Abstract
We previously reported a novel fusion between TEL and FGFR3 in a patient with peripheral T-cell lymphoma with t(4; 12)(p16;p13). Disease in this patient subsequently progressed to acute myelogenous leukemia (AML) with the same translocation. Sequence analysis of TEL-FGFR3 fusion transcripts suggested that these diseases originated from the same multipotent stem cell. To determine the transforming property of TEL-FGFR3, we established transfectants of this chimeric fusion gene and investigated the major signal pathways of TEL-FGFR3–induced transformation using various signal transduction inhibitors including SU5402 (fibroblast growth factor tyrosine kinase [FGFR TK] inhibitor). Our results indicated that (1) the expression of TEL-FGFR3 but not ΔHLH-TEL-FGFR3 resulted in efficient focus formation in NIH/3T3 cells and conferred interleukin 3 independence to Ba/F3 cells by a constitutive tyrosine kinase activity probably through oligomerization by the HLH domain of TEL; (2) although effector proteins including classical mitogen-activated protein kinase (MAPK), p38 MAPK, phosphatidylinositol 3-kinase (PI3-K), mammalian target or rapamycin (mTOR), signal transducer and activator of transcription 3 (STAT-3) and STAT-5 were activated in TEL-FGFR3 transformants, the growth of the transformants was inhibited by SU5402 (concentration that inhibits 50% [IC50] = 5 μM) and the PI3-K inhibitor, LY294002 (IC50 = 10 μM) and wortmannin (IC50 = 5 μM), but not by U0126, SB203580, or rapamycin; and (3) injection of TEL-FGFR3 transformants induced lethal leukemia into syngeneic mice. Taken together, the leukemogenic potential of TEL-FGFR3 may be mediated in part through PI3-K.
Introduction
Chronic myeloid leukemia (CML) is a clonal hematopoietic stem cell disorder characterized by t(9;22)(q34;q11) that results in the BCR-ABL fusion gene. Many in vitro and in vivo studies have shown that BCR-ABL alone is sufficient to cause CML, and mutational analyses have indicated that constitutive tyrosine kinase (TK) activity is required for its transforming properties as leukemogenesis.1,2 In a recent clinical study, imatinib mesylate, a selective inhibitor of BCR-ABL TK, resulted in excellent hematologic and cytogenetic responses in more than 90% of patients with CML.3 However, a small percentage of patients present with clinical and hematologic features suggestive of CML, but lack the BCR-ABL rearrangement. This BCR-ABL- CML has been classified as a myeloproliferative disease (MPD).4 Most cases of BCR-ABL- CML have a normal karyotype, but a few patients have a reciprocal translocation that results in TEL-PDGFβR, TEL-ABL, TEL-JAK2, ZNF198-fibroblast growth factor receptor 1 (FGFR1), FOP-FGFR1, CEP-FGFR1, or BCR-FGFR1 that generates constitutively activated TK fusions.5-15 In particular, chimera fusion proteins involving FGFR1 have been observed in patients with MPD with eosinophilia and lymphadenopathy and in a high percentage of cases of T-cell non-Hodgkin lymphoma that progresses to acute myelogenous leukemia (AML).16-18 With the advent of targeted signal transduction therapy, making an accurate clinical diagnosis and a molecular approach toward hematopoietic malignancies have become increasingly important. We previously reported a novel TEL-FGFR3 fusion gene in a patient with peripheral T-cell lymphoma (PTCL) with t(4;12)(p16;p13).19 Subsequently, this PTCL progressed to AML with the same chromosomal translocation. We suspected that this fusion transcript had originated from the same clone as the neoplastic multipotent stem cell. In this report, we demonstrate the transforming property of TEL-FGFR3 chimera fusion protein and that it displays constitutive kinase activity that is probably mediated through the oligomerization of the TEL HLH domain [also known as the pointed (PNT) or a sterile α-motif (SAM) domain]. This constitutive activation seems to deregulate various signaling pathways including mitogen-activated protein kinases (MAPKs), phosphatidylinositol 3-kinase (PI3-K), p38 MAPK, and signal transducer and activator of transcription (STAT) pathways that elicit biologic processes including cell cycle progression, cellular differentiation, and apoptosis. We clarify the major signal transduction pathways of TEL-FGFR3–induced transformation and characterize the effect of various inhibitors on these pathways, and in particular the FGFR TK inhibitor, SU5402, to establish a promising approach for a novel molecular target chemotherapy.
Patient, materials, and methods
Informed consent for all procedures was obtained from the patient.
Case report
Informed consent for all procedures was obtained from the patient. Approval for this study was obtained from the Saitama Medical School institutional review board.
The clinical history of the patient has been described previously.19 A 63-year-old woman was admitted with a clinical diagnosis of PTCL stage IVB. A bone marrow (BM) aspiration revealed that 2.8% of the BM nuclear cells were lymphoma cells with the same phenotype as that of the lymph node biopsy specimen and cytogenetic analysis showed 46, XX, t(4;12)(p16; p13) [16], 46, XX [4]. The patient was treated with chemotherapy according to the modified CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisolone) protocol. After initial chemotherapy, her lymph node swelling generally improved but cytogenetic analysis of the BM showed 46,XX,t(4;12)(p16;p13)[2],46,XX[18]. She was treated with 6 courses of modified CHOP therapy and achieved complete hematologic remission (CR). Eleven months later, however, the patient developed minimally differentiated AML-M0 according to the French-American-British (FAB) classification. Complete blood examination revealed a hemoglobin level of 108 g/L, platelet count of 62 × 109/L, and white blood cell (WBC) count of 15.2 × 109/L with the following differential: neutrophils 23%, lymphocytes 13%, monocytes 16%, meta-myelocytes 4%, myelocytes 4%, and blast cells 39%. A biochemical study revealed an elevated serum lactase dehydrogenase (LDH) level of 519 IU/mL (normal, 107-220 IU/mL). A BM aspirate showed normocellularity, with 43.8% blasts that were negative on peroxidase staining, 0.9% matured eosinophils, 12.9% monocytes, and reduced percentage of erythroblasts (1.9%). Flow cytometric analysis showed that her blasts were positive for CD7, CD13, CD33, CD34, and HLA-DR, but negative for CD4, CD14, CD19, and CD56. BM cytogenetics revealed t(4;12)(p16;p13) in 20 of 20 metaphases. The first CR was induced by combination chemotherapy with idarubicin and cytarabine (Ara-C), and CR was maintained for 6 months. In her first relapse, she developed AML and BM aspiration showed 16.5% blasts with fibrosis. Repeated cytogenetic analyses of her peripheral blood (PB) revealed that almost all of her peripheral blood cells had 46,XX, t(4;12)(p16;p13) with additional chromosomal abnormalities. The patient failed to respond to Ara-C, etoposide, and vincristine, and she died of progressive pneumonia and sepsis 7 months after receiving reinduction therapy.
RT-PCR
Nested reverse transcription-polymerase chain reaction (RT-PCR) analysis for TEL-FGFR3 chimeric transcripts was performed with the RNA LA PCR kit (Takara Bio, Ohtsu, Japan). Using TriZol (Gibco, Grand Island, NY), RNAs were prepared from 1 × 107 BM or PB mononuclear cells that had been obtained from the patient at each stage of her disease and from a healthy donor as a negative control. After purification, 1 μg RNA was used to generate cDNA using reverse transcriptase and random hexamer. The first PCR was performed with the following primers: TELE5 (5′-TGTCCAGAGGACCCCCAGGC-3′), which is specific for the 5′-untranslated region (UTR) of TEL, in combination with FGF-RV0 (5′-CTCACATTGTTGGGGACC-3′), which is specific for the 3′-UTR of FGFR3. The PCR conditions were as follows: initial denaturation at 94°C for 1 minute, followed by 35 cycles of 94°C for 20 seconds, 58°C for 20 seconds, and 72°C for 1.5 minutes, and a final elongation step at 72°C for 10 minutes. The second PCR was performed as described previously19 using Taq DNA polymerase high fidelity (Invitrogen, Carlsbad, CA). Direct sequence analysis was performed on these PCR products.
Vector constructs
For easy detection of the fusion proteins, the mammalian expression C-terminal fusion vector, which contains the V5 epitope, pTracer-EF/V5-HisC (Invitrogen), was chosen. To construct the vector expressing wild-type TEL-FGFR3 with the C-terminal V5 peptide tag, nested PCR was performed again using the first PCR product mentioned (see “RT-PCR”) with the sense primer, E93F1 (5′-GGAAAAACCTGAGAACTT-3′), and the antisense primer, FGF-RV1mut (5′-GACCAGTGGCCCTACACG-3′), which had been converted from the antisense primer FGF-RV1 (mutated base is underlined). Furthermore, to identify the function of the HLH domain of TEL, which is known as a factor necessary for autophosphorylation of TEL-FGFR3 through oligomerization, an HLH deletion (amino acids 39-114) mutant (ΔHLH) was generated by the PCR method using PfuUltra high-fidelity DNA polymerase (Stratagene, La Jolla, CA).
These PCR products were cloned directly into the TA-cloning vector, pCR2.1-TOPO, and amplified using the TOPO TA cloning kit (Invitrogen), and the fidelity of the sequences was verified by direct sequencing of the PCR-amplified DNA. It was next digested from the TOPO vector by the EcoRI restriction enzyme and then subcloned into the same site in the pTracer-EF/V5-HisC by using a DNA ligation kit version 2 (Takara Bio) to result in the plasmid pTracer-EF/V5-HisC/TEL-FGFR3 (TF-V5) and pTracer-EF/V5-HisC/ΔHLH-TEL-FGFR3 (ΔHLH-TF-V5). All clones used for expression studies were verified by sequencing of both strands to have the correct inserts and no mutation and were purified by using the EndoFree Plasmid Maxi Kit (Qiagen, Hilden, Germany).
Cell cultures and electroporation
The interleukin 3 (IL-3)–dependent murine pro-B cell line, Ba/F3 (Riken Cell Bank, Tsukuba, Japan), was maintained in RPMI 1640 (Gibco Invitrogen, Carlsbad, CA) with 10% fetal calf serum (FCS) and 5 ng/mL recombinant murine IL-3 (R&D Systems, Minneapolis, MN). The murine embryo fibroblast cell line, NIH/3T3 (clone 5611; JCRB0615), was purchased from Health Science Research Resources Bank (Sennan, Japan) and was maintained in Dulbecco modified Eagle medium (DMEM; ICN Biomedicals, Aurora, OH) with 10% FCS. All cells were cultured at 37°C in a5%CO2 humidified incubator. To express TF-V5 as well as ΔHLH-TF-V5 in Ba/F3 cells, 106 Ba/F3 cells were electroporated at 250 V/960 μF in a Gene Pulser (Bio-Rad, Hercules, CA) with 20 μg plasmid DNA. To establish polyclonal transfectants, cells were maintained in IL-3–supplemented culture medium with 800 μg/mL zeocin (Invitrogen) for 14 days. Mock-transfected polyclonal cells and parent cells were used as controls.
Focus formation assay
NIH/3T3 cells were seeded at 5 × 105 cells/10-cm plate and incubated overnight. Cells were transfected by the calcium phosphate method with the mammalian transfection kit (Stratagene). A total amount of plasmid plus carrier DNA equal to 30 μg was used in each experiment. Forty-eight hours after transfection, cells were seeded 1 × 105 cells/10-cm plate and cultured with 200 μg/mL zeocin. The culture medium with zeocin was replaced every 3 days. Fourteen days after transfection, the plates were fixed and stained with Giemsa and focus formation was assessed. The experiments were performed in triplicate and at least 2 independent experiments were performed.
Immunofluorescence staining
Transfected NIH/3T3 cells were trypsinized, replated in ethanol-sterilized microscope slides, and fixed with 4% paraformaldehyde-PBS for 25 minutes. They were stained with anti-V5 antibody (1:200; Invitrogen) and then with affinity-purified, rhodamine-conjugated antimouse secondary antibody (1:200; Chemicon International, Temecula, CA). DAPI I (4′6-diamidino-2-phenylindole-2HCl) counterstain (Vysis, Downers Grove, IL) plus antifade solution (Vysis) and 20 × standard saline citrate (SSC) buffer was applied to the cells to allow visualization of the nuclei. The slides were analyzed by fluorescence microscopy (Eclipse E800; Nikon, Tokyo, Japan) using the red (rhodamine), green (green fluorescent protein [GFP]), and blue (DAPI) channels. Images were captured with a CCD camera (Cohu, San Diego, CA) attached to the microscope with a Nikon Plan Apo 100 ×/1.40 oil objective lens (Nikon) and cropped in Leica QFISH software (Leica Imaging Systems, Cambridge, United Kingdom).
Cell proliferation assay
For growth curves, using CellTiter 96 AQueous One Solution (Promega, Madison, WI), the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay was performed in 96-well plates according to the manufacturer's instructions. Briefly, the cells (2000/well) were plated in triplicate in 100 μL medium with or without IL-3, and then incubated in 20 μL MTS/phenazine ethosulfate (PES) solution for 1 hour, after which the amount of soluble formazan (reduced MTS tetrazolium) was measured by optical density at 490 nm using ImmunoReader NJ-2000 (Nalge Nunc International KK, Tokyo, Japan). For the inhibitor assays, inhibitors were added at increasing concentrations to examine their effects on IL-3– and factor-independent proliferation. Cells were used for experiments within 10 passages and all experiments were repeated at least 3 times.
Immunoprecipitation and Western blotting
Cells (1 × 107) were washed once in cold phosphate-buffered saline (PBS) and lysed in 1 mL NP-40 lysis buffer (0.2% NP-40, 50 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.3, 250 mM NaCl, 5 mM EDTA [ethylenediaminetetraacetic acid] plus Halt protease inhibitor cocktail, EDTA-free [Pierce Chemical, Rockford, IL], and phosphatase inhibitor cocktail 2 [Sigma, St Louis, MO]). Cell lysates were cleared by centrifugation at 27 000g at 4°C for 15 minutes. For immunoprecipitation (IP), 150 μL total cell lysate was precleared with 25 μL protein G-Sepharose (Amersham Biosciences, Buckinghamshire, United Kingdom) and incubated with anti-V5-Tag (MBL, Nagoya, Japan) at 4°C for 1 hour and then 30 μL protein G-Sepharose was added at 4°C for 30 minutes. The precipitates were washed 3 times in cold lysis buffer and then boiled in loading buffer for 5 minutes. The lysates or immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a Hypond-P polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences). After blocking with 5% dry milk in PBS containing 0.1% Tween-20 (PBST), the blots were incubated for 1 hour at room temperature with the primary antibodies, followed by incubation with IgG-conjugated horseradish peroxidase (HRP) secondary antibodies, and visualized using an enhanced chemiluminescence (ECL) detection system (Amersham Biosciences). To test the effect of the inhibitors, cells were pretreated with each inhibitor for 5 minutes to 6 hours, and lysed; then, Western blotting was performed with the appropriate antibodies and the phosphorylation status of specific downstream substrates was detected. K562 cells were used as a positive control for phosphorylated MAPK,20 Akt,21 and STAT-5,22 and a negative control for phosphorylated p38 MAPK and STAT-3.
Leukemogenesis in syngeneic mice
Seven-week-old BALB/c mice (CLEA Japan, Tokyo, Japan) were injected through a tail vein with 3 × 106 cells from monoclonal transfected Ba/F3 cell lines (6 in each group). These cell lines had been obtained from the polyclonal transfectants by the limiting dilution method. Cells in an early passage were used. The survival of the injected mice was monitored until 8 weeks. Terminally sick mice were humanely killed by cervical dislocation and examined for the development of hematologic malignancy. The spleen was subjected to histopathologic examination. This study was approved by the Animal Care Committee of our institution.
Other reagents
For multiplex Western blotting, the following antibodies including Multiplex Western Cocktail 1 (Cell Signaling Technology, Beverly, MA) were used: anti-Akt, anti–phospho-Akt (Ser473), anti–phospho-S6 ribosomal protein (S6RP; Ser235/236), anti–phospho-p44/42 MAPK (Thr202/Tyr204), and anti–phospho-p90RSK (Ser380). Other antibodies used included anti–phospho-p38 MAPK (Thr180/Tyr182), anti–phospho-STAT-3 (Tyr705), anti–phospho-STAT-3 (Ser727), and anti–phospho-STAT-5 (Tyr694) antibodies (Cell Signaling Technology). Rabbit polyclonal antibody ERK-1, p38 (Santa Cruz Biotechnology, Santa Cruz, CA), anti–STAT-3, anti–STAT-5 (BD Transduction Laboratories, Lexington, KY), antiphosphotyrosine, clone 4G10 (Upstate Biotechnology, Lake Placid, NY), anti–β-actin (Abcam, Cambridge, United Kingdom), sheep anti–mouse IgG HRP, and donkey anti–rabbit IgG HRP (Amersham Biosciences) were used.
The following inhibitors were used: SU5402, rapamycin, SB203580 (Calbiochem, San Diego, CA), AG1296, U0126, LY294002 (Biomol Research Laboratories, Plymouth Meeting, PA), and wortmannin (Sigma). All inhibitors were resolved in dimethyl sulfoxide (DMSO) and used at a maximum concentration of 0.1% in culture medium. Target molecules for these inhibitors are shown in Figure 4B.
Activation of downstream effector proteins in TEL-FGFR3 transfectants. (A) Multiplex Western blot analysis of lysates of polyclonal TF-V5–transfected Ba/F3 cells that had been incubated in the presence or absence of SU5402. Western blotting of lysates of cells that were transfected with mock and incubated with IL-3 (lane 1), mock driven with IL-3 for 30 minutes (lane 2), ΔHLH-TF-V5 (lane 3), TF-V5 (lane 4), or TF-V5 and incubated in the presence of 0.1% DMSO (lane 5), 5 μM SU5402 (lane 6), 10 μM SU5402 (lane 7), or 25 μM SU5402 (lane 8) for 1 hour was performed. K562 cells were used as a positive control for phosphorylated MAPK, Akt, and STAT-5, and a negative control for phosphorylated p38 MAPK and STAT-3 (lane 9). (B) Hypothetical schema of TEL-FGFR3–induced signal transduction pathway and the effects of the pharmacologic inhibitors used in this study.
Activation of downstream effector proteins in TEL-FGFR3 transfectants. (A) Multiplex Western blot analysis of lysates of polyclonal TF-V5–transfected Ba/F3 cells that had been incubated in the presence or absence of SU5402. Western blotting of lysates of cells that were transfected with mock and incubated with IL-3 (lane 1), mock driven with IL-3 for 30 minutes (lane 2), ΔHLH-TF-V5 (lane 3), TF-V5 (lane 4), or TF-V5 and incubated in the presence of 0.1% DMSO (lane 5), 5 μM SU5402 (lane 6), 10 μM SU5402 (lane 7), or 25 μM SU5402 (lane 8) for 1 hour was performed. K562 cells were used as a positive control for phosphorylated MAPK, Akt, and STAT-5, and a negative control for phosphorylated p38 MAPK and STAT-3 (lane 9). (B) Hypothetical schema of TEL-FGFR3–induced signal transduction pathway and the effects of the pharmacologic inhibitors used in this study.
Results
Consistent expression of TEL-FGFR3 chimeric transcripts during the course of the disease
To verify the presence of TEL-FGFR3 chimeric transcripts throughout all stages of the disease in the patient, RT-PCR was performed. As expected from the results of karyotype analyses, TEL-FGFR3 chimeric transcripts (predicted PCR product size, 1767 bp) were detected by RT-PCR in all samples obtained from the patient (Figure 1A). The time at which each PCR sample was obtained is shown in Table 1. Sequence analyses revealed that all of the resulting PCR products had the same fusion breakpoint and no mutations except for a CAG trinucleotides insertion (data not shown). These results suggest that all of the hematologic diseases of the patient were partly caused by TEL-FGFR3. Furthermore, all of the hematologic diseases of the patient were thought to have originated from the same clone, although additional chromosome abnormalities may have occurred during the course of the disease from PTCL to AML.
TEL-FGFR3 transcripts and vector constructs. (A) RT-PCR analysis for detection of TEL-FGFR3 transcripts. Nested RT-PCR was performed using 1 μg RNA from a series of samples of bone marrow (BM) and peripheral blood (PB) obtained from the patient with t(4;12)(p16;p13). The final product was analyzed by electrophoresis on a 1.5% agarose gel in Tris (tris(hydroxymethyl)aminomethane)–borate/EDTA (TBE) buffer, soaked in ethidium bromide, and visualized under UV light. The black arrow indicates the single PCR product (1767 bp). The PCR products from all samples had the same TEL-FGFR3 fusion breakpoint by sequence analysis. The materials (BM or PB) and dates on which the materials were obtained are as follows: BM cells on December 5, 2000 (2 months after the patient was diagnosed with PTCL; lane 1), BM cells on September 12, 2001 (when the patient was diagnosed with AML; lane 2), PB on September 14, 2001 (lane 3), PB on January 8, 2003 (lane 4), and BM cells of a healthy donor as a negative control (lane C). M indicates the molecular size markers. The numbers on the left of the figure indicate the molecular size (in base pair [bp]). (B) Schematic representation of proteins encoded by vector constructs of TEL-FGFR3 and TEL-FGFR3 lacking the HLH domain. (i) Full-length TEL protein of 452 amino acids (aa). The positions of the HLH domain and E26 transformation-specific (ETS) DNA-binding domain are indicated by dark shaded boxes. (ii) Full-length FGFR3 protein of 806 amino acids, which contains 3 extracellular immunoglobulin-like domains, a transmembrane domain, and TK 1 and 2 subdomains interrupted by a kinase insert. (iii) Full-length TEL-FGFR3 fusion protein of 589 amino acids, with a V5 epitope tag (TF-V5). This translocation fused nucleotide (nt) 543 of TEL to nt 1270 of FGFR3 as described previously.16 Closed arrow indicates the t(4;12) fusion breakpoint. Open arrow indicates the CAG trinucleotides insert that was consistently found in the fusion transcripts in all samples from the patient. (iv) ΔHLH-TF-V5, with an in-frame deletion of 76 amino acids (Δ amino acids 39-114 of TF-V5) and which lacks most of the TEL-HLH domain. HLH indicates helix-loop-helix domain; ETS, ETS DNA-binding domain; Ig, immunoglobulin-like domain; TM, transmembrane domain; TK, tyrosine kinase; KI, kinase insert; V5, V5 epitope.
TEL-FGFR3 transcripts and vector constructs. (A) RT-PCR analysis for detection of TEL-FGFR3 transcripts. Nested RT-PCR was performed using 1 μg RNA from a series of samples of bone marrow (BM) and peripheral blood (PB) obtained from the patient with t(4;12)(p16;p13). The final product was analyzed by electrophoresis on a 1.5% agarose gel in Tris (tris(hydroxymethyl)aminomethane)–borate/EDTA (TBE) buffer, soaked in ethidium bromide, and visualized under UV light. The black arrow indicates the single PCR product (1767 bp). The PCR products from all samples had the same TEL-FGFR3 fusion breakpoint by sequence analysis. The materials (BM or PB) and dates on which the materials were obtained are as follows: BM cells on December 5, 2000 (2 months after the patient was diagnosed with PTCL; lane 1), BM cells on September 12, 2001 (when the patient was diagnosed with AML; lane 2), PB on September 14, 2001 (lane 3), PB on January 8, 2003 (lane 4), and BM cells of a healthy donor as a negative control (lane C). M indicates the molecular size markers. The numbers on the left of the figure indicate the molecular size (in base pair [bp]). (B) Schematic representation of proteins encoded by vector constructs of TEL-FGFR3 and TEL-FGFR3 lacking the HLH domain. (i) Full-length TEL protein of 452 amino acids (aa). The positions of the HLH domain and E26 transformation-specific (ETS) DNA-binding domain are indicated by dark shaded boxes. (ii) Full-length FGFR3 protein of 806 amino acids, which contains 3 extracellular immunoglobulin-like domains, a transmembrane domain, and TK 1 and 2 subdomains interrupted by a kinase insert. (iii) Full-length TEL-FGFR3 fusion protein of 589 amino acids, with a V5 epitope tag (TF-V5). This translocation fused nucleotide (nt) 543 of TEL to nt 1270 of FGFR3 as described previously.16 Closed arrow indicates the t(4;12) fusion breakpoint. Open arrow indicates the CAG trinucleotides insert that was consistently found in the fusion transcripts in all samples from the patient. (iv) ΔHLH-TF-V5, with an in-frame deletion of 76 amino acids (Δ amino acids 39-114 of TF-V5) and which lacks most of the TEL-HLH domain. HLH indicates helix-loop-helix domain; ETS, ETS DNA-binding domain; Ig, immunoglobulin-like domain; TM, transmembrane domain; TK, tyrosine kinase; KI, kinase insert; V5, V5 epitope.
Hematologic and cytogenetic data obtained during the clinical course of the patient with t(4;12)(p16;p13)
Date . | October 2000 . | December 2000 . | September 2000 . | December 2001 . | February 2002 . | June 2002 . | January 2003 . |
|---|---|---|---|---|---|---|---|
| Clinical diagnosis | NHL PTCL stage IVb | NHL in CR | AML M0 | AML in CR | AML in CR | AML M0 with fibrosis | AML M0 with fibrosis |
| Disease status | PTCL onset | CR | AML onset | CR | CR | AML relapse | NR |
| WBC count, × 103/μL | 5.74 | 1.61 | 15.24 | 3.50 | 2.15 | 1.64 | 0.89 |
| Neutrophils, % | 57.0 | 21.0 | 32.0 | 60.0 | 45.0 | 36.0 | 20.0 |
| Monocytes, % | 8.0 | 23.0 | 16.0 | 16.0 | 18.0 | 13.0 | 3.0 |
| Eosinophils, % | 7.0 | 2.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.0 |
| Blasts, % | 0.0 | 0.0 | 39.0 | + | + | 3.0 | 70.0 |
| BM aspirate blasts, % | 2.8 | 2.7 | 43.8 | 0.5 | 1.2 | 16.5 | ND |
| Karyotype* | 46,XX, t(4;12) (p16;p13)[3] 49,XX, +i(1)(q10),t(4;12) (p16;p13), +10, +19[10] 49,XX, +i(1)(q10), +11, +19[3] 46,XX[4] | 46,XX, t(4;12) (p16;p13)[2] 46,XX,inv(3) (q21;q26)[1] 46,XX[17] | 46,XX,t(3;8)(q27;q24),t(4;12) (p16;p13)[20] | 46,XX,t(4;12) (p16;p13)[2] 46,XX[18] | 46,XX,t(4;12) (p16;p13), inc[1]46,XX[39] | 46,XX,t(4;12)(p16;p13) [2]46,XX, t(3;8)(q27;q24), t(4;12)(p16;p13)[1] | 46,XX, t(2;2)(p21;p25), t(3;8)(q27;q24), t(4;12)(p16;p13) add(7)(q22) [20] |
| Chemotherapy | THP-COP(THP+CPA+VCR+PSL) | THP-COP (THP+CPA+VCR+PSL) | IDA+Ara-C MIT+Ara-C | DNR+Ara-C c+PSL ACR+Ara-C c+PSL | Ara-C+VP-16+PSL,VCR,VDS | Ara-C+VP-16+PSL,VCR,VDS | Palliative therapy |
| RT-PCR for TEL-FGFR3 | +, BM | +, BM | +, BM; +, PB | ND | ND | ND | +, PB |
Date . | October 2000 . | December 2000 . | September 2000 . | December 2001 . | February 2002 . | June 2002 . | January 2003 . |
|---|---|---|---|---|---|---|---|
| Clinical diagnosis | NHL PTCL stage IVb | NHL in CR | AML M0 | AML in CR | AML in CR | AML M0 with fibrosis | AML M0 with fibrosis |
| Disease status | PTCL onset | CR | AML onset | CR | CR | AML relapse | NR |
| WBC count, × 103/μL | 5.74 | 1.61 | 15.24 | 3.50 | 2.15 | 1.64 | 0.89 |
| Neutrophils, % | 57.0 | 21.0 | 32.0 | 60.0 | 45.0 | 36.0 | 20.0 |
| Monocytes, % | 8.0 | 23.0 | 16.0 | 16.0 | 18.0 | 13.0 | 3.0 |
| Eosinophils, % | 7.0 | 2.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.0 |
| Blasts, % | 0.0 | 0.0 | 39.0 | + | + | 3.0 | 70.0 |
| BM aspirate blasts, % | 2.8 | 2.7 | 43.8 | 0.5 | 1.2 | 16.5 | ND |
| Karyotype* | 46,XX, t(4;12) (p16;p13)[3] 49,XX, +i(1)(q10),t(4;12) (p16;p13), +10, +19[10] 49,XX, +i(1)(q10), +11, +19[3] 46,XX[4] | 46,XX, t(4;12) (p16;p13)[2] 46,XX,inv(3) (q21;q26)[1] 46,XX[17] | 46,XX,t(3;8)(q27;q24),t(4;12) (p16;p13)[20] | 46,XX,t(4;12) (p16;p13)[2] 46,XX[18] | 46,XX,t(4;12) (p16;p13), inc[1]46,XX[39] | 46,XX,t(4;12)(p16;p13) [2]46,XX, t(3;8)(q27;q24), t(4;12)(p16;p13)[1] | 46,XX, t(2;2)(p21;p25), t(3;8)(q27;q24), t(4;12)(p16;p13) add(7)(q22) [20] |
| Chemotherapy | THP-COP(THP+CPA+VCR+PSL) | THP-COP (THP+CPA+VCR+PSL) | IDA+Ara-C MIT+Ara-C | DNR+Ara-C c+PSL ACR+Ara-C c+PSL | Ara-C+VP-16+PSL,VCR,VDS | Ara-C+VP-16+PSL,VCR,VDS | Palliative therapy |
| RT-PCR for TEL-FGFR3 | +, BM | +, BM | +, BM; +, PB | ND | ND | ND | +, PB |
NHL indicates non-Hodgkin lymphoma; CR, complete remission; ND, not done; NR, no response; THP, pirarubicin; COP, cyclophosphamide, Oncovin (vincristine sulfate), prednisone; CPA, cyclophosphamide; VCR, vincristine; PSL, prednisolone; IDA, idarubicin; Ara-C, cytarabine; MIT, mitoxantrone; DNR, daunorubicin; ACR, aclarubicin; VP-16, etoposide; VDS, vindesine.
The karyotypes were described according to the International System for Human Cytogenetic Nomenclature, ISCN23
Transforming activity of TEL-FGFR3 in Ba/F3 and NIH/3T3 cells
To test whether the TEL-FGFR3 chimeric transcripts have transforming activity, we constructed a fusion cDNA in the expression vector pTracerEF/V5. Further, to test whether the HLH domain of TEL-FGFR3 is necessary for its biologic activity, we constructed an HLH deletion mutant expression vector (ΔHLH-TF-V5; Figure 1B). The IL-3–dependent cell line, Ba/F3, which does not express FGFR3 (data not shown), was electroporated with TF-V5, ΔHLH-TF-V5, or vector alone as a negative control. IL-3–independent cell growth was observed in Ba/F3 cells transfected with TF-V5, but not in Ba/F3 cells transfected with ΔHLH-TF-V5 or vector alone (mock; Figure 2Aii). TF-V5–transfected cells grew at the same rate when maintained with or without IL-3 (Figure 2Ai-ii), indicating that the chimeric protein efficiently substituted for the cytokine that controls the proliferation of Ba/F3 cells. To confirm that TEL-FGFR3 has transforming activity, the focus formation assay was performed using NIH/3T3 cells. Cells transfected with TF-V5 formed a significantly higher number of foci than cells transfected with ΔHLH-TF-V5 or mock (Figure 3A-B). These results revealed that TEL-FGFR3 has transforming activity and that the HLH domain of TEL is indispensable for its transforming activity.
TEL-FGFR3 promotes IL-3–independent cell growth of Ba/F3. Inhibitory effect of SU5402 on TEL-FGFR3 transformants. (A) Ba/F3 cells were transfected with TF-V5 (□), ΔHLH-TF-V5 (○), or empty vector (mock; ⋄). Then, 2 × 103 cells were plated (i) in the presence of IL-3 or (ii) in the absence of IL-3. The effects of SU5402 (iii) and AG1296 (iv) on the growth of polyclonal transformants are shown. At daily intervals, 20 μL MTS was added to the cultures and incubated for 1 hour. In (iii), □ indicates 0 μM SU5402; ○,5 μM; ▵,10 μM;  , 25 μM; ⋄, DMSO; and
, 25 μM; ⋄, DMSO; and  , mock 25 μM with IL-3. In (iv), □ indicates 0 μM AG1296; ○, 1 μM; ▵, 5 μM;
, mock 25 μM with IL-3. In (iv), □ indicates 0 μM AG1296; ○, 1 μM; ▵, 5 μM;  , 10 μM;
, 10 μM;  25 μM; ⋄, DMSO; and ⊕, mock 25 μM with IL-3. Each value is the mean of 3 experiments done in triplicate plus the SD. (B) Protein expression and autotyrosine phosphorylation of TEL-FGFR3 in polyclonal transfectants. Ba/F3 cells (2 × 106) were transfected with empty vector (lane 1), ΔHLH-TF-V5 clone (lane 2), TF-V5 clone (lane 3), or TF-V5 clone and incubated in the presence of 0.1% DMSO (lane 4), 5 μM SU5402 (lane 5), 10 μM SU5402 (lane 6), or 25 μM SU5402 (lane 7) for 1 hour or with 5 μM AG1296 (lane 8), 10 μM AG1296 (lane 9), or 25 μM AG1296 (lane 10) for 1 hour. Cells were lysed, immunoprecipitated with anti-V5 antibody, and analyzed by Western blotting using antiphosphotyrosine antibody (4G10) as indicated. The same membrane was reprobed with anti-V5 antibody and showed that nearly equivalent amounts of fusion protein were loaded in each lane.
25 μM; ⋄, DMSO; and ⊕, mock 25 μM with IL-3. Each value is the mean of 3 experiments done in triplicate plus the SD. (B) Protein expression and autotyrosine phosphorylation of TEL-FGFR3 in polyclonal transfectants. Ba/F3 cells (2 × 106) were transfected with empty vector (lane 1), ΔHLH-TF-V5 clone (lane 2), TF-V5 clone (lane 3), or TF-V5 clone and incubated in the presence of 0.1% DMSO (lane 4), 5 μM SU5402 (lane 5), 10 μM SU5402 (lane 6), or 25 μM SU5402 (lane 7) for 1 hour or with 5 μM AG1296 (lane 8), 10 μM AG1296 (lane 9), or 25 μM AG1296 (lane 10) for 1 hour. Cells were lysed, immunoprecipitated with anti-V5 antibody, and analyzed by Western blotting using antiphosphotyrosine antibody (4G10) as indicated. The same membrane was reprobed with anti-V5 antibody and showed that nearly equivalent amounts of fusion protein were loaded in each lane.
TEL-FGFR3 promotes IL-3–independent cell growth of Ba/F3. Inhibitory effect of SU5402 on TEL-FGFR3 transformants. (A) Ba/F3 cells were transfected with TF-V5 (□), ΔHLH-TF-V5 (○), or empty vector (mock; ⋄). Then, 2 × 103 cells were plated (i) in the presence of IL-3 or (ii) in the absence of IL-3. The effects of SU5402 (iii) and AG1296 (iv) on the growth of polyclonal transformants are shown. At daily intervals, 20 μL MTS was added to the cultures and incubated for 1 hour. In (iii), □ indicates 0 μM SU5402; ○,5 μM; ▵,10 μM;  , 25 μM; ⋄, DMSO; and
, 25 μM; ⋄, DMSO; and  , mock 25 μM with IL-3. In (iv), □ indicates 0 μM AG1296; ○, 1 μM; ▵, 5 μM;
, mock 25 μM with IL-3. In (iv), □ indicates 0 μM AG1296; ○, 1 μM; ▵, 5 μM;  , 10 μM;
, 10 μM;  25 μM; ⋄, DMSO; and ⊕, mock 25 μM with IL-3. Each value is the mean of 3 experiments done in triplicate plus the SD. (B) Protein expression and autotyrosine phosphorylation of TEL-FGFR3 in polyclonal transfectants. Ba/F3 cells (2 × 106) were transfected with empty vector (lane 1), ΔHLH-TF-V5 clone (lane 2), TF-V5 clone (lane 3), or TF-V5 clone and incubated in the presence of 0.1% DMSO (lane 4), 5 μM SU5402 (lane 5), 10 μM SU5402 (lane 6), or 25 μM SU5402 (lane 7) for 1 hour or with 5 μM AG1296 (lane 8), 10 μM AG1296 (lane 9), or 25 μM AG1296 (lane 10) for 1 hour. Cells were lysed, immunoprecipitated with anti-V5 antibody, and analyzed by Western blotting using antiphosphotyrosine antibody (4G10) as indicated. The same membrane was reprobed with anti-V5 antibody and showed that nearly equivalent amounts of fusion protein were loaded in each lane.
25 μM; ⋄, DMSO; and ⊕, mock 25 μM with IL-3. Each value is the mean of 3 experiments done in triplicate plus the SD. (B) Protein expression and autotyrosine phosphorylation of TEL-FGFR3 in polyclonal transfectants. Ba/F3 cells (2 × 106) were transfected with empty vector (lane 1), ΔHLH-TF-V5 clone (lane 2), TF-V5 clone (lane 3), or TF-V5 clone and incubated in the presence of 0.1% DMSO (lane 4), 5 μM SU5402 (lane 5), 10 μM SU5402 (lane 6), or 25 μM SU5402 (lane 7) for 1 hour or with 5 μM AG1296 (lane 8), 10 μM AG1296 (lane 9), or 25 μM AG1296 (lane 10) for 1 hour. Cells were lysed, immunoprecipitated with anti-V5 antibody, and analyzed by Western blotting using antiphosphotyrosine antibody (4G10) as indicated. The same membrane was reprobed with anti-V5 antibody and showed that nearly equivalent amounts of fusion protein were loaded in each lane.
Focus-forming activity of TEL-FGFR3 and subcellular localization of TEL-FGFR3 chimeric protein in NIH/3T3 cells. (A) Representative results of the focus-forming assay in NIH/3T3 cells. NIH/3T3 cells were transfected with 30 μg of the indicated constructs and cultured for 2 weeks. Giemsa staining was performed for visualization of foci. (B) Comparison of the number of foci per dish. NIH/3T3 cells were transfected with 20 μg or 30 μg of the indicated DNA constructs. Colonies larger than 1 mm in diameter were counted. The results are expressed as the means plus SD of triplicate plates. Similar results were obtained in 2 independent experiments. (C) Subcellular localization of TEL-FGFR3 protein in NIH/3T3 cells by immunofluorescence staining. NIH/3T3 cells were transfected with (i) TF-V5, (ii) ΔHLH-TF-V5, or (iii) empty vector (mock), and (iv) nontransfected NIH/3T3 cells. Cells were cultured on microscope slides for 48 hours, fixed, and then subjected to permeabilization and indirect staining, as described in “Materials and methods.” DAPI was used as a nuclear counterstain. Expression of V5-associated protein derived from the EF-1 α promoter of TF-V5, ΔHLH-TF-V5, or mock and GFP-zeocin fusion protein derived from the independent cytomegalovirus (CMV) promoter in the same vectors was analyzed with fluorescence microscopy using the red (rhodamine), green (GFP), and blue (DAPI) channels. Images were captured with a CCD camera attached to a Nikon Eclipse E800 microscope with a Nikon Plan Apo 100×/1.40 oil objective lens (Nikon), and cropped and merged in Leica QFISH software (original magnification × 1000).
Focus-forming activity of TEL-FGFR3 and subcellular localization of TEL-FGFR3 chimeric protein in NIH/3T3 cells. (A) Representative results of the focus-forming assay in NIH/3T3 cells. NIH/3T3 cells were transfected with 30 μg of the indicated constructs and cultured for 2 weeks. Giemsa staining was performed for visualization of foci. (B) Comparison of the number of foci per dish. NIH/3T3 cells were transfected with 20 μg or 30 μg of the indicated DNA constructs. Colonies larger than 1 mm in diameter were counted. The results are expressed as the means plus SD of triplicate plates. Similar results were obtained in 2 independent experiments. (C) Subcellular localization of TEL-FGFR3 protein in NIH/3T3 cells by immunofluorescence staining. NIH/3T3 cells were transfected with (i) TF-V5, (ii) ΔHLH-TF-V5, or (iii) empty vector (mock), and (iv) nontransfected NIH/3T3 cells. Cells were cultured on microscope slides for 48 hours, fixed, and then subjected to permeabilization and indirect staining, as described in “Materials and methods.” DAPI was used as a nuclear counterstain. Expression of V5-associated protein derived from the EF-1 α promoter of TF-V5, ΔHLH-TF-V5, or mock and GFP-zeocin fusion protein derived from the independent cytomegalovirus (CMV) promoter in the same vectors was analyzed with fluorescence microscopy using the red (rhodamine), green (GFP), and blue (DAPI) channels. Images were captured with a CCD camera attached to a Nikon Eclipse E800 microscope with a Nikon Plan Apo 100×/1.40 oil objective lens (Nikon), and cropped and merged in Leica QFISH software (original magnification × 1000).
Autotyrosine phosphorylation of TEL-FGFR3 chimeric proteins
We hypothesized that homo-oligomerization by the HLH domain of TEL-FGFR3 might induce constitutive activation of TK through autotyrosine phosphorylation of FGFR3. To explore this possibility, the chimeric proteins derived from the TF-V5– or the ΔHLH-TF-V5–transfected cells were immunoprecipitated by anti-V5 antibody and analyzed by Western blotting with antiphosphotyrosine antibody. As a negative control, Ba/F3 cells transfected with the empty vector (mock) were also analyzed. On probing the blot with anti-V5 antibody, there were nearly equivalent amounts of proteins in the TF-V5–transfected Ba/F3 cells (68.6 kDa) and ΔHLH-TF-V5–transfected Ba/F3 cells (55.0 kDa). Reprobing by antiphosphotyrosine antibody showed that there was a high level of tyrosine-phosphorylated protein (68.6 kDa) in the TF-V5–transfected Ba/F3 cells. However, tyrosine-phosphorylated protein was not detected in the Mock nor in the ΔHLH-TF-V5–transfected Ba/F3 cells (Figure 2B). These results suggest that the expression of TEL-FGFR3 resulted in tyrosine phosphorylation of FGFR3 through oligomerization by the HLH domain of TEL, which induced constitutive TK activity and resulted in IL-3–independent growth of Ba/F3 cells. However, there remains a possibility that lack of autotyrosine phosphorylation in ΔHLH-TF-V5 transfectants may be due to poor folding of the chimeric protein.
SU5402 is a specific growth inhibitor of TEL-FGFR3–transfected cells
To identify a TK inhibitor that has an antiproliferative effect on TEL-FGFR3–transfected cells, we investigated the effect of SU5402 and AG1296, which have previously been reported to be FGFR1- and FGFR2-specific TK inhibitors, respectively.24,25 In the presence of SU5402, the growth of TF-V5 transfectants was inhibited in a dose-dependent manner (concentration that inhibits 50% [IC50] = 5 μM). In contrast, the growth of Ba/F3 cells transfected with an empty vector (mock) was not inhibited even at a high concentration of SU5402 (10-25 μM) in the presence of IL-3 (Figure 2Aiii). AG1296 inhibited the growth of TEL-FGFR3 transformants only at a high concentration (25 μM) and not in a dose-dependent manner, suggesting that AG1296 is not a specific TK inhibitor for FGFR3 (Figure 2Aiv). Protein analysis revealed that tyrosine phosphorylation of TEL-FGFR3 was inhibited by SU5402 in a dose-dependent manner (Figure 2B), and this effect appeared rapidly within 5 minutes (data not shown). AG1296 weakly inhibited the phosphorylation of tyrosine residues in TEL-FGFR3 (Figure 2B). Taken together, these data suggest that SU5402 is a specific inhibitor of TEL-FGFR3.
Cytoplasmic localization of TEL-FGFR3 chimeric proteins
Immunofluorescence staining was performed with the antibody specific for the V5 epitope to verify the localization of the TEL-FGFR3 chimeric proteins in NIH/3T3 cells. Both chimeric proteins TF-V5 and ΔHLH-TF-V5 were located exclusively in the cytoplasm of cells transfected with the respective DNA construct (Figures 3Ci-ii). Slight or no expression of protein associated with V5 was observed in mock-transfected cells and in nontransfected NIH/3T3 cells (as negative control), respectively (Figures 3Ciii-iv).
TEL-FGFR3 activates various signaling pathways and PI3-K inhibitors suppressed the growth of TEL-FGFR3 transfectants
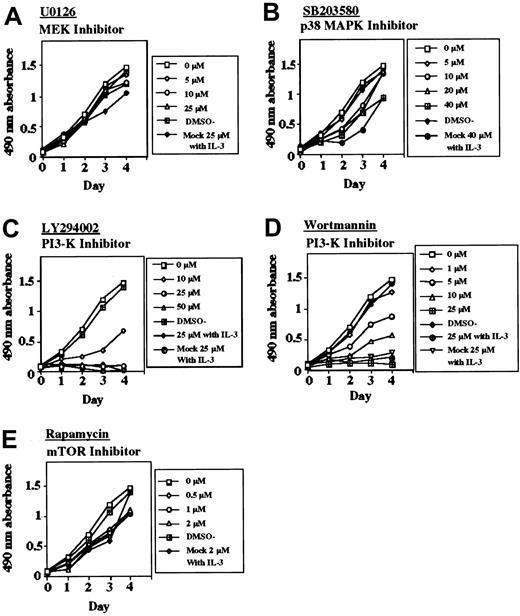
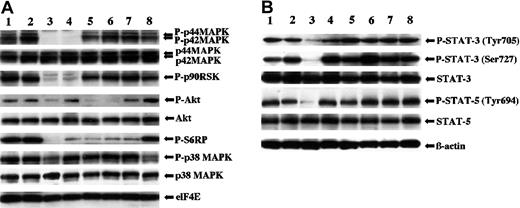
It has been shown that activation of FGFRs results in the stimulation of multiple signaling pathways including MAPKs, STATs, and PI3-K/Akt, which could confer the transforming activity.13,26,27 To determine which signaling pathways might be required for the transformation, we performed multiplex Western blot analysis of the lysates of TF-V5 transfectants. The levels of the phosphorylated forms of p42/p44 MAPK, Akt, p38 MAPK, STAT-3, and STAT-5 were higher in the TF-V5–transfected Ba/F3 cells than in the ΔHLH-TF-V5–transfected Ba/F3 cells and mock-transfected cells (Figure 4A). SU5402 reduced the levels of the phosphorylated forms of these proteins in a dose-dependent manner (Figure 4A). As shown in Figure 4, the levels of activation of p42/p44 MAPK, Akt, and STAT-5 were similar between TF-V5 transfectants and K562 cells (for positive control). Next, to clarify the major signal transduction pathways for the transformation, we investigated the antiproliferative effect of various inhibitors of downstream signaling (Figure 5). To test if the classical MAPK pathway is required for the transformation, we used the MEK inhibitor, U0126. This compound at 25 μM inhibited p42/p44 MAPK phosphorylation and its downstream p90RSK phosphorylation (Figure 6A). However, U0126 had no effect on the proliferation of the TEL-FGFR3 transformants (Figure 5A). U0126 partially inhibited S6RP phosphorylation, although U0126 did not affect Akt phosphorylation. These results suggested the presence of cross-talk between MAPK and Akt for S6RP activation, which was previously reported by Iijima et al.28 In contrast, both PI3-K inhibitors, LY294002 and wortmannin, completely suppressed the proliferation of TEL-FGFR3 transformants at 25 μM (Figure 5C-D), these concentrations of PI3-K inhibitors that inhibit the phosphorylation of Akt and its downstream pS6RP (Figure 6A). IL-3 could not rescue the growth suppression of TEL-FGFR3–transfected cells induced by these PI3-K inhibitors at 25 μM (Figure 5C-D). Rapamycin is an inhibitor of mammalian target of rapamycin (mTOR), which is one of the downstream targets of Akt. Rapamycin did not affect the proliferation of TEL-FGFR3 transformants, even though it completely blocked S6RP phosphorylation (Figures 5E and 6A). These results suggest that PI3-K activates some unknown targets, rather than mTOR, for TEL-FGFR3 transformation. It has been shown that p38 MAPK is implicated in the differentiation and proliferation of hematopoietic cells.15,29,30 We demonstrated the expression and phosphorylation of p38 MAPK in the TEL-FGFR3 transformants. In the TEL-FGFR3 transformants, there was a high level of phosphorylated p38 MAPK, and the phosphorylation of p38 MAPK was inhibited by SU5402 (Figure 4A). However, SB203580, an inhibitor of p38 MAPK, did not affect the proliferation of TEL-FGFR3 transformants, even though it completely inhibited p38 MAPK activity (Figures 4A and 5B). Therefore, p38 MAPK did not play a major role in TEL-FGFR3 transformation. Finally, the levels of phosphorylated STATs were investigated. There were high levels of expression of STAT-3 and STAT-5 in TEL-FGFR3 transformants compared with those of STAT-3 and STAT-5 in the IL-3–driven mock-transfected cells (Figure 4A). SU5402, but not U0126, LY294002, nor SB203580, reduced the levels of the phosphorylated forms of STATs in the transformants. These results showed that the activation of STAT-3 and STAT-5 by TEL-FGFR3 was not mediated through the MAPK, PI3-K, and p38 MAPK pathways.
Effects of various inhibitors on the cell growth of TEL-FGFR3 transfectants. The responses of Ba/F3 cells that were transfected with TF-V5 or mock to increasing concentrations of (A) U0126, (B) SB203580, (C) LY294002 in the presence or absence of IL-3, (D) wortmannin in the presence or absence of IL-3, or (E) rapamycin are shown. Similar results were obtained in several experiments.
Effects of various inhibitors on the cell growth of TEL-FGFR3 transfectants. The responses of Ba/F3 cells that were transfected with TF-V5 or mock to increasing concentrations of (A) U0126, (B) SB203580, (C) LY294002 in the presence or absence of IL-3, (D) wortmannin in the presence or absence of IL-3, or (E) rapamycin are shown. Similar results were obtained in several experiments.
Effects of various inhibitors on the level of activation of downstream effector proteins in TEL-FGFR3 transfectants. (A) Western blotting using multiple antibodies for the phosphorylated forms of classical MAPK, p90 RSK, Akt, S6RP, and p38 MAPK in polyclonal TF-V5 transfectants (lane 1) and in cells transfected with TF-V5 that were incubated in the presence of 0.1% DMSO (lane 2), 25 μM SU5402 (lane 3), 25 μM U0126 (lane 4), 25 μM LY294 002 (lane 5), 25 μM wortmannin (lane 6), 2 μM rapamycin (lane 7), or 40 μM SB203580 (lane 8) for 6 hours. The eukaryotic initiation factor 4E (elF4E) was used for internal control. (B) Western blot analysis for STATs in polyclonal TF-V5 transfectants that were incubated in the absence of inhibitors (lane 1), and in cells transfected with TF-V5 that were incubated in the presence of 0.1% DMSO (lane 2), 25 μM SU5402 (lane 3), 25 μM U0126 (lane 4), 25 μM LY294002 (lane 5), 25 μM wortmannin (lane 6), 2 μM rapamycin (lane 7), or 10 μM SB203580 (lane 8). β-Actin was used for internal control.
Effects of various inhibitors on the level of activation of downstream effector proteins in TEL-FGFR3 transfectants. (A) Western blotting using multiple antibodies for the phosphorylated forms of classical MAPK, p90 RSK, Akt, S6RP, and p38 MAPK in polyclonal TF-V5 transfectants (lane 1) and in cells transfected with TF-V5 that were incubated in the presence of 0.1% DMSO (lane 2), 25 μM SU5402 (lane 3), 25 μM U0126 (lane 4), 25 μM LY294 002 (lane 5), 25 μM wortmannin (lane 6), 2 μM rapamycin (lane 7), or 40 μM SB203580 (lane 8) for 6 hours. The eukaryotic initiation factor 4E (elF4E) was used for internal control. (B) Western blot analysis for STATs in polyclonal TF-V5 transfectants that were incubated in the absence of inhibitors (lane 1), and in cells transfected with TF-V5 that were incubated in the presence of 0.1% DMSO (lane 2), 25 μM SU5402 (lane 3), 25 μM U0126 (lane 4), 25 μM LY294002 (lane 5), 25 μM wortmannin (lane 6), 2 μM rapamycin (lane 7), or 10 μM SB203580 (lane 8). β-Actin was used for internal control.
Taken together, the leukemogenic potential of TEL-FGFR3 may be mediated at least in part through PI3-K (Figure 4B).
TEL-FGFR3–transfected Ba/F3 cells induce leukemia in mice
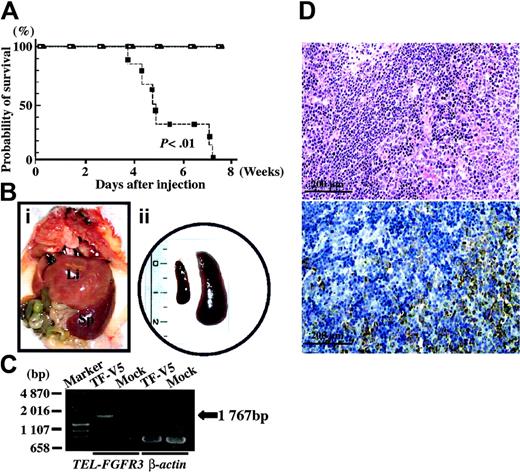
To investigate whether TEL-FGFR3 induces leukemia in syngeneic mice, TF-V5–, ΔHLH-TF-V5–, or mock-transfected Ba/F3 cells were intravenously injected into BALB/c mice. All 6 mice that had been given TF-V5–transfected cells died within 8 weeks after injection (median survival, 37.0 days). All mice that had received ΔHLH-TF-V5–transfected cells or mock-transfected cells remained free of disease at 8 weeks after injection (Figure 7A). All 6 mice that had received TF-V5–transfected cells developed a disease resembling leukemia with enlargement of the spleen (Figure 7B). RT-PCR analysis showed expression of TEL-FGFR3 in the spleen of the leukemic mice (Figure 7C). Histopathologic examination of the spleen of leukemic mice revealed infiltration of blastlike cells (Figure 7D).
In vivo leukemogenic potential of TEL-FGFR3–transfected Ba/F3 cells. (A) Kaplan-Meier survival curves of BALB/c mice that had been injected with clonal populations of TF-V5– (▪), mock- (□), or ΔHLH-TF-V5–transfected Ba/F3 cells (▴). (Bi) Macroscopic view of the abdominal organs of a mouse that had been injected with TF-V5–transfected Ba/F3 cells. The terminally sick mouse was humanely killed 8 weeks after injection; Li indicates liver; Sp, spleen. (ii) Whole spleen specimens isolated from a mouse that had been injected with TF-V5–transfected Ba/F3 cells (right) and a mouse that had been injected with mock-transfected Ba/F3 cells (left). The mice were humanely killed 8 weeks after injection. Scale is in centimeters. (C) RT-PCR for TEL-FGFR3 using RNA that had been extracted from the spleens of mice injected with TF-V5–transfected Ba/F3 cells or mock-transfected Ba/F3 cells. RT-PCR for β-actin was performed as a positive control. (D) Histopathology of the spleen of a mouse that had been injected with TF-V5–transfected Ba/F3 cells. The same paraffin sections are shown. Photographs are taken on a light microscope (BHT-321; Olympus, Tokyo, Japan) with an Olympus NC SPlan 100×/1.40 dry objective lens (Olympus). (i) Hematoxylin and eosin staining. Infiltration of a massive number of leukemia cells is observed. (ii) Staining with anti-V5 antibody. Infiltration of leukemia cells is observed. Normal residual lesions in the spleen were not stained. Similar results were obtained in 3 experiments using 3 independent monoclonal transfectants.
In vivo leukemogenic potential of TEL-FGFR3–transfected Ba/F3 cells. (A) Kaplan-Meier survival curves of BALB/c mice that had been injected with clonal populations of TF-V5– (▪), mock- (□), or ΔHLH-TF-V5–transfected Ba/F3 cells (▴). (Bi) Macroscopic view of the abdominal organs of a mouse that had been injected with TF-V5–transfected Ba/F3 cells. The terminally sick mouse was humanely killed 8 weeks after injection; Li indicates liver; Sp, spleen. (ii) Whole spleen specimens isolated from a mouse that had been injected with TF-V5–transfected Ba/F3 cells (right) and a mouse that had been injected with mock-transfected Ba/F3 cells (left). The mice were humanely killed 8 weeks after injection. Scale is in centimeters. (C) RT-PCR for TEL-FGFR3 using RNA that had been extracted from the spleens of mice injected with TF-V5–transfected Ba/F3 cells or mock-transfected Ba/F3 cells. RT-PCR for β-actin was performed as a positive control. (D) Histopathology of the spleen of a mouse that had been injected with TF-V5–transfected Ba/F3 cells. The same paraffin sections are shown. Photographs are taken on a light microscope (BHT-321; Olympus, Tokyo, Japan) with an Olympus NC SPlan 100×/1.40 dry objective lens (Olympus). (i) Hematoxylin and eosin staining. Infiltration of a massive number of leukemia cells is observed. (ii) Staining with anti-V5 antibody. Infiltration of leukemia cells is observed. Normal residual lesions in the spleen were not stained. Similar results were obtained in 3 experiments using 3 independent monoclonal transfectants.
Discussion
In this report, we describe data on a patient who presented with PTCL that subsequently progressed to AML (M0), both of which were associated with TEL-FGFR3 fusion transcripts that had resulted from t(4;12)(p16;p13). On sequence analysis, we found that all TEL-FGFR3 fusion transcripts had the same fusion breakpoint, suggesting that both the PTCL and AML had originated from the same clone. In our previous report, although we could not perform fluorescence in situ hybridization (FISH) and sequence analysis on her lymph node specimen obtained at the time of PTCL, immunohistologic studies revealed that her lymphoma cells strongly expressed FGFR3, even though normal lymph node tissue does not express FGFR3.15 It may be considered that her PTCL cells had ectopically expressed TEL-FGFR3. Therefore, we had suspected that the t(4;12)(p16;p13) in her BM cells was derived from involvement of her BM in PTCL. However, the t(4;12)(p16;p13) remained in her BM cells after PTCL cells disappeared from her BM by chemotherapy. It seems more likely that a neoplastic multipotent stem cell had the t(4;12)(p16;p13) and then the lineage switch from PTCL to AML occurred during the course of her disease.
These clinical and pathologic features are similar to those of the 8p11 syndrome, which is characterized by eosinophilia and a high incidence of T-cell non-Hodgkin lymphoma that progresses to MPD/AML (M4 or M5).16-18 A recent study revealed that FGFR1 on 8p11 is a responsible gene of t(8;13)(p11;q12), t(6;8)(q27;p12), and t(8;9)(p12;q33), which result in expression of ZNF198-FGFR1, FOP-FGFR1, and CEP110-FGFR1, respectively.9-14 In this patient, eosinophilia was not recognized and the T-cell lymphoma progressed to AML (M0) without a T-cell phenotype. Because the complication of T-cell malignancy is extremely rare in MPD, it is interesting that the FGFR family might contribute to the development of T-cell malignancy.11,12,19,31 On the other hand, T-cell malignancy has not been reported among patients with BCR-FGFR1 and it rarely occurs in patients with BCR-ABL to our knowledge.15 TEL-ABL and TEL-JAK2 have been reported to induce T-cell acute lymphoblastic leukemia (T-ALL) in humans and in transgenic mice.32-34 The partner gene of FGFRs, that is, TEL, ZNF198, FOP, or CEP110, might play an important role in the development of T-cell malignancy. To elucidate whether TEL-FGFR3 actually elicits these disease phenotype, we will have to study the transformation potential of the TEL-FGFR3 at the hematopoietic stem cell level as described by Lavau et al.35
Our in vitro and in vivo studies showed that there was a good association between constitutive activation of FGFR3 and the development of leukemia. FGFR3 has been reported to contribute to tumorigenesis in a large proportion of bladder and cervical carcinoma cases with variable active mutations that cause constitutive TK activity.36 Chesi et al37 reported that FGFR3 has oncogenic potential when overexpressed in t(4;14) multiple myeloma, and subsequent activating mutations in FGFR3 led to constitutive activation of FGFR3 TK during tumor progression. In our study, sequence analysis of the TF-V5 expression vector showed no mutations although there was a trinucleotide (CAG) insertion in the juxtamembrane region of FGFR3. This region is of interest because it corresponds to the FRS2-binding region in FGFR1.38 FRS2, which is known as a docking protein of FGFRs, plays a major role in mediating signals from the activated receptors to the downstream pathways.39 In our preliminary analysis, Ba/F3 cells transfected with TEL-FGFR3 without the CAG insertion did not lose factor-independent cell growth (data not shown). Therefore, in our model, the constitutive activation of FGFR3 was caused by the phosphorylation of tyrosine residues through oligomerization of the TEL HLH domain. A similar mechanism of activation of FGFR1 was reported in various FGFR1 fusion proteins including ZNF198-FGFR1, FOP-FGFR1, CEP110-FGFR1, and BCR-FGFR1.8-15 FGFRs have been reported to have different capacities to promote FGF-dependent proliferation when expressed in Ba/F3 cells.40 Whereas FGFR1 elicits a strong proliferative response, FGFR3 elicits a weaker response.39,41 The intracellular region of FGFRs contains a bipartite TK domain and a kinase insert sequence, the latter of which is responsible for signal transduction.42 Wang and Goldfarb43 reported that the important factor that distinguishes the mitogenic potentials of the FGFRs in Ba/F3 cells is the number of tyrosine residues within the kinase insert region. We demonstrated that TEL-FGFR3 confers similar transforming activity to Ba/F3 cells and the same IC50 value of SU5402 for cell proliferation (5 μM), compared with the results on BCR-FGFR1 and ZNF198-FGFR1 reported previously.15 This indicates that FGFR3 and FGFR1 have the same transforming abilities when their TK is constitutively activated.
Our experiments on the downstream signaling cascades show that TEL-FGFR3 activates various effector proteins including MAPK, PI3-K, mTOR, STAT-3, STAT-5, and p38 MAPK. The results of selective inhibitor analysis indicated that PI3-K, but not MAPK or p38 MAPK, is critical for the transformation of Ba/F3 cells by TEL-FGFR3. The PI3-K inhibitors, LY294002 and wortmannin, dramatically inhibited the proliferation of TEL-FGFR3 transformants, whereas the mTOR inhibitor had little effect. Therefore, the downstream signal cascade of PI3-K/Akt, except for mTOR, might participate in its transforming property. The results described are consistent with those of previously reported studies on BCR-FGFR1 and ZNF198-FGFR1.15 Nevertheless, inhibition of p38 MAPK with various concentrations of SB203580 had little effect on the transformants in our study. Demiroglu et al15 reported that 40 μM SB203580 completely suppressed the proliferation of both BCR-FGFR1–and ZNF198-FGFR1–transformed Ba/F3 cells. However, they did not show the dose-dependent effects of SB203580. We suspected that this discrepancy might be due to differences in the interpretation of results. Hunt et al44 suggested that SB203580 at higher concentrations affects not only p38 MAPK but also other unknown targets. It should be considered that no definite conclusions can be drawn as to the involvement of p38 MAPK, unless the biologic effects of SB203580 can be seen at a dose of less than 0.5 μM. Our data indicate that p38 MAPK does not play an important role in the proliferation of TEL-FGFR3 transformants. Recently, Wong et al45 reported that the expression of BCR-ABL in embryonic stem cells contributes to the expansion of hematopoietic progenitor cells having BCR-ABL through down-regulation of p38 MAPK. As TEL-FGFR3 prominently activated p38 MAPK, the difference in the degree of activation of p38 MAPK might contribute to differences in disease phenotype including T-cell malignancy.
The activation of STATs by cytokine/growth factors and nonreceptor TK has been shown to play diverse roles in cell proliferation, differentiation, and apoptosis.46,47 Transforming studies using a dominant-positive form of STAT showed that constitutive activation of STAT-3 contributes to cellular transformation.48-50 We found phosphorylation of STAT-3 and STAT-5 in TEL-FGFR3 transformants, in which the level of phosphorylation of STAT-3 was approximately 10 times higher than that in mock-transfected cells. These results suggest that STAT-3 and STAT-5 might play important roles in the cellular transformation by TEL-FGFR3. The phosphorylation of STAT-3 and STAT-5 was completely inhibited by SU5402 but not by U0126, LY294002, or SB203580. These results suggested that STAT-3 and STAT-5 are directly activated by TEL-FGFR3 and not through the MAPK or PI3-K pathway. It should be noted that when TF-V5–transfected cells were treated with 25 μM SU5402, their proliferation was completely inhibited, although Akt was weakly phosphorylated at nearly the same level as that in mock-transfected cells. Therefore, SU5402 may have inhibited the proliferation of TF-V5–transfected cells by inactivating other signaling pathways including STAT-3 and STAT-5. Future studies will address the role of STATs and the PI3-K pathway in the TEL-FGFR3–transforming property.
In conclusion, a hematopoietic malignancy associated with TEL-FGFR3 is considered to be a stem cell disorder that can progress to both T-cell lymphoma and AML. TEL-FGFR3 transformed NIH/3T3 and Ba/F3 cells in vitro and TEL-FGFR3–transfected Ba/F3 cells induced leukemia in vivo. This transforming property is mediated through constitutive FGFR3 TK activity, which activates a variety of signaling pathways. Activation of the PI3-K pathway is required for optimum transformation. SU5402 is a specific inhibitor of TEL-FGFR3 and this presents a possible therapeutic modality for the treatment of FGFR3-associated malignancies.
Prepublished online as Blood First Edition Paper, October 28, 2004; DOI 10.1182/blood-2003-12-4290.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Dr Masahiro Kaneshige for valuable comments and support of the deletion mutants design. We thank Yumiko Uchida and Kanae Sakate for technical assistance and Dr Itsuro Jinnai, Dr Akiko Yamamoto, and Dr Toshimitsu Matsui (Kobe University School of Medicine, Japan) for helpful discussions.


![Figure 1. TEL-FGFR3 transcripts and vector constructs. (A) RT-PCR analysis for detection of TEL-FGFR3 transcripts. Nested RT-PCR was performed using 1 μg RNA from a series of samples of bone marrow (BM) and peripheral blood (PB) obtained from the patient with t(4;12)(p16;p13). The final product was analyzed by electrophoresis on a 1.5% agarose gel in Tris (tris(hydroxymethyl)aminomethane)–borate/EDTA (TBE) buffer, soaked in ethidium bromide, and visualized under UV light. The black arrow indicates the single PCR product (1767 bp). The PCR products from all samples had the same TEL-FGFR3 fusion breakpoint by sequence analysis. The materials (BM or PB) and dates on which the materials were obtained are as follows: BM cells on December 5, 2000 (2 months after the patient was diagnosed with PTCL; lane 1), BM cells on September 12, 2001 (when the patient was diagnosed with AML; lane 2), PB on September 14, 2001 (lane 3), PB on January 8, 2003 (lane 4), and BM cells of a healthy donor as a negative control (lane C). M indicates the molecular size markers. The numbers on the left of the figure indicate the molecular size (in base pair [bp]). (B) Schematic representation of proteins encoded by vector constructs of TEL-FGFR3 and TEL-FGFR3 lacking the HLH domain. (i) Full-length TEL protein of 452 amino acids (aa). The positions of the HLH domain and E26 transformation-specific (ETS) DNA-binding domain are indicated by dark shaded boxes. (ii) Full-length FGFR3 protein of 806 amino acids, which contains 3 extracellular immunoglobulin-like domains, a transmembrane domain, and TK 1 and 2 subdomains interrupted by a kinase insert. (iii) Full-length TEL-FGFR3 fusion protein of 589 amino acids, with a V5 epitope tag (TF-V5). This translocation fused nucleotide (nt) 543 of TEL to nt 1270 of FGFR3 as described previously.16 Closed arrow indicates the t(4;12) fusion breakpoint. Open arrow indicates the CAG trinucleotides insert that was consistently found in the fusion transcripts in all samples from the patient. (iv) ΔHLH-TF-V5, with an in-frame deletion of 76 amino acids (Δ amino acids 39-114 of TF-V5) and which lacks most of the TEL-HLH domain. HLH indicates helix-loop-helix domain; ETS, ETS DNA-binding domain; Ig, immunoglobulin-like domain; TM, transmembrane domain; TK, tyrosine kinase; KI, kinase insert; V5, V5 epitope.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/5/10.1182_blood-2003-12-4290/6/m_zh80050574920001.jpeg?Expires=1765899747&Signature=fTywh91vuQEGPB9BRp64kdRGGInIjMljrmYc40uh0AQiJiGTrdkA2TBQ9JBdIAFMNvVkiVifp3yVGahtjwSf3ZFCLiMy0DoNXH7H-SVjCx8kXy8-CQfdF9g5gjnn9jvY~tfpTe111KSfJdvdeQzM53g1jEFCkCZM7DERkBbbzRelYMiGqriC06~6z4e2nyVbUCRJVzD4F4YPbAqaKpgyekmH2UoWmMPGbIDwplLqs~4azifkGigIhiKlFyiwmeNZQUQStMpeCCruDSb2T56StTq-zgprjkbmrn9fxDL87HcvvkEM3w5eXsyQ536KeIKiZgr1utuiaS35UFOUkomtIw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal