Abstract
Fanconi anemia (FA) is a genetic disease characterized by bone marrow failure and cancer predisposition. Here we have identified Spanish Gypsies as the ethnic group with the world's highest prevalence of FA (carrier frequency of 1/64-1/70). DNA sequencing of the FANCA gene in 8 unrelated Spanish Gypsy FA families after retroviral subtyping revealed a homozygous FANCA mutation (295C>T) leading to FANCA truncation and FA pathway disruption. This mutation appeared specific for Spanish Gypsies as it is not found in other Gypsy patients with FA from Hungary, Germany, Slovakia, and Ireland. Haplotype analysis showed that Spanish Gypsy patients all share the same haplotype. Our data thus suggest that the high incidence of FA among Spanish Gypsies is due to an ancestral founder mutation in FANCA that originated in Spain less than 600 years ago. The high carrier frequency makes the Spanish Gypsies a population model to study FA heterozygote mutations in cancer.
Introduction
Fanconi anemia (FA) is a rare genetic syndrome characterized by various congenital abnormalities, bone marrow failure, and cancer predisposition.1 Recent results of somatic cell hybridization, protein association, and other functional complementation studies indicate that there are at least 11 complementation groups in FA (A, B, C, D1, D2, E, F, G, I, J, and L), each connected with a distinct disease gene.2,3 The products of the genes FANCA, C, E, F, G, L, and possibly I interact in a nuclear core complex required for FANCD2 activation by monoubiquitination at residue K561, a key posttranslational modification in the FA pathway.4 Active FANCD2 then functions downstream to maintain the genome integrity by mechanisms that are still not well understood.5-7
FA is a rare autosomal recessive disease with an overall prevalence of 1 to 5 per million and an estimated carrier frequency of 1 in 200 to 1 in 300 in most populations.8 This is also true in Spain, which has about 40 million inhabitants and 122 patients with FA registered in the Spanish FA Research Network (SFARN) register. Some ethnic groups have a higher prevalence of FA due to founder effects and isolation. Two previously reported examples are the Afrikaner population of South Africa and the Ashkenazi Jewish population with a carrier frequency of 1 in 77 and 1 in 90, respectively.9 Some geographic clusters also exist in Italy with a similar high prevalence.10,11 Here we report an even higher prevalence of FA in Spanish Gypsies. A total of 31 Gypsy patients with FA are currently registered in the SFARN register, indicating that one quarter of all Spanish patients with FA belong to this ethnic group. As there are approximately 500 000 to 600 000 Gypsies in Spain,12 the estimated frequency of carriers is 1 in 64 to 1 in 70, the highest ever reported in the world. Here we report the genetic characterization of FA in this ethnic group.
Study design
In order to genetically characterize FA in the Spanish Gypsy ethnic group, blood samples were obtained from patients with FA and from carriers, all previously diagnosed on the basis of their clinical features and chromosome hypersensitivity to diepoxybutane (DEB), essentially as described elsewhere.13-15 This study was duly approved by the Universitat Autònoma de Barcelona Ethical Committee for Human Research and informed consent was obtained according to the Declaration of Helsinki.
Results and discussion
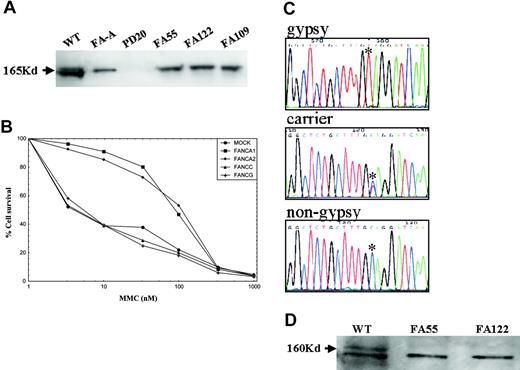
Patients were characterized by FANCA and FANCD2 immunoblotting, retroviral subtyping, FANCA gene sequencing, and haplotype analysis. The affected families were not clustered in any specific geographic region but scattered throughout Spain. All of the Spanish Gypsy patients studied by FANCD2 immunoblotting lacked monoubiquitinated FANCD2 (Figure 1A), indicating that the genetic defect was upstream of FANCD2 in the FA core complex. Genetic subtyping by retrovirus-mediated gene transfer showed that all the Spanish Gypsy patients examined belong to the complementation group FA-A (Figure 1B). Quantitative fluorescence multiplex polymerase chain reaction (PCR) analysis17,18 indicated that no large deletions involving the most frequently deleted FANCA exons (5, 11, 17, 21, and/or 31) were present in Spanish Gypsy patients (data not shown) although this type of mutation has been found in a quarter of the Spanish non-Gypsy patients with FA-A previously analyzed.18 DNA sequencing of FANCA uncovered a novel homozygous truncating mutation in exon 4 (295C>T; Q99X) in 8 unrelated Spanish Gypsy patients with FA (Figure 1C; Table 1). One of these Spanish Gypsy families was of Portuguese origin (Table 1). The 295C>T mutation was not found in a Spanish Gypsy patient of Romanian descent, in Spanish non-Gypsy patients with FA-A, or in healthy individuals, and all FA obligate carriers analyzed were heterozygous for this mutation (Figure 1C, Table 1, and data not shown). The truncating nature of the mutation was confirmed by FANCA immunoblotting with antibodies detecting the C-terminus of FANCA (Figure 1D). While approximately 20% of all Spanish patients with FA show somatic mosaicism (data not shown), none of the Gypsy patients were mosaic, suggesting that the pathogenic mutation is difficult to revert by a mutation in cis, as expected for a base substitution.
Genetic characterization of FA in Spanish Gypsy patients. (A) FANCD2 immunoblotting revealed 2 isoforms in a wild-type (WT) cell, no signal in a FANCD2-deficient cell line (PD20), and a nonubiquitinated isoform in an FA-A cell line or in cell lines derived from Spanish Gypsy patients nos. 55, 122, and 109, indicating that the genetic defect is upstream of FANCD2 monoubiquitination. Immunoblotting experiments were performed following a standard Western blot method with a commercially available antibody against FANCD2 (Santa Cruz Biotech, Santa Cruz, CA). (B) Subtyping by retrovirus-mediated cDNA gene transduction in a cell line derived from a Spanish Gypsy patient with FA. The complementation group was determined by retroviral transduction of cDNA of FA genes essentially as previously reported.16 Briefly, peripheral blood T cells were proliferated in the presence of monoclonal antibodies anti-CD3 and anti-CD28. Afterward, cells were transduced with retroviral vectors containing FANCA (2 vectors), FANCC, FANG cDNA, and control vector (MOCK) and then evaluated for MMC hypersensitivity to detect phenotypic cellular correction. Only retrovirus-encoding wild-type FANCA cDNA led to reversion of the cellular hypersensitivity to mitomycin C (MMC), indicating that the patient belonged to complementation group FA-A. (C) Spanish Gypsy patients with FA with a 295C>T base change, as detected by DNA sequencing, leading to a stop codon (Q99X). All obligate carriers were heterozygote for this mutation. (D) FANCA immunoblotting with an affinity-purified antibody against the C-terminus of the FANCA protein in 2 Spanish Gypsy patients (no. 55 and no. 122) with FA, and a wild-type cell line. FANCA signal (upper band) is not present in the Gypsy patients, consistent with the truncating nature of the Spanish Gypsy mutation. The lower band is an unspecific signal that serves as an internal control and is equally present in all cell extracts.
Genetic characterization of FA in Spanish Gypsy patients. (A) FANCD2 immunoblotting revealed 2 isoforms in a wild-type (WT) cell, no signal in a FANCD2-deficient cell line (PD20), and a nonubiquitinated isoform in an FA-A cell line or in cell lines derived from Spanish Gypsy patients nos. 55, 122, and 109, indicating that the genetic defect is upstream of FANCD2 monoubiquitination. Immunoblotting experiments were performed following a standard Western blot method with a commercially available antibody against FANCD2 (Santa Cruz Biotech, Santa Cruz, CA). (B) Subtyping by retrovirus-mediated cDNA gene transduction in a cell line derived from a Spanish Gypsy patient with FA. The complementation group was determined by retroviral transduction of cDNA of FA genes essentially as previously reported.16 Briefly, peripheral blood T cells were proliferated in the presence of monoclonal antibodies anti-CD3 and anti-CD28. Afterward, cells were transduced with retroviral vectors containing FANCA (2 vectors), FANCC, FANG cDNA, and control vector (MOCK) and then evaluated for MMC hypersensitivity to detect phenotypic cellular correction. Only retrovirus-encoding wild-type FANCA cDNA led to reversion of the cellular hypersensitivity to mitomycin C (MMC), indicating that the patient belonged to complementation group FA-A. (C) Spanish Gypsy patients with FA with a 295C>T base change, as detected by DNA sequencing, leading to a stop codon (Q99X). All obligate carriers were heterozygote for this mutation. (D) FANCA immunoblotting with an affinity-purified antibody against the C-terminus of the FANCA protein in 2 Spanish Gypsy patients (no. 55 and no. 122) with FA, and a wild-type cell line. FANCA signal (upper band) is not present in the Gypsy patients, consistent with the truncating nature of the Spanish Gypsy mutation. The lower band is an unspecific signal that serves as an internal control and is equally present in all cell extracts.
Mutational and haplotype analysis of Gypsy patients with Fanconi anemia
Patient . | Residence . | Origin . | Tribal group* . | Mutational analysis . | Haplotype† . |
|---|---|---|---|---|---|
| FA55 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA87 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA103 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA108 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA109 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA113 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA122 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA123 | Spain | Portugal | Cales | Homozygote 295C > T | 1/1 |
| FA163 | Spain | Romania | Roma | ND/ND‡ | 2/2 |
| VU483 | Holland | Hungary | Roma | 1263delF/ND | 2/3 |
| VANA | Slovakia | Romania | Roma | Homozygote IVS11 + 2 del TAGG | 4/4 |
| KZFE | Germany | Germany | Sinti | Homozygote 1267C > T | 5/5 |
| EJM-737 | Ireland | Ireland | Irish Travellers | Homozygote ex11-14 deletion | 6/6 |
| JM-512 | Ireland | Ireland | Irish Travellers | Homozygote ex11-14 deletion | 6/6 |
| KMM-336§ | Ireland | Ireland | Irish Travellers | Homozygote ex11-14 deletion | 6/6 |
Patient . | Residence . | Origin . | Tribal group* . | Mutational analysis . | Haplotype† . |
|---|---|---|---|---|---|
| FA55 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA87 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA103 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA108 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA109 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA113 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA122 | Spain | Spain | Cales | Homozygote 295C > T | 1/1 |
| FA123 | Spain | Portugal | Cales | Homozygote 295C > T | 1/1 |
| FA163 | Spain | Romania | Roma | ND/ND‡ | 2/2 |
| VU483 | Holland | Hungary | Roma | 1263delF/ND | 2/3 |
| VANA | Slovakia | Romania | Roma | Homozygote IVS11 + 2 del TAGG | 4/4 |
| KZFE | Germany | Germany | Sinti | Homozygote 1267C > T | 5/5 |
| EJM-737 | Ireland | Ireland | Irish Travellers | Homozygote ex11-14 deletion | 6/6 |
| JM-512 | Ireland | Ireland | Irish Travellers | Homozygote ex11-14 deletion | 6/6 |
| KMM-336§ | Ireland | Ireland | Irish Travellers | Homozygote ex11-14 deletion | 6/6 |
The genealogy of Gypsies is complex, but they are divided into 3 principal tribal groups, the Roma, the Cales (or Gitanos), and the Munush (or Sinti). Irish Travellers are often confused with Roma Gypsies but they remain a separate ethnic group (see “Results and discussion”).
Arbitrary numbers based on the analysis of 4 highly polymorphic microsatellite markers flanking FANCA. Each chromosome/allele is defined by a haplotype block including the 4 markers. All Spanish patients except the one of Romanian origin were homozygous for a given haplotype block (1) and all the other patients but VU483 were homozygous for a specific but different haplotype block (2, 4, etc..).
ND indicates mutation not detected; the 295C > T mutation was disregarded in the last 7 patients shown in the table.
This patient corresponds to cell line EUFA/VU544.
Afro-American patients with FA from Ghana also have a truncating mutation in exon 4 of FANCA but involving a different codon. The Ghana mutation is associated with a severe clinical phenotype (Arleen D. Auerbach, Rockefeller University; personal communication, July 15, 2004). However, analysis of the registered clinical and cellular data obtained from all Spanish patients with FA in an SFARN standardized manner, the clinical phenotype of the Spanish Gypsy patients is similar to other Spanish patients with FA-A with a similar average number of congenital abnormalities and age of onset of impaired hematopoiesis. Only the severity of the congenital abnormalities was slightly higher in Spanish Gypsies. At the cellular level, the phenotype is also quite similar. This is true for the spontaneous frequency of breaks per cell (0.36 ± 0.16 in 5 Spanish Gypsy patients versus 0.22 ± 0.03 in 18 non-Gypsy Spanish FA-A nonmosaic patients), the number of breaks per cell after DEB (3.1 ± 0.8 in 7 Spanish Gypsy patients versus 3.6 ± 0.4 in 20 non-Gypsy Spanish FA-A nonmosaic patients), and the percentage of aberrant cells after DEB (76.7 ± 17.3 in 7 Spanish Gypsy patients versus 74.6 ± 14.6 in 20 non-Gypsy Spanish FA-A patients). The survival of blood T cells at 33 nM of mitomycin C (MMC) is even higher (P = .014) in Gypsy patients (33.1% ± 1.87 % in 5 Gypsy patients versus 21.2%± 1.5% in 27 non-Gypsy patients with FA-A). The fact that a similar mutation in FA patients of Ghanese decent is associated with a severe phenotype suggests that other ethnically related genetic factors such as trans-acting polymorphic genes are probably modulating the clinical evolution in patients with FA. Such effect has been shown even for an identical mutation, IVS4 + 4 in FANCC, in Ashkenazi and Japanese populations.
There is genetic evidence that the Gypsies originally migrated from Northern India to the West in the early middle ages, and then in a second 14th century migration, known as the Aresajipi migration, from Eastern Europe to Western Europe.12 They arrived in Spain through the Pyrenees in the early 15th century, with the first documented presence in Barcelona in 1425.19 The genealogy of Gypsies is highly complex but they are divided into 3 principal tribal groups, all genetically related: the Roma, the Munush (also called Sinti), and the Gitanos (also known as Cales), the latter mainly concentrated in Spain and France.19,20 In order to define whether the identified mutation is unique in Spain or present in the whole ethnic group and brought to Spain, we performed a worldwide search of Gypsy patients with FA outside Spain. Although millions of Gypsies live in neighboring countries such as France, Germany, Italy, and especially in Eastern European countries such as Romania, Slovakia, or Hungary, only 3 non-Spanish Gypsy patients with FA-A were found in Hungary (1 family), Germany (1 family), and Slovakia (1 family). In addition, 3 patients from Ireland belonging to the Irish Travellers ethnic group were also identified. The Irish Travellers have been often confused with traditional Gypsies. However, in spite of cultural exchange and some intermarriage, the Irish Travellers remain a distinct people. The fact that FA is usually associated to consanguinity and the previously mentioned intermarriage of Irish Travellers with Roma Gypsies made us include these Irish patients in this study. DNA sequencing revealed that none of these patients share the mutation with the Spanish Gypsies (Table 1). Therefore, the FANCA 295C>T mutation is specific of Spanish Gypsy patients.
We finally performed an extensive haplotype analysis to determine a common ancestral origin of the Spanish Gypsy mutation. Haplotype analysis was carried out by PCR-based techniques as previously described, by using 4 polymorphic microsatellite markers flanking FANCA: D16S3026, D16S3407, D16S3121, and D16S303.9,21 The results indicate that all Spanish Gypsy patients, including the one of Portuguese origin, had exactly the same homozygote haplotype that is clearly distinguishable from the haplotype of Spanish Gypsy carriers, the non-Gypsy Spanish FA-A patients, and the non-Spanish Gypsy patients or Irish Travellers (Table 1 and data not shown). This result clearly indicates the Spanish Gypsy mutation is associated to a specific haplotype and that all Spanish Gypsy patients studied share a common ancestry.
In conclusion, the data presented here strongly suggest that the high incidence of FA among Spanish Gypsies is due to a founder ancestral mutation in FANCA, leading to protein truncation and FA pathway disruption. This mutation probably originated less than 600 years ago in a Gypsy family that migrated to Spain and spread by consanguinity all over the Iberian peninsula, including Spain and Portugal. Consanguinity and a common ancestry are wellknown features of this ethnic group.22 The extremely high incidence of FA carriers among Spanish Gypsies and the easy detection of their associated mutation/haplotype makes this ethnic group a population model to study the role of heterozygous FA mutations and the FA pathway in both sporadic and hereditary cancer. Several studies are underway in our laboratory in this context.
Prepublished online as Blood First Edition Paper, November 2, 2004; DOI 10.1182/blood-2004-07-2588.
Supported in part supported by the Generalitat de Catalunya (project SGR-00197-2002), the Spanish Ministry of Health and Consumption (SMHC; projects FIS PI020145 and FIS-Red G03/073), the Spanish Ministry of Science and Technology (SMCT; projects SAF 2002-03234, SAF2002-11833-E, and SAF 2003-00328), the Commission of the European Union (projects FIGH-CT-2002-00217, FI6R-CT-2003-508842, and HPMF-CT-2001-01330 and FEDER funds), and a “Ramón y Cajal” contract (J.S.) entitled “Genome stability and DNA repair,” cofinanced by the SMCT and the Universitat Autònoma de Barcelona.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are greatly indebted to all patients who participated in this study. Many thanks are due to A.D. Auerbach (New York, NY), E. Gluckman (Paris, France), and A. Savoia (Napoli, Italy) for sharing their data. Also thanks to Maureen Hoatlin for sharing the anti-FANCA antibody and Antonio Fontdevila for helpful comments. This research has been done in the framework of the Spanish Fanconi Anemia Research Network under the auspices of the Spanish Ministry of Health and Consumption (SMHC).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal