Abstract
Recent reports have indicated that human immunodeficiency virus (HIV) and murine leukemia virus (MLV) vectors preferentially integrate into active genes. Here, we used a novel approach based on genetic trapping to rapidly score several thousand integration sites and found that MLV vectors trapped cellular promoters more efficiently than HIV vectors. Remarkably, 1 in 5 MLV integrations trapped an active promoter in different cell lines and primary hematopoietic cells. Such frequency was even higher in growth-stimulated lymphocytes. We show that the different behavior of MLV and HIV vectors was dependent on a different integration pattern within transcribed genes. Whereas MLV-based traps showed a strong bias for promoter-proximal integration leading to efficient reporter expression, HIV-based traps integrated throughout transcriptional units and were limited for expression by the distance from the promoter and the reading frame of the targeted gene. Our results indicate a strong propensity of MLV to establish transcriptional interactions with cellular promoters, a behavior that may have evolved to enhance proviral expression and may increase the insertional mutagenesis risk. Promoter trapping efficiency provides a convenient readout to assess transcriptional interactions between the vector and its flanking genes at the integration site and to compare integration site selection among different cell types and in different growth conditions.
Introduction
Integration of a transgene into the cell chromatin may ensure stable expression of the gene product in the target cell and its progeny. Because of this feature, integrative vectors, such as retrovirusbased vectors, have been the preferred choice for gene delivery into hematopoietic cells, including lymphocytes, hematopoietic progenitors (HPCs), and hematopoietic stem cells (HSCs). However, the occurrence of adverse events consequent to vector integration in an otherwise successful human gene therapy trial has prompted a reassessment of the insertional mutagenesis risk by retroviral vectors.1-4 In one X-linked severe combined immunodeficiency trial, 2 patients developed a leukemia-like syndrome after transplantation with gamma-retrovirus–transduced HSCs.2,5 In both leukemic clones, the gamma-retroviral vector (RV) integrated close to the promoter of a proto-oncogene and up-regulated its expression. Recent reports have challenged the notion that retroviral integration occurs randomly in the target cell genome and indicated specific biases for integration into transcriptionally active genes both for murine leukemia virus (MLV)–and HIV-based vectors.6-8 Better understanding and comparative analysis of integration site selection among different integrating vectors are thus critically needed to properly evaluate the risk-benefit ratio in gene therapy applications and to develop safer vectors. Toward these aims, several groups have engaged in high-throughput retrieval of vector integration sites. Here, we describe a complementary approach, based on promoter traps, that allows quantitative and comparative assessment of some vector-specific integration biases by probing hundred thousand integrations at once in different types of target cells, including primary cells tested in different conditions, with a simple, cost-effective procedure.
By comparing cells carrying equal amounts of integrated vectors, we show that MLV vectors trapped cellular promoters much more efficiently, and likely targeted more active genes, than HIV vectors. By comparing the average transcript length of MLV and HIV traps and by modifying the trap design to make it independent of the reading frame of the targeted gene, we show that the observed differences between MLV and HIV vectors were due to a different integration pattern within transcribed genes. Expression of MLV-based traps was highly efficient because of a strong bias for promoter-proximal integration. Expression of HIV-based traps, however, was limited by the distance from the promoter and the reading frame of the targeted gene because the vector integrated throughout the transcriptional unit. The different mechanism of expression of MLV- and HIV-based traps was confirmed by mapping the integration site in selected reporter-expressing cells. These results, obtained in different types of cell lines and primary cells, highlighted the propensity of MLV to establish transcriptional interactions with cellular promoters, a behavior that may have evolved to enhance proviral expression, and which likely increases the risk associated with insertional mutagenesis.
Materials and methods
Generation of HIV-based and MLV-based promoter traps
A puromycin resistance–green fluorescent protein (PuroR-GFP) fusion gene was cloned into the promoter trap pROSA-GFNR9 in place of GFNR.PGK-neo, and the resulting expression cassette (Splice Acceptor site.PuroR-GFP.polyA) was excised by AflII/NheI digestion and cloned in reverse orientation into the HIV-based self-inactivating (SIN) lentiviral vector pRRL.SIN.cPPT.PGK.GFP in place of PGK.GFP or into the MLV-based SIN retroviral vector pRkat43.3.PGK.YFP10 in place of PGK.YFP to generate lentiviral trap (LT) and retroviral trap (RT) vectors, respectively. The encephalomyocarditis virus internal ribosome entry site (IRES) element11 was polymerase chain reaction (PCR)–amplified from plasmid pRRL.sin.PPT.CMV.Luciferase.IRES-EMCVwt.GFP.wPRE12 using oligonucleotides designed to add stop codons in all 3 reading frames in the 5′ portion of the element. The resulting PCR product was in-frame cloned upstream to the PuroR-GFP fusion protein. The woodchuck posttranscriptional regulatory element (wPRE)13 was cloned downstream to the PuroR-GFP cDNA. The resulting expression cassettes were cloned into the HIV-based and the MLV-based vectors to generate IRES-, wPRE-, and IRES.wPRE-LT and -RT. The HIV-based pRRL.SIN.cPPT.PGK.GFP. wPRE14 lentiviral (L) and the MLV-based pRkat 43.3.PGK.GFP retroviral (R) vectors were used as controls.
Vesicular stomatitis virus (VSV)–pseudotyped vector stocks were produced by transient cotransfection of the selected transfer vector construct, the packaging construct pCMVΔR8.74 (for HIV-based vectors) and pCMV.GAG.POL (for MLV-based vectors), and the pMD2.G construct in 293T cells and concentrated hundred-fold by ultracentrifugation as previously described.14 Vector stocks were titered on 293T cells by integration titer, end-point green fluorescent protein (GFP) and PuroR expression titer, as previously described.15 Briefly, vector integration titers were calculated measuring the vector DNA content of transduced in reference to known standards, such as a curve of plasmid DNA or cell clones carrying known amounts of integrated vector, by Southern blot, or real-time PCR analysis, or both. For Southern blot, the DNA was digested with AflII, which excises a fragment containing the reporter gene from each vector type. Real-time PCR analysis was performed as previously described16 using oligonucleotides and probe complementary to the GFP sequence common to all vector types (forward primer, 5′-CAGCTCGCCGACCACTA-3′; reverse primer, 5′-GGGCCGTCGCCGAT-3′; 6-carboxylfluorescein [FAM] reporter probe, 5′-CCAGCAGAACACCCCC-3′; Applied Biosystem, Foster City, CA).
Integration titers were in the range of 108 to 109 vector copies/mL (transducing units293T/mL) for all vector types. GFP expression titers were calculated from the frequency of GFP-positive cells by fluorescence activated cell sorting (FACS) analysis of cultures transduced with high dilutions of vector stocks. PuroR expression titers were determined by plating transduced cells at different dilutions in medium containing 1 μg/mL puromycin and scoring colonies after 1 to 2 weeks by crystal violet staining.
Transduction of cell lines
293T, MLP29, and H5V cells were maintained in Iscove modified Dulbecco medium (IMDM; Sigma Chemical, Milan, Italy) with 10% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY) and glutamine. Cells were transduced with normalized amounts of the different vectors to obtain equal levels of integration and grown for at least 4 days before FACS analysis to reach steady-state PuroR/GFP expression. Selection of puromycin-resistant cells was performed by using 1 μg/mL puromycin for 5 to 14 days.
Transfection of linear plasmid forms of vector traps
Linear plasmid forms (0.2, 1, and 2 μg) of both HIV and MLV traps, spanning from the 5′ to the 3′ long terminal repeat (LTR), were prepared by AflIII/SfiI digestion and transfected into 293T cells by calcium-phosphate DNA precipitation. Transfectants were analyzed for GFP expression 3 days later or cultured in medium containing 1 μg/mL puromycin for 1 week and scored for colony growth by crystal violet staining.
Mapping of trap integration sites
Integration site mapping was performed on DNA extracted from puromycin-resistant 293T cells. The genomic-proviral junction sequence was identified using linear amplification-mediated (LAM)–PCR, as previously described17 (using the following biotinylated primer for the retroviral trap: 5′-CGACCCTGTTCCATCTGTTCCTGACC-3′). PCR products were cloned into pCR2.1-TOPO (Invitrogen, Carlsbad CA) and sequenced. Sequences of LAM-PCR products were mapped using BLAST (basic local alignment search tool) analysis against the human genomic sequence at the ENSEMBL database (September 2004 freeze).
Transduction of primary hematopoietic cells
Cells from human subjects were obtained with informed consent according to the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donors according to a protocol approved by the H. San Raffaele Bioethical Committee by leukophoresis and Lymphoprep gradient separation (Axis-Shiel PoC AS, Oslo, Norway), stimulated for 2 days with anti-CD3 (1 μg/mL) and anti-CD28 (0.5 μg/mL) antibodies (both from DAKO, Glostrup, Denmark) and phytohemagglutinin (1 μg/mL), and grown in RPMI with 5% human serum (Sigma) at 1 × 106 cells/mL in the presence of human recombinant interleukin 2 (IL-2; 10 U/mL; EuroCetus Italia S.r.l., Milan, Italy). Activated cells were transduced using 106 and 107 transducing units293T/mL vector. Cells were analyzed by FACS 1 week after transduction or cultured in the presence of puromycin (2 μg/mL) for an additional week before FACS analysis.
Bone marrow was harvested from femurs and tibias of 6-week-old C57Bl/6 mice. Hematopoietic progenitors were isolated using a kit (StemCell Technologies, Vancouver, Canada) and stimulated for 24 hours with 20 ng/mL recombinant mouse (rm)–IL-3, 100 ng/mL rm–stem cell factor (SCF), rm–FMS-like tyrosine kinase 3 (FLT-3) ligand, and rm-thrombopoietin (TPO), all from PeproTech (Rocky Hill, NH) in StemSpan serum-free medium (StemCell Technologies). Cells (106/mL) were transduced using 5 × 107 transducing units293T/mL.
Mononuclear cells were obtained from human cord blood scheduled for discard according to a protocol approved by the H. San Raffaele Bioethical Committee, by gradient centrifugation over Lymphoprep, and CD34+ cells were isolated using the Miltenyi magnetic cell sorting (MACS) kit (Miltenyi Biotec, Gladbach, Germany) and stimulated for 36 hours with 20 ng/mL recombinant human (rh)–IL-6, 100 ng/mL rh-SCF, 100 ng/mL rh-FLT-3 ligand, and 20 ng/mL rh-TPO, all from PeproTech, in StemSpan serum-free medium,17 before transduction with 107 transducing units293T/mL. For liquid cultures, cells were maintained in medium containing 20 ng/mL rh–IL-6, 100 ng/mL rh-SCF, and 20 ng/mL rh–IL-3, all from PeproTech, for 7 days before analysis. For puromycin selection, puromycin (1 μg/mL) was added to the medium 3 days after transduction. For FACS analysis, cells were stained with propidium iodide (PI) and analyzed by FACS. Only viable, PI-negative cells were used for the analysis. Colony-forming cell (CFC) assays and GFP-specific PCR were performed as previously described.17 For scoring puromycin-resistant CFCs, cells were plated 24 hours after transduction on methylcellulose-based medium (StemCell Technologies), with or without 1 μg/mL puromycin.
Results
MLV vectors trap cellular promoters more efficiently than HIV vectors in target cell lines
We introduced a promoter trapping construct9 into HIV and MLV vectors and compared their trapping efficiency by using a puromycin resistance-GFP (PuroR-GFP) reporter. We cloned the PuroR-GFP gene between a strong cellular splice acceptor (SA) and a polyadenylation (polyA) site9 and inserted the resulting expression cassette in reverse orientation into SIN MLV10 and HIV14 vectors, immediately upstream to the modified 3′ LTR (Figure 1A). The reverse orientation prevents reporter expression from the 5′ LTR of the transduced vector when the LTR is not fully inactivated, as in the case of the SIN MLV vector (described in Figure 2). Reporter expression is dependent on vector integration downstream to a transcriptionally active cellular promoter. If the vector integrates with the appropriate orientation within a promoter-proximal intron or exon of a transcribed gene, a fusion messenger transcript will be produced that is likely to be processed to express the reporter. The placement of the expression cassette close to the 3′ end of the vector leaves most viral sequences outside of the fusion transcript, reducing their potential influence on reporter expression. The frequency and average level of reporter expression may thus read out the frequency of vector integration into active genes, their average expression level, and may uncover preferential integration in the proximity of cellular promoters. If target site selection differs between MLV and HIV according to these parameters, it will also result in quantifiable differences in their promoter trapping efficiency.
Transduction of 293T and MLP29 cells by HIV-based lentiviral (LT) and MLV-based retroviral (RT) promoter traps. (A) A schematic of the vector trap integrated within the first intron of a cellular gene. The expression cassette is placed in reverse transcriptional orientation with respect to the vector framework. Ψ indicates viral encapsidation signal, including the 5′ portion of gag gene (GA); RRE, rev responsive element, cPPT, central polypurine tract (LT only); polyA, polyadenylation signal; SD and SA, splice donor and acceptor sites. (B, left) Southern blot analysis of 293T cells transduced with LT, RT, MLV-based retroviral vector (R), and HIV-based lentiviral vector (L). Matched vector amounts were used that yielded an average of 1 integrated vector copy per cell. A standard curve of plasmid DNA was used to calculate the vector copy-number. DNA was digested with AflII and the filter probed for puromycinR-GFP sequences. (Right) FACS analysis (dot plot) of GFP expression in transduced cells. The percentage and mean fluorescence intensity (MFI) of GFP-positive cells, and the vector copy-number per cell (calculated by Southern blot), are indicated for each cell culture. (C) FACS analysis of GFP expression in MLP29 cells transduced with the indicated traps and left untreated (dot plots on the left; the MFI of GFP-positive cells is indicated) or treated with puromycin (dot plots on the right, and histograms; the MFI of the total population is indicated). UC indicates untransduced untreated cells. (D) FACS analysis (dot plot) of 293T cells transfected with the indicated amounts of linear plasmid forms of each vector trap. The results shown are representative of at least 3 experiments performed.
Transduction of 293T and MLP29 cells by HIV-based lentiviral (LT) and MLV-based retroviral (RT) promoter traps. (A) A schematic of the vector trap integrated within the first intron of a cellular gene. The expression cassette is placed in reverse transcriptional orientation with respect to the vector framework. Ψ indicates viral encapsidation signal, including the 5′ portion of gag gene (GA); RRE, rev responsive element, cPPT, central polypurine tract (LT only); polyA, polyadenylation signal; SD and SA, splice donor and acceptor sites. (B, left) Southern blot analysis of 293T cells transduced with LT, RT, MLV-based retroviral vector (R), and HIV-based lentiviral vector (L). Matched vector amounts were used that yielded an average of 1 integrated vector copy per cell. A standard curve of plasmid DNA was used to calculate the vector copy-number. DNA was digested with AflII and the filter probed for puromycinR-GFP sequences. (Right) FACS analysis (dot plot) of GFP expression in transduced cells. The percentage and mean fluorescence intensity (MFI) of GFP-positive cells, and the vector copy-number per cell (calculated by Southern blot), are indicated for each cell culture. (C) FACS analysis of GFP expression in MLP29 cells transduced with the indicated traps and left untreated (dot plots on the left; the MFI of GFP-positive cells is indicated) or treated with puromycin (dot plots on the right, and histograms; the MFI of the total population is indicated). UC indicates untransduced untreated cells. (D) FACS analysis (dot plot) of 293T cells transfected with the indicated amounts of linear plasmid forms of each vector trap. The results shown are representative of at least 3 experiments performed.
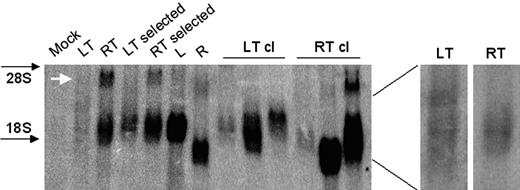
Northern blot analysis of cells transduced by HIV-based (LT) and MLV-based (RT) promoter traps and control MLV-based (R) and HIV-based (L) vectors. Analysis was performed on 293T cells transduced with the indicated vectors, before or after puromycin selection, and in representative puromycin-resistant clones. Cells were transduced with matched vector doses yielding multicopy integration (average of 10) per cell in the unselected cells (LT and RT), and average single-copy integration in the selected populations (LT/RT selected), clones (LT/RT cl), and in cells transduced by control vectors (L, R). The white arrow indicates the LTR-driven antisense transcript of RT. Note that this transcript is expressed to low extent because of the SIN modification and is better detected in cells containing high numbers of vector integrants. The LT and RT lanes are shown enlarged on the right and after a shorter exposure time for the RT lane to better compare the hybridization pattern.
Northern blot analysis of cells transduced by HIV-based (LT) and MLV-based (RT) promoter traps and control MLV-based (R) and HIV-based (L) vectors. Analysis was performed on 293T cells transduced with the indicated vectors, before or after puromycin selection, and in representative puromycin-resistant clones. Cells were transduced with matched vector doses yielding multicopy integration (average of 10) per cell in the unselected cells (LT and RT), and average single-copy integration in the selected populations (LT/RT selected), clones (LT/RT cl), and in cells transduced by control vectors (L, R). The white arrow indicates the LTR-driven antisense transcript of RT. Note that this transcript is expressed to low extent because of the SIN modification and is better detected in cells containing high numbers of vector integrants. The LT and RT lanes are shown enlarged on the right and after a shorter exposure time for the RT lane to better compare the hybridization pattern.
We transduced human 293T cells with VSV-pseudotyped stocks of each vector trap and of control standard vectors expressing GFP from an internal phosphoglycerate kinase promoter. Whereas the control MLV and HIV vectors expressed the reporter gene to similarly high efficiency, with expression titers approaching integration titers,15 we observed significant differences between the 2 types of trapping vector. By analyzing cell populations containing similar amounts of integrated vector, we found that promoter trapping efficiency, calculated as the ratio between expression and integration, was significantly higher (4- to 5-fold) for the MLV than the HIV trap (Figure 1B). In this and other experiments, integration was measured in transduced cells by Southern analysis or real-time PCR using a probe specific for the GFP sequence contained in all types of constructs. Trapping efficiency was calculated from cultures expressing the reporter gene in less than 20% of the cells to avoid the possibility that multiple trapping events per cell led to nonlinear dose-response between integration and expression. In 6 independent experiments, promoter trapping efficiency was 20% ± 3% for the MLV trap and 5% ± 1% for the HIV trap with a highly significant statistical difference (P < .005). We reproduced these findings in murine cell lines, such as liver progenitor MLP29 cells (Figure 1C) and endothelial H5V cells. In the latter cells, trapping efficiency was 17% ± 2% for the MLV trap and 5% ± 2% for the HIV trap (P < .05).
Remarkably, the expression titer of the MLV trap was on average only 5-fold lower than its integration titer, indicating that 1 of 5 integrations occurred downstream to a transcriptionally active promoter and led to reporter expression in the cells tested. Intriguingly, the mean fluorescence intensity (MFI) of GFP was higher in transduced cells expressing the MLV trap (1.6-fold on average in 293T cells, n = 6, P < .05) as compared with cells expressing the HIV trap. This difference was maintained after puromycin selection of the trap-expressing cells (Figure 1C).
A possible explanation for these findings was that the MLV vector framework was more permissive to gene expression following integration than the HIV one, either because the 3′ LTR and flanking sequences enhanced expression of the fusion messenger or because of the residual promoter activity in the 5′ LTR. The latter, however, drives antisense transcription of the reporter cassette and may thus inhibit rather than promote its expression. To investigate this point, we transfected linear plasmid forms of both vector traps, spanning the distance from the 5′ to the 3′ LTR, into 293T cells by calcium-phosphate DNA precipitation. By this approach, insertion of the vector DNA into the target cell chromatin occurs by the cellular recombination machinery, thus independently from the viral proteins that govern the efficient integration of retroviral genomes. As in the case of viral delivery, expression of the reporter gene was dependent on vector DNA insertion downstream to an active promoter. Contrary to the findings observed after viral transduction, we found that the trapping efficiencies of the transfected MLV and HIV traps were not statistically different (P = .6; Figure 1D and other data not shown). These results indicated that the higher promoter trapping efficiency observed for the MLV trap after transduction was dependent on the retroviral integration machinery and not on differences in the vector backbone. The higher GFP MFI observed for HIV traps as compared with MLV traps after DNA transfection suggested that, in the absence of viral components affecting integration site selection, the HIV trap was even slightly more permissive to reporter expression than the latter, possibly because of the complete absence of antisense transcription from the vector 5′ LTR. Taken together, infection and transfection experiments indicated that the MLV mechanism of integration specifically favored promoter trapping and suggested that it preferentially targeted more active genes than those targeted by HIV vectors.
A bias for promoter-proximal integration accounts for the high promoter trapping efficiency of MLV vectors
We analyzed reporter expression in 293T cells transduced with matched amounts of each vector trap by Northern blot (Figure 2). Consistently with the protein expression data, the hybridization signal was much weaker in cells transduced by the HIV trap than in cells transduced by the MLV trap. In addition, the HIV-derived transcripts displayed a much wider size range as compared with MLV-derived transcripts. The majority of MLV-derived transcripts did not exceed 2 kb in length, except for a distinct 4-kb band, representing the transcript originating from the 5′ LTR. Since the reporter transcript encompasses approximately 1.8 kb of vector sequence, these results showed that the large fraction of MLV integrations resulting in reporter expression (1 of 5) occurred in close proximity to the transcription start site of active genes. Such a strong bias for promoter-proximal insertion within active transcription units represents an advantage for promoter trapping, since lengthy fusion mRNAs originating from promoter-distal insertions are unlikely to express the reporter protein. Interestingly, Northern analysis of transduced cells, performed after puromycin selection, showed strong enrichment in HIV-derived transcripts of similar size as that observed for the MLV traps, indicating that the fraction of HIV integrations resulting in efficient reporter protein expression were those occurring proximal to a promoter or within the first intron. The average reporter expression level of these 2 cell populations, selected for similar placement of the reporter cassette within the transcription unit, remained higher for MLV traps, further suggesting that, on average, MLV vectors targeted more active genes than those targeted by HIV vectors within the same cells.
We then isolated a panel of HIV and MLV trap clones by puromycin selection and found by Northern analysis that they were representative of the 2 parental bulk-selected cell populations (Figure 2). We retrieved the vector-genome junction from the puromycin-selected populations and clones by LAM-PCR18 and mapped the integration site on the human genome by BLAST analysis using the ENSEMBL search engine (September 2004 freeze). A list of the mapped integration sites for both types of vector is shown in Table 1. Representative examples of vector orientation and placement within the targeted loci are shown in Figure 3.
Mapping of integration sites
Int. ID . | Chromosomal band . | Chromosomal position, bp . | Trapped gene name/ref seq gene ID . | Trapped gene description . | Trap orientation relative to gene transcription . | Distance from TSS, kb . | Position within transcription unit . | Putative reporter transcript . |
|---|---|---|---|---|---|---|---|---|
| LT8 | 5q23.3 | 131666449 | PDLIM4 | LIM protein RIL (reversion-induced LIM protein) | Sense | 3 | Intron 1 | 5′ UTR fusion |
| NM_003687 | ||||||||
| LT1 | 15q22.2 | 57692244 | BNIP2 | BCL2/adenovirus E1B 19-kDa interacting protein 2 | Sense | 5 | Intron 1 | 5′ UTR fusion |
| NM_004330 | ||||||||
| LTA4 | 17p1 3.2 | 4470248 | UBE2G1 | Ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, Caenorhabditis elegans) | Sense | 5 | Intron 1 | In-frame fusion with protein coding mRNA |
| NM_003342 | ||||||||
| NM_182682 | ||||||||
| LT2 | 3p21.1 | 49785970 | IHPK1 | Inositol hexaphosphate kinase 1 | Sense | 13 | Intron 1 | In-frame fusion with protein coding mRNA |
| NM_153273 | ||||||||
| LTA3 | 6p21.31 | 35685569 | FKBP5 | FK506 binding protein 5 | Sense | 16 | Intron 1 or intron 3, depending on alternative promoter | In-frame fusion with protein coding mRNA |
| NM_004117 | ||||||||
| LT3 | 2q31.1 | 172195652 | TLK1 | Tousled-like kinase 1 | Sense | 24 | Intron 1 | In-frame fusion with protein coding mRNA |
| NM_022644 | ||||||||
| LTA10 | 10q26.11 | 12014001 | C10orf46 | Chromosome 10 open reading frame 46 | Sense | 40 | Intron 3 | Fusion with an untranslated splicing isoform (C10orf46003) |
| NM_153810 | ||||||||
| LTA1 | 4q13.3 | 7230830 | MOBKL1A | MOB1, Mps One Binder kinase activator-like 1A (yeast) | Antisense* | 40* | Intron 4* | Fusion with natural antisentisense transcripts |
| NM_173468 | ||||||||
| LT10 | 17q23.3 | 62422791 | CSH2 | Chorionic somatomammotropin hormone 2 | Antisense* | 1.7* | Exon 4* | Fusion with natural antisentisense transcripts |
| NM_022646 | ||||||||
| NM_022645 | ||||||||
| NM_022644 | ||||||||
| NM_020991 | ||||||||
| LTA13 | 19q13.43 | 62696928 | FLJ23233 | Hypothetical protein FLJ23233 | Antisense* | 16* | Exon 5* | Fusion with natural antisentisense transcripts |
| NM_024691 | ||||||||
| RT15.8 | 6p21.31 | 35745703 | FKBP5 | FK506 binding protein 5 | Sense | -2 | Upstream TSS, downstream predicted promoter† | Reporter transcript without fusion to endogenous mRNA |
| NM_004117 | ||||||||
| RT15.10 | Xq21.1 | 75907912 | DKFZp564K142 | Implantation-associated protein | Sense | -1.1 | Upstream TSS, downstream predicted promoter† | Reporter transcript without fusion with endogenous mRNA |
| NM_032121 | ||||||||
| RT16.2 | 20q13.32 | 57911395 | STK16 | Serine/threonine kinase 16 | Sense | -0.4 | Upstream TSS, downstream predicted promoter† | Reporter transcript without fusion with endogenous mRNA |
| NM_003691 | ||||||||
| RT13 | 3q11.2 | 98965659 | EST: | NA | Sense | 0.1 | Unknown | EST fusion |
| BM676895.1 | ||||||||
| RT5.6 | 19p13.3 | 2895544 | ZNF77 | Zinc finger protein 77 (pT1) | Sense | 0.2 | Intron 1 | In-frame fusion protein |
| NM_021217 | ||||||||
| RT6.1 | 16p13.3 | 679892 | DKFZp434F054 | Hypothetical protein DKFZp434F054 | Sense | 0.5 | Exon 1 or intron 2 depending on alternative splicing | 5′ UTR fusion or inframe fusion protein depending on alternative splicing |
| NM_032259 | ||||||||
| RT25 | 20q13.33 | 62983740 | GMEB2 | Glucocorticoid modulatory element binding protein 2 | Sense | 1 | Intron 1 | 5′ UTR fusion |
| NM_012384 | ||||||||
| RT7.6 | 19p13.3 | 1215668 | UNIGENE ID: | NA | Sense | 1.5 | Putative Exon 4 | Fusion with untranslated ESTs |
| Hs79706 | ||||||||
| Hs322473 | ||||||||
| Hs383245 | ||||||||
| RT12 | 22q12.3 | 34330975 | MB | Myoglobin | Sense | 6 | Intron 2 | In-frame fusion protein |
| NM_203377 | ||||||||
| NM_005368 | ||||||||
| NM_203378 | ||||||||
| RT23 | 22q12.1 | 27383168 | cB42E1.c22.1 | NA | Antisense* | 17* | Intron 1 | Fusion with natural antisentisense transcripts? |
Int. ID . | Chromosomal band . | Chromosomal position, bp . | Trapped gene name/ref seq gene ID . | Trapped gene description . | Trap orientation relative to gene transcription . | Distance from TSS, kb . | Position within transcription unit . | Putative reporter transcript . |
|---|---|---|---|---|---|---|---|---|
| LT8 | 5q23.3 | 131666449 | PDLIM4 | LIM protein RIL (reversion-induced LIM protein) | Sense | 3 | Intron 1 | 5′ UTR fusion |
| NM_003687 | ||||||||
| LT1 | 15q22.2 | 57692244 | BNIP2 | BCL2/adenovirus E1B 19-kDa interacting protein 2 | Sense | 5 | Intron 1 | 5′ UTR fusion |
| NM_004330 | ||||||||
| LTA4 | 17p1 3.2 | 4470248 | UBE2G1 | Ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, Caenorhabditis elegans) | Sense | 5 | Intron 1 | In-frame fusion with protein coding mRNA |
| NM_003342 | ||||||||
| NM_182682 | ||||||||
| LT2 | 3p21.1 | 49785970 | IHPK1 | Inositol hexaphosphate kinase 1 | Sense | 13 | Intron 1 | In-frame fusion with protein coding mRNA |
| NM_153273 | ||||||||
| LTA3 | 6p21.31 | 35685569 | FKBP5 | FK506 binding protein 5 | Sense | 16 | Intron 1 or intron 3, depending on alternative promoter | In-frame fusion with protein coding mRNA |
| NM_004117 | ||||||||
| LT3 | 2q31.1 | 172195652 | TLK1 | Tousled-like kinase 1 | Sense | 24 | Intron 1 | In-frame fusion with protein coding mRNA |
| NM_022644 | ||||||||
| LTA10 | 10q26.11 | 12014001 | C10orf46 | Chromosome 10 open reading frame 46 | Sense | 40 | Intron 3 | Fusion with an untranslated splicing isoform (C10orf46003) |
| NM_153810 | ||||||||
| LTA1 | 4q13.3 | 7230830 | MOBKL1A | MOB1, Mps One Binder kinase activator-like 1A (yeast) | Antisense* | 40* | Intron 4* | Fusion with natural antisentisense transcripts |
| NM_173468 | ||||||||
| LT10 | 17q23.3 | 62422791 | CSH2 | Chorionic somatomammotropin hormone 2 | Antisense* | 1.7* | Exon 4* | Fusion with natural antisentisense transcripts |
| NM_022646 | ||||||||
| NM_022645 | ||||||||
| NM_022644 | ||||||||
| NM_020991 | ||||||||
| LTA13 | 19q13.43 | 62696928 | FLJ23233 | Hypothetical protein FLJ23233 | Antisense* | 16* | Exon 5* | Fusion with natural antisentisense transcripts |
| NM_024691 | ||||||||
| RT15.8 | 6p21.31 | 35745703 | FKBP5 | FK506 binding protein 5 | Sense | -2 | Upstream TSS, downstream predicted promoter† | Reporter transcript without fusion to endogenous mRNA |
| NM_004117 | ||||||||
| RT15.10 | Xq21.1 | 75907912 | DKFZp564K142 | Implantation-associated protein | Sense | -1.1 | Upstream TSS, downstream predicted promoter† | Reporter transcript without fusion with endogenous mRNA |
| NM_032121 | ||||||||
| RT16.2 | 20q13.32 | 57911395 | STK16 | Serine/threonine kinase 16 | Sense | -0.4 | Upstream TSS, downstream predicted promoter† | Reporter transcript without fusion with endogenous mRNA |
| NM_003691 | ||||||||
| RT13 | 3q11.2 | 98965659 | EST: | NA | Sense | 0.1 | Unknown | EST fusion |
| BM676895.1 | ||||||||
| RT5.6 | 19p13.3 | 2895544 | ZNF77 | Zinc finger protein 77 (pT1) | Sense | 0.2 | Intron 1 | In-frame fusion protein |
| NM_021217 | ||||||||
| RT6.1 | 16p13.3 | 679892 | DKFZp434F054 | Hypothetical protein DKFZp434F054 | Sense | 0.5 | Exon 1 or intron 2 depending on alternative splicing | 5′ UTR fusion or inframe fusion protein depending on alternative splicing |
| NM_032259 | ||||||||
| RT25 | 20q13.33 | 62983740 | GMEB2 | Glucocorticoid modulatory element binding protein 2 | Sense | 1 | Intron 1 | 5′ UTR fusion |
| NM_012384 | ||||||||
| RT7.6 | 19p13.3 | 1215668 | UNIGENE ID: | NA | Sense | 1.5 | Putative Exon 4 | Fusion with untranslated ESTs |
| Hs79706 | ||||||||
| Hs322473 | ||||||||
| Hs383245 | ||||||||
| RT12 | 22q12.3 | 34330975 | MB | Myoglobin | Sense | 6 | Intron 2 | In-frame fusion protein |
| NM_203377 | ||||||||
| NM_005368 | ||||||||
| NM_203378 | ||||||||
| RT23 | 22q12.1 | 27383168 | cB42E1.c22.1 | NA | Antisense* | 17* | Intron 1 | Fusion with natural antisentisense transcripts? |
Int. ID indicates integration ID; NA, not available.
Relative to the known gene at the targeted locus. Because of the occurrence of natural antisense transcripts (NATs) for this gene, reporter expression may be driven by these transcripts for which no information is available on TSS and intron/exon organization
Integration occurred upstream of the putative TSS of the targeted gene. First EF and eponine algorithms identified putative promoters in the region, some of which may drive reporter expression
Mapping of HIV-based (LT) and MLV-based (RT) promoter traps integration site in selected 293T cells. Puromycin-resistant cell populations and clones were isolated from 293T cells transduced with LT or RT at very low vector input to ensure single-copy integration. Genomic DNA from selected bulk populations, 15 LT and 15 RT clones was subjected to vector-specific LAM-PCR to retrieve the proviral-genomic junction. Ten LT and 10 RT integrations were unambiguously mapped after BLAST analysis against the human genome database using the ENSEMBL search engine (September 2004 freeze). All characterized integrations are listed in Table 1. Three representative LT (A-C) and RT (D-F) integrations are shown here in detail. The genomic region around the integration site (length in kb indicated on top) were obtained from the ENSEMBL output and graphically simplified. Blue thick bars represent the selected chromosomal region. Query sequence from LAM-PCR product is placed in the middle of the genomic interval and is represented by a red mark (BLAST hit) above or below the chromosomal bar depending on the provirus orientation. Because LAM-PCR products from the LT and RT were obtained using the 3′ LTR and 5′ LTR as template, respectively, the orientation of the trapping cassette on the ENSEMBL output was opposite for LT and RT. For clarity, proviral integration and the direction of transcription of the PuroR-GFP cassette are represented by a green arrow. Transcripts, protein alignments, and genomic annotations were retrieved by searching different databases as displayed on the left side of each picture. Genes displayed above the blue bar are transcribed from left to right; genes displayed below the bar are transcribed from right to left. (A) LTA4 integration on 17p13.2 landed in the first intron of the UBE2G1 gene before the first coding ATG. Promoters (circles with arrows) are identified by algorithms Eponine and FirstEF (red marks). (B) LTA3 integration on 6p21.31 landed in the third intron of the FKBP5 gene before the first coding ATG. Fusion transcript could be generated by 2 different promoters (circles with arrows) as identified by algorithms Eponine and FirstEF (red marks)19 and presence of a CpG island (purple mark). The distance from the closest promoter is 16 kb. (C) LT1 integration on 15q22.2 landed into the first intron of the BNIP gene after the first noncoding exon. Distance from transcription start site (TSS) was +5.5 kb. Reporter expression can be explained by a trap/5′ untranslated region (UTR) fusion transcript. (D) Integration RT16.2 on 20q13.32 landed into a CpG island, 350 base pair (bp) upstream of the putative transcription start site of the sintaxin 16 gene. Both First EF and Eponine algorithms identified several putative promoter regions, one of which may drive reporter expression (circle with arrow). (E) RT25 integration on 20q13.33 landed into the first intron of the GMEB2 gene after the first noncoding exon. Distance from TSS was +1 kb. First EF and Eponine algorithms identified putative promoter regions (circle with arrow). (F) RT6.1 Integration on 16p13.3 landed into the DKFZp434F054 gene. Depending on the splicing, the trap can be fused to a 5′ UTR exon. Alternatively, the same 5′ UTR portion can be spliced to generate an in-frame protein coding fusion transcript with the reporter. Distance from TSS was +0.4 kb.
Mapping of HIV-based (LT) and MLV-based (RT) promoter traps integration site in selected 293T cells. Puromycin-resistant cell populations and clones were isolated from 293T cells transduced with LT or RT at very low vector input to ensure single-copy integration. Genomic DNA from selected bulk populations, 15 LT and 15 RT clones was subjected to vector-specific LAM-PCR to retrieve the proviral-genomic junction. Ten LT and 10 RT integrations were unambiguously mapped after BLAST analysis against the human genome database using the ENSEMBL search engine (September 2004 freeze). All characterized integrations are listed in Table 1. Three representative LT (A-C) and RT (D-F) integrations are shown here in detail. The genomic region around the integration site (length in kb indicated on top) were obtained from the ENSEMBL output and graphically simplified. Blue thick bars represent the selected chromosomal region. Query sequence from LAM-PCR product is placed in the middle of the genomic interval and is represented by a red mark (BLAST hit) above or below the chromosomal bar depending on the provirus orientation. Because LAM-PCR products from the LT and RT were obtained using the 3′ LTR and 5′ LTR as template, respectively, the orientation of the trapping cassette on the ENSEMBL output was opposite for LT and RT. For clarity, proviral integration and the direction of transcription of the PuroR-GFP cassette are represented by a green arrow. Transcripts, protein alignments, and genomic annotations were retrieved by searching different databases as displayed on the left side of each picture. Genes displayed above the blue bar are transcribed from left to right; genes displayed below the bar are transcribed from right to left. (A) LTA4 integration on 17p13.2 landed in the first intron of the UBE2G1 gene before the first coding ATG. Promoters (circles with arrows) are identified by algorithms Eponine and FirstEF (red marks). (B) LTA3 integration on 6p21.31 landed in the third intron of the FKBP5 gene before the first coding ATG. Fusion transcript could be generated by 2 different promoters (circles with arrows) as identified by algorithms Eponine and FirstEF (red marks)19 and presence of a CpG island (purple mark). The distance from the closest promoter is 16 kb. (C) LT1 integration on 15q22.2 landed into the first intron of the BNIP gene after the first noncoding exon. Distance from transcription start site (TSS) was +5.5 kb. Reporter expression can be explained by a trap/5′ untranslated region (UTR) fusion transcript. (D) Integration RT16.2 on 20q13.32 landed into a CpG island, 350 base pair (bp) upstream of the putative transcription start site of the sintaxin 16 gene. Both First EF and Eponine algorithms identified several putative promoter regions, one of which may drive reporter expression (circle with arrow). (E) RT25 integration on 20q13.33 landed into the first intron of the GMEB2 gene after the first noncoding exon. Distance from TSS was +1 kb. First EF and Eponine algorithms identified putative promoter regions (circle with arrow). (F) RT6.1 Integration on 16p13.3 landed into the DKFZp434F054 gene. Depending on the splicing, the trap can be fused to a 5′ UTR exon. Alternatively, the same 5′ UTR portion can be spliced to generate an in-frame protein coding fusion transcript with the reporter. Distance from TSS was +0.4 kb.
Concerning HIV integrations, 6 of 10 insertions occurred within the first intron of an identified gene, with the trap inserted in the proper orientation for reporter expression. These data provided experimental evidence that selection for reporter expression strongly enriched for HIV integrations occurring within the first intron of an expressed gene. Notably, the distance of these integrations from the transcription start site (TSS) ranged from 3 to 24 kb, in sharp contrast with MLV integrations that were clustered close to the promoter. Three insertions were mapped within known transcription units but apparently in the wrong orientation for expression. Searching of EST databases revealed natural antisense transcripts (NATs19 ) for these loci, providing a possible mechanism of reporter expression.
Concerning MLV integrations, 8 of 10 mapped sites occurred in close proximity (< 1.5 kb) to established or putative transcriptional promoter regions, as identified by the upstream location to a RefSeq transcript, the content of CpG islands, Eponine and FirstEF annotations for putative promoter/transcription start sites.20 These data strongly supported the proposed mechanism that promoter-proximal integration accounted for the high promoter trapping efficiency by the MLV vector.
MLV vectors trap cellular promoters more efficiently than HIV vectors in primary hematopoietic cells
We compared the behavior of MLV and HIV vector traps in a panel of hematopoietic cells representing relevant targets for gene therapy. Human PBMCs were activated by phytohemagglutinin (PHA) and anti-CD3/anti-CD28 treatment for 2 days, transduced with 2 matched doses of HIV and MLV traps, and analyzed for GFP expression by FACS analysis and for vector integration by real-time PCR 7 to 10 days later. As shown in Table 2, the HIV trap integrated into the chromatin of PBMCs with higher efficiency than the MLV trap, as expected after a single round of infection with these 2 types of vectors. However, MLV trapping efficiency was on average 4 times higher than that of HIV (P < .01). Remarkably, up to 37% of integrated MLV traps expressed the reporter gene in these target cells. As seen with continuous cell lines, the GFP MFI after puromycin selection was higher for the MLV than the HIV trap (1.3-fold; P < .05).
Transduction of PBMCs by HIV- and MLV-based traps
. | HIV trap, LT†‡ . | . | . | . | MLV trap, RT†‡ . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Expression . | . | . | . | Expression . | . | . | . | ||||||
| Donor . | % GFP+ . | MFI* . | Integration, CpC . | % trapping efficiency, expr/integr . | % GFP+ . | MFI* . | Integration, CpC . | % Trapping efficiency, expr/integr . | ||||||
| A | ||||||||||||||
| 106 TU/mL | 1.0 | 16.0 | 0.12 | 8.3 | 0.6 | 19.1 | ND | ND | ||||||
| 107 TU/mL | 1.7 | 13.8 | 0.20 | 8.5 | 1.5 | 19.2 | 0.04 | 37.5 | ||||||
| B | ||||||||||||||
| 106 TU/mL | 2.8 | 12.6 | 0.32 | 8.7 | 0.8 | 14.1 | 0.02 | 37.7 | ||||||
| 107 TU/mL | 5.5 | 11.5 | 0.69 | 8.0 | 3.3 | 15.0 | 0.09 | 36.6 | ||||||
. | HIV trap, LT†‡ . | . | . | . | MLV trap, RT†‡ . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Expression . | . | . | . | Expression . | . | . | . | ||||||
| Donor . | % GFP+ . | MFI* . | Integration, CpC . | % trapping efficiency, expr/integr . | % GFP+ . | MFI* . | Integration, CpC . | % Trapping efficiency, expr/integr . | ||||||
| A | ||||||||||||||
| 106 TU/mL | 1.0 | 16.0 | 0.12 | 8.3 | 0.6 | 19.1 | ND | ND | ||||||
| 107 TU/mL | 1.7 | 13.8 | 0.20 | 8.5 | 1.5 | 19.2 | 0.04 | 37.5 | ||||||
| B | ||||||||||||||
| 106 TU/mL | 2.8 | 12.6 | 0.32 | 8.7 | 0.8 | 14.1 | 0.02 | 37.7 | ||||||
| 107 TU/mL | 5.5 | 11.5 | 0.69 | 8.0 | 3.3 | 15.0 | 0.09 | 36.6 | ||||||
TU indicates 293T transducing units; ND, not determined.
Mean fluorescence intensity is measured after puromycin selection
Paired Student ttest calculated on MFI of GFP-positive cells (% GFP+), with P < .05
Unpaired Student ttest calculated on trapping efficiency, with P < .01
We then analyzed the behavior of the traps in HPCs. Murine HPCs were enriched from bone marrow cells by lineage marker depletion, stimulated with a combination of cytokines for 24 hours to reach efficient transduction by both vector types, and transduced with doses of HIV and MLV traps matched to yield comparable vector integration amounts (not shown). FACS analysis performed after 5 days of culture indicated a sharp difference in reporter gene expression, with the cultures transduced by the MLV trap containing a higher frequency of GFP-expressing cells and reaching a higher GFP MFI (Figure 4A).
Transduction of primary hematopoietic progenitors by HIV-based (LT) and MLV-based (RT) promoter traps. (A) FACS analysis of GFP expression in murine hematopoietic progenitors transduced with matched amounts of the indicated vector traps and analyzed 5 days after transduction. The frequency and MFI of GFP-positive cells are indicated. Representative results of 2 experiments performed. (B) FACS analysis of GFP expression in human cord blood CD34+ cells stimulated for 36 hours with a cytokine cocktail, transduced with matched vector amounts yielding low-copy (< 1) integration per cell, and analyzed after 1 week in liquid culture. The percentage of GFP-positive cells is indicated. Representative results of 2 experiments performed. (C) FACS analysis of GFP expression (histogram) of puromycin-selected cells from the populations shown in panel B. UC indicates unselected cells.
Transduction of primary hematopoietic progenitors by HIV-based (LT) and MLV-based (RT) promoter traps. (A) FACS analysis of GFP expression in murine hematopoietic progenitors transduced with matched amounts of the indicated vector traps and analyzed 5 days after transduction. The frequency and MFI of GFP-positive cells are indicated. Representative results of 2 experiments performed. (B) FACS analysis of GFP expression in human cord blood CD34+ cells stimulated for 36 hours with a cytokine cocktail, transduced with matched vector amounts yielding low-copy (< 1) integration per cell, and analyzed after 1 week in liquid culture. The percentage of GFP-positive cells is indicated. Representative results of 2 experiments performed. (C) FACS analysis of GFP expression (histogram) of puromycin-selected cells from the populations shown in panel B. UC indicates unselected cells.
Human HPCs were purified from cord blood by CD34+ selection and stimulated for 36 hours in medium containing a combination of early-acting cytokines before transduction.17 We then treated the transduced cells with or without puromycin and analyzed them by FACS for GFP expression and by clonogenic assays to determine the frequency of puromycin-resistant hematopoietic colonies. In a first experiment, cells were transduced with matched amounts of either vector to yield an average 20% transduction frequency, to allow calculating trapping efficiency from the assumption that vector-positive CFCs obtained in these conditions contain a single vector copy. Consistently with the results obtained in other cell types, the frequency of GFP-positive cells in liquid culture (Figure 4B), and the frequency of puromycin-resistant CFCs (Table 3) were significantly higher for cells transduced by the MLV than the HIV trap. PCR analysis for vector-specific sequences in hematopoietic colonies confirmed similar frequency of integration by both vectors at the predicted level (Table 3), showing that also in HPCs the MLV vector trapped cellular promoters more efficiently than HIV. Furthermore, the MLV trap was expressed to a higher extent both in the absence of puromycin selection (not shown), or following selection (Figure 4C).
CFC assays of transduced CD34+ cells
Experiment . | Vector-positive CFCs, % . | Puromycin-resistant CFCs, % . | Trapping efficiency, % . |
|---|---|---|---|
| LT exp 1 | 23 | 0.5 | 2 |
| RT exp 1 | 18 | 3.3 | 18 |
| LT exp 2 | 96 | 7.1 | < 7* |
| RT exp 2 | 23 | 4.9 | 21 |
Experiment . | Vector-positive CFCs, % . | Puromycin-resistant CFCs, % . | Trapping efficiency, % . |
|---|---|---|---|
| LT exp 1 | 23 | 0.5 | 2 |
| RT exp 1 | 18 | 3.3 | 18 |
| LT exp 2 | 96 | 7.1 | < 7* |
| RT exp 2 | 23 | 4.9 | 21 |
Colony-forming cell (CFC) assays of the same cells shown in Figure 4B (exp1) and from a different experiment that used a higher input of LT vector (exp2), plated in semisolid medium with or without puromycin. The frequency of vector- positive CFCs was determined by PCR performed on unselected CFCs. The frequency of puromycin-resistant CFCs was calculated from the ratio between CFCs grown in medium with and without puromycin. The frequency of promoter trapping was calculated as the ratio between puromycin-resistant CFCs and vector-positive CFCs.
In the presence of multicopy vector integration, the frequency of trapping (trapping efficiency) is overestimated
In a second experiment, we increased the HIV vector input and found that cultures matching the frequency of reporter expression obtained with the MLV trap had multicopy HIV integration in the CFCs (Table 3). Interestingly, also in these latter transduction conditions (low-copy MLV versus multicopy HIV trap integration), puromycin-selected MLV-transduced cells expressed the reporter gene to a higher extent than HIV-transduced cells (not shown).
Improved trap design increases trapping efficiency of HIV vector by rescuing expression from promoter-distal integration
As discussed above, we ascribed the different trapping efficiency of MLV and HIV traps to a different bias in integration site selection within transcriptional units, in agreement with recently reported studies.6-8 Promoter-proximal integrations, as observed more frequently with MLV, will express the reporter protein up to 50% of the time, depending on the orientation of insertion, and whether it occurred upstream of the first coding ATG. Instead, integrations occurring downstream of the first coding ATG, as observed more frequently with HIV, will express the reporter protein in no more than 1 in 6 times, due to the need for correct orientation (1 of 2) and in-frame fusion (1 of 3). Furthermore, lengthy fusion transcript and chimeric protein resulting from promoter-distal integration may be unstable and further decrease the overall trapping efficiency.
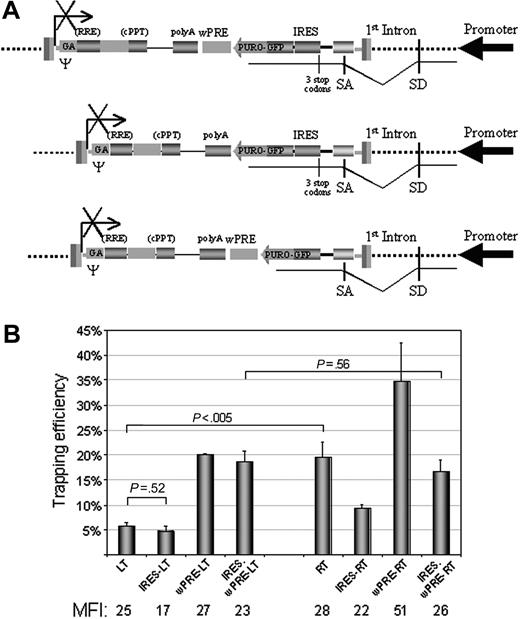
To verify this explanation, we modified both vector traps to make reporter expression insensitive to out-of-frame fusion and to increase the stability of fusion mRNA. We inserted 3 tandem stop codons in all reading frames followed by the IRES11 of the mouse encephalomyocarditis virus between the SA site and the PuroR-GFP transgene to terminate translation of the endogenous gene in fusion transcripts and drive downstream reporter expression in mRNAcap-independent way. We then added the wPRE13 between the PuroR-GFP transgene and the polyA site to increase fusion transcript stability and enhance IRES-dependent expression, which has been shown to be less efficient than that driven by the mRNAcap.21 We cloned the modified trapping cassette into the MLV and HIV vector backbone to generate IRES.wPRE-RT and IRES.wPRE-LT traps, respectively, and compared the performance of all traps in 293T cells (Figure 5A).
Trapping efficiency of IRES/wPRE modified HIV-based (LT) and MLV-based (RT) promoter traps. (A) Schematic of the modified trapping cassettes; legend as in Figure 1A. IRES indicates internal ribosome entry site; wPRE, woodchuck post-transcriptional regulatory element. (B) Histogram showing the trapping efficiency of the indicated vector traps. Bars indicate the percentage of integrations that express the reporter gene (trapping efficiency, mean ± SEM, n = 4). Statistical significance by Student t test of the difference between the indicated experimental groups is shown. For each trap, the mean fluorescence intensity (MFI) of GFP-positive cells (calculated from cell cultures with frequency of GFP+ cells≤ 15%) is indicated (mean, n = 2).
Trapping efficiency of IRES/wPRE modified HIV-based (LT) and MLV-based (RT) promoter traps. (A) Schematic of the modified trapping cassettes; legend as in Figure 1A. IRES indicates internal ribosome entry site; wPRE, woodchuck post-transcriptional regulatory element. (B) Histogram showing the trapping efficiency of the indicated vector traps. Bars indicate the percentage of integrations that express the reporter gene (trapping efficiency, mean ± SEM, n = 4). Statistical significance by Student t test of the difference between the indicated experimental groups is shown. For each trap, the mean fluorescence intensity (MFI) of GFP-positive cells (calculated from cell cultures with frequency of GFP+ cells≤ 15%) is indicated (mean, n = 2).
Remarkably, the modified IRES.wPRE-LT trap was significantly more expressed than the original HIV trap (P < .01), with an average increase in trapping efficiency of at least 3-fold. However, no significant change was observed for the modified MLV trap (Figure 5B). To assess the relative contribution of frame-independence and increased RNA stability in the rescue of HIV trap expression by the IRES.wPRE-LT construct, we generated traps containing either the IRES or the wPRE element. Inclusion of IRES alone was detrimental to reporter gene expression, as expected from other studies22 and as clearly observed for the MLV trap, which showed a statistically significant reduction in trapping efficiency and GFP MFI in transduced cells. Notably, the HIV trap did not show such an effect, suggesting that the detrimental effect of IRES on gene expression level was balanced by an increase in the proportion of transcripts leading to reporter expression. As expected, inclusion of the wPRE alone significantly enhanced expression of both MLV and HIV traps, however, maintaining significant differences in trapping efficiency and GFP MFI between the 2 vector types.
In conclusion, the differential effect of IRES on HIV versus MLV trap expression indicated that HIV integrations were much more frame-dependent for expression than MLV; ie, they occurred within transcribed genes but downstream of the first coding ATG. However, the frame-independence of reporter expression by MLV integrations strongly supported our previous conclusion that they occurred in close proximity to promoters. Thus, the differences in trapping efficiency observed for the original constructs (RT and LT) were due to a specific bias of MLV in integration site selection, ie, close proximity to active promoters, and not to a more frequent integration into transcribed genes or to vector backbone-dependent effects on reporter expression.
Discussion
Here we used a promoter trap built into HIV and MLV vectors to evaluate integration site selection in the target cell genome. We found a highly significant difference in trapping efficiency between the 2 vectors and showed by several lines of evidence that this effect was dependent on a different integration pattern of the 2 viruses within transcribed genes.
Our study provides direct, functional evidence that MLV vectors preferentially integrate close to highly active promoters in the different cell types tested and can efficiently exploit them for proviral expression. Such results are consistent with a recent study that analyzed 903 MLV integration sites by PCR-mediated cloning and reported preferential occurrence within ± 5 kb from transcription start sites of cellular genes (20% of the sites), and within ± 1 kb from CpG islands (16% of the sites).7 Notably, with the promoter trapping approach used here, we were able to screen several hundred thousand integrations in different target cell types, including primary hematopoietic cells, for both MLV and HIV vectors. By using a functional readout, our analysis did not rely on the sometimes uncertain “in silico” identification of genes and transcription regulatory elements and on our current knowledge of the overall extent of genome transcription,23 and avoided possible biases as a result of the vector-genome junction retrieval procedures. Thus, our results confirm and extend the conclusions of the study by Wu et al.7
Remarkably, from 1 of 7 up to 1 of 3 MLV integrations resulted in reporter gene expression. Because only integrations occurring downstream from a promoter and with the proper orientation will express the reporter, the MLV bias for integration proximal to active promoters may even be higher than previously estimated.7 This surprisingly high trapping efficiency may be dependent on specific experimental conditions, such as strong synchronous T-cell receptor (TCR) activation in PBMCs, a low vector input, and the inclusion of the wPRE element in the trap that enhances reporter gene expression by stabilizing the transcript.13 Target cell stimulation may increase the number or the avidity of the preferred genomic sites available to a low vector input and highlight a nonrandom saturable integration behavior that tethers the viral preintegration complex to transcriptionally active promoters. In agreement with this hypothesis, we noticed that the MLV promoter trapping efficiency decreased by increasing the vector input in the cell lines tested (data not shown).
In addition, by comparing MLV and HIV integration in similarly infected cells, we consistently found that the MLV trap was expressed to a slightly but significantly higher level than the HIV trap, suggesting preferential MLV integration into a subset of more actively transcribed genes. However, we cannot exclude that the different integration pattern of MLV and HIV within transcription units may play a role in this finding, given the complex interplay of factors affecting reporter protein expression, including mRNA and protein stability. Notably, addition of the wPRE element enhanced expression of both MLV and HIV traps while maintaining the difference in GFP MFI between the 2 vector types. A comparative analysis of gene expression at the trapped locus and at the wild-type allele within isolated clones would clarify these issues. If verified, the finding of MLV integration into a subset of more actively transcribed genes will highlight the specificity of the rules governing integration site selection by different integrating viruses, making a strong case for virus-specific interactions with cellular proteins tethering the integration complex to selected gene sets.24 Targeting of a highly transcribed gene subset by MLV may reflect tethering of its integration complex by cellular components recruited at highly active or induced (growth-responsive) promoters.
Promoter targeting may have evolved in the parental retroviruses to provide a fraction of their progeny with a supplementary cellular promoter, less susceptible to silencing by genome surveillance mechanisms than the one embedded in the LTR,25 and possibly enhancing env gene expression by alternative splicing. The proximity and functional relationship frequently established between MLV proviruses and flanking cellular promoters in our study indicate that integration of enhancer-rich wild-type MLV LTR is likely to up-regulate transcription from cellular promoters and, depending on proviral position and orientation, overexpress the cognate genes, including proto-oncogenes. The observed MLV integration behavior thus explains the well-known oncogenicity of wild-type gamma-retroviruses26,27 and underscores the risk of insertional mutagenesis by MLV-derived vectors.2,3,28
HIV- and HIV-derived vectors have been reported to integrate more frequently within transcribed genes than outside of them.6 However, since they insert throughout the transcriptional unit without showing a bias for promoter-proximal integration7 (our data in “Results”), the promoter trapping efficiency of HIV traps was much lower than that observed for MLV traps. When we modified the trap design to make reporter expression potentially insensitive to promoter-distal integration, HIV traps expressed the reporter in up to 1 of 5 integrations. This figure is consistent with a strongly favored HIV integration into active genes, if one considers that trap expression occurs only upon integration in the proper orientation, that expression of IRES-containing traps is suboptimal, and that only a fraction of the genome is transcribed in target cells. Preferential intragenic integration of HIV vectors was very recently reported in primary hematopoietic cells,29,30 and integration intensity was shown to positively correlate with transcriptional intensity in one study.29 Preferential insertion throughout transcriptionally permissive chromatin may benefit the lentivirus lifestyle, which spread horizontally in the cognate host species, target interphase chromatin crossing the nucleopore, and express their own transcription regulators. Whether this behavior of the parental virus, together with the advanced vector engineering that fully inactivates the HIV LTR, provide a safer integrating tool for gene therapy remains, however, to be directly demonstrated.
Promoter trapping efficiency may provide a convenient readout to test whether genetic engineering of the vector framework, ie, the incorporation of chromatin insulators, can disrupt transcriptional interactions with the flanking genome. Furthermore, this approach can be used to compare integration site selection among different target cell types and between cells transduced under different growth conditions. In addition, the improved HIV traps described in this study provide powerful new tools to perform gene expression studies in target cells which are poorly transfected by MLV-based vectors or other means, such as primary and quiescent cells, stem cells, and embryos.
Prepublished online as Blood First Edition Paper, November 12, 2004; DOI 10.1182/blood-2004-03-0798.
Supported by grants from Telethon (TIGET), Associazione Italiana per la Ricerca sul Cancro (AIRC) and the Italian Ministry of Scientific Research (MIUR) (L.N.) and from the Italian Ministry of Health (R.F. 02/184) (E. Medico).
M.DeP., E. Montini, and F.R.S.d.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Lucia Sergi Sergi and Andrea Verhovez for technical help.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal