and PNA– (expression of α2,3-sialylated epitope). (B) Complementation of Lec2 with wild-type CMP–sialic acid transporter gene and lack of complementation with either of the A1 or A2 patient's cDNA alleles. The transduction efficiency was of about 50% as illustrated by the MAA-FITC staining of half of the cells transduced with the normal human wild-type transcript. Left, fluorescein-labeled MAA; right, rhodamine-labeled PNA; middle, superimposition of the green and red pictures. A complete change of phenotype (from MAA–, PNA+ to MAA+, PNA–) can be seen in the transducted cells.
Fluorescent staining with PNA and MAA lectins of CHO, Lec2, and complemented Lec2 cells with the 2 patient alleles. (A) All cells are MAA– and PNA+ in the Lec2 cell line (expression of nonsialylated precursor). Unlike this, the opposite phenotype is seen with the parental CHO cell line, where all cells are MAA+ and PNA– (expression of α2,3-sialylated epitope). (B) Complementation of Lec2 with wild-type CMP–sialic acid transporter gene and lack of complementation with either of the A1 or A2 patient's cDNA alleles. The transduction efficiency was of about 50% as illustrated by the MAA-FITC staining of half of the cells transduced with the normal human wild-type transcript. Left, fluorescein-labeled MAA; right, rhodamine-labeled PNA; middle, superimposition of the green and red pictures. A complete change of phenotype (from MAA–, PNA+ to MAA+, PNA–) can be seen in the transducted cells.
The hamster CMP–sialic acid transporter gene has been cloned19 and shown to encode for a 336–amino acid protein that has more than 98% identity with the human sequence. Previous works have validated this model since the human, murine, and hamster CMP–sialic acid transporters have either been cloned or characterized by complementation.14,19,22
The 2 alleles from the patient and the normal human wild-type allele were incorporated in adenoviral vectors and used to complement Lec2 cells. The transduction gave a 50% yield. Lectin analysis of the transfected cells with PNA and MAA revealed lack of complementation phenotype (PNA+, MAA–) for the 2 patient transcripts (alleles A1 and A2) and a positive complementation phenotype (PNA–, MAA+) for the normal wild-type transcript used as a positive control (Figure 7B). The double staining illustrates that the reversion of the phenotype is total in the cells complemented with wild-type CMP–sialic acid transporter cDNA. The lack of MAA staining in the cells complemented with the 2 patient CMP–sialic acid transporter transcripts illustrates that neither of these mutated transcripts is able to restore the correct sialylation phenotype.
The positive expression of the A1 and A2 cDNA alleles into the transduced cells was verified by specific RT-PCR, using the human TRN-2s and NTR-4s as specific primers. In addition, the proteins were tagged with a flag-tag at their N-terminus. The wild-type–tagged protein was easily detected in the Golgi apparatus of the transfected cells, but the A1 and the A2 mutants could only be transiently detected in the endoplasmic reticulum. Since they have premature stop codons, they are probably rapidly degraded. This instability of the mutant proteins makes the quantification at the protein level very difficult.
Sialylation pattern of plasma proteins
The usual biochemical diagnostic tests for CDGs are isoelectric focusing of plasma transferrin and Western blot analysis of plasma glycoproteins (transferrin, α1-antitrypsin, haptoglobin, and α1-acid glycoprotein). The plasma of the patient gave a normal sialylation pattern of transferrin, with 2 major bands containing 4 or more sialic acids, corresponding to a fully glycosylated protein. The control CDG-I samples had partial or total desialyalation patterns of the 2 N-glycan chains.
The Western blot analysis of the patient's plasma proteins also gave normal migration profiles for transferrin, α1-antitrypsin, haptoglobin, and α1-acid glycoprotein compared with the healthy controls.
Discussion
The Lec2 mutant cell line represents the hamster CDG counterpart of our patient's leukocytes. These cells have a homozygous point mutation in the donor splice site of intron 6 of the CMP–sialic acid transporter, leading to exon 6 skipping and the complete inactivation of the gene.
The patient had received from his mother a CACT insertion in intron 6 that creates a new U2 snRNA site, which is in competition with the putative normal U2 snNRA site (Figure 4) and can induce splice alterations leading to deletions of exon 6 (Figure 6). The gene frequency for the CACT mutated allele in the screened population is 34/106 = 0.32, and 2 normal blood donors were homozygous mutated like the mother of the patient. Therefore, this mutation must be leaky, as are many CDG mutations,25 allowing for the expression of enough wild-type transcripts in the homozygous mutated individuals to avoid the disease. But, the addition of this maternal leaky splice mutation to the other completely inactivated allele of paternal origin induced the disease in the patient. The 2 major alleles found in the patient were unable to complement the deficient Lec2 cells, suggesting that they are both inactive. Only these 2 alleles were fully characterized. However, the ulterior analysis of the other minor splice variants present in a heterozygous CACT individual suggests that other minor variants of the patient transcripts might also have been present. However, we expect these minor short transcript variants to also be inactive, since skipping of the entire exon 6 has been shown to be inactive in Lec2 cells and skipping of exons 5 and 6 or 3 and 6 creates frameshifts with severe truncations of the enzyme.
From the lack of activity of the 2 alleles of the CMP–sialic acid transporter of this patient, we expected to see a hyposialylation pattern of plasma glycoproteins. However, since all tests showed normal glycosylation patterns, we originally thought that this could be due to contamination of the sample by the frequent transfusions that the patient underwent, but it may also be due to different degrees of altered splicing in different cells or to the leaky character of the maternal splice mutation. The phenotype of this patient can be explained by the lack of some sialylated oligosaccharide structures, mainly that of sialyl-Lex, which has considerable roles in cell-cell interactions that are defective in this patient (infections, megakaryocyte immaturity). In addition, the surfaces of megakaryocytes and platelets are heavily sialylated, and a decrease of sialylation or the lack of specific types of sialylated glycoconjugates might impair the blebbing process at the origin of the platelet formation.
We have identified by cDNA sequencing and complementation studies the first human case of inactivation of the CMP–sialic acid transporter gene due to genetic alterations caused by a double microdeletion in 1 allele and a 130-bp deletion in the other allele, resulting in the partial skipping of exon 6. Therefore, we propose that the disease might be caused by these inactivating mutations and corresponds to a new type of CDG of type-IIf. Alternatively, it can also be considered as a congenital disorder of sialylation (CDS) as are some other recently described diseases.26,27 The lack of sialyl-Lex in PMNs, with normal ABH blood group expression and normal sialyltransferase and fucosyltransferase activities, should make clinicians suspect a CDG with alteration of the CMP–sialic acid transporter. The usual laboratory tests for detection of the liver hyposialylation of plasma proteins in CDGs are not adequate for the diagnosis of this disease, mainly affecting the sialylation in α2,3-linkage of sia-Lex on PMNs.
This patient died more than 10 years ago and unfortunately there are no other tissue samples or cells or genomic DNA left in order to verify these different hypotheses and to complete the study now that we have a candidate gene.
Prepublished online as Blood First Edition Paper, December 2, 2004; DOI 10.1182/blood-2004-09-3509.
Supported by Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM), Université de Paris-XI and Association de la Recherche sur le Cancer (ARC) 3611, and the French Research Network on CDG GIS-IMR0308.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Christiane Le Bizec for DNA sequencing.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal