Abstract
The erythrocyte colony-forming unit (CFU-E) is a rare bone marrow (BM) progenitor that generates erythrocyte colonies in 48 hours. The existence of CFU-Es is based on these colonies, but CFU-Es have not been purified prospectively by phenotype. We have separated the “nonstem,” “nonlymphoid” compartment (lineage marker [lin]–c-Kit+Sca-1–IL-7Rα–) into interleukin 3 receptor α negative (IL-3Rα–) and IL-3Rα+ subsets. Within IL-3Rα– but not IL-3Rα+ cells we have identified TER119–CD41–CD71+ erythrocyte-committed progenitors (EPs). EPs generate CFU-E colonies at about 70% efficiency and generate reticulocytes in vivo. Depletion of EPs from BM strongly reduces CFU-E frequencies. EPs lack potential for erythrocyte burst-forming unit, megakaryocyte, granulocyte (G), and monocyte (M) colonies, and for spleen colony-forming units. Chronically suppressed erythropoiesis in interferon consensus sequence-binding protein (ICSBP)–deficient BM is associated with reduced frequencies of both the EP population and CFU-E colonies. During phenylhydrazine-induced acute anemia, numbers of both the EP population and CFU-E colonies increase. Collectively, EPs (lin–c-Kit+Sca-1–IL-7Rα–IL-3Rα–CD41–CD71+) account for most, if not all, CFU-E activity in BM. As a first molecular characterization, we have compared global gene expression in EPs and nonerythroid GM progenitors. These analyses define an erythroid progenitor-specific gene expression pattern. The prospective isolation of EPs is an important step to analyze physiologic and pathologic erythropoiesis.
Introduction
Hematopoietic stem cells (HSCs) give rise to lineage (lin)–restricted, intermediate progenitors which, in turn, generate single-lineage committed progenitors.1,2 Multipotent cells might commit themselves stochastically into certain lineage fate,3 visible as single-lineage progeny. Therefore, it is difficult to assign, in retrospect, the origin of hematopoietic colonies to stem cells, intermediate, or single-lineage committed progenitors. Clonogenic developmental potential must be prospectively predictable. This requires the physical isolation of lineage-committed progenitors based on a unique cell surface phenotype.
Myeloid progenitors lack stem cell activity and lymphoid potential but possess erythrocyte, megakaryocyte, or granulocyte-monocyte (GM) potential. Several myeloid progenitors have been isolated, which satisfy the criteria of committed progenitors. These include mast cell progenitors (c-KithiThy-1lo),4 megakaryocyte progenitors (MPs; CD9+CD41+FcγRloCD34+CD38+),5 bipotent GM progenitors (GMPs; lin–c-Kit+Sca-1–CD34+FcγRhi), and megakaryocyte-erythrocyte progenitors (MEPs; lin–c-Kit+Sca-+–CD34–FcγRlo).6 Common myeloid progenitors (CMPs) have been reported as lin–c-Kit+Sca-1–CD34+FcγRlo cells.6 Erythrocyte-committed progenitors (EPs) have not been prospectively isolated by cell surface phenotype. The isolation of the erythroid colonyforming “unit” is a prerequisite to gain direct experimental access to this hallmark stage of erythropoiesis.
We have analyzed the expression pattern of a major stem/progenitor cell growth factor receptor, the interleukin 3 receptor α (IL-3Rα) chain (CD123),7-9 in the myeloid progenitor compartment. IL-3Rα expression divides the lin–c-Kit+IL-7Rα–Sca-1– cells into IL-3Rα– and IL-3Rα+ subpopulations. By combining IL-3Rα expression with further markers (FcγR [CD16/CD32], CD41, CD71), we obtained a high-resolution map of the “non-stem,” “nonlymphoid” progenitor compartment in mouse bone marrow (BM). This map reveals the position in the hematopoietic hierarchy and developmental potential of 6 separate progenitors. One of these progenitors is the elusive EP. EPs belong to the IL-3Rα– but not the IL-3Rα+ subset and are defined as lin–c-Kit+IL-7Rα–Sca-1–CD41–CD71+ cells. EPs have exclusively erythrocyte colony-forming unit (CFU-E) potential in vitro and can contribute significantly to reticulocytes in peripheral blood when transplanted into myeloablated mice. EPs correspond quantitatively to the colony-forming “unit” in BM. Therefore, EPs represent a missing link in the hematopoietic “tree” from HSCs via erythrocyte burst-forming units (BFU-Es) to erythrocytes. We have also taken advantage of the EP purification to gain molecular insight into the CFU-E stage of erythropoiesis by analyzing global gene expression in EPs. The isolation and gene expression profile of EPs provides a useful foundation for further analyses of physiologic and pathologic erythropoiesis.
Materials and methods
Mice, monoclonal antibodies, and cell sorting
C57Bl/6 mice were analyzed at 4 to 8 weeks of age. Interferon consensus sequence-binding protein, (ICSBP)+/+ and ICSBP–/–, mice10 were analyzed in age-matched groups between 3 and 6 months of age when the myeloproliferative syndrome was prominent. Lineage antibodies were B220 (clone RA3-6B2), CD3ϵ (clones 145-2C11 or 17A2), Gr-1 (clone RB6-8C5), Mac1 (clone M1/70), and TER119 (all labeled with fluorescein isothiocyanate [FITC]). Further antibodies were phycoerythrin (PE)–labeled anti-IL-3Rα (clone 5B11), biotinylated anti-FcγRII/III (clone 2.4G2), FITC-labeled anti-CD41 (clone MWreg30), PE-labeled CD71 (clone C2), allophycocyanin (APC)–labeled anti-CD117 (clone 2B8), FITC-labeled or biotinylated anti-Sca-1 (clone D7) (all from Becton Dickinson, Heidelberg, Germany), and FITC-labeled anti-IL-7Rα (clone A7R34) (a gift from S. Nishikawa, RIKEN Center for Developmental Biology, Kobe, Japan). Second-step reagents were streptavidin-tricolor (Caltag, Burlingame, CA), or streptavidin-APC (Molecular Probes, Eugene, OR). Flow cytometric analyses and cell sorting were done as described.11 EPs showed the highest cloning efficiency after sorting in phosphate-buffered saline (PBS) sheath fluid, and collection in 70% fetal calf serum (FCS) in PBS. To analyze frequencies of CFU-E colonies in EP-depleted BM, the total non-EP BM fraction was collected by cell sorting and assayed for CFU-E colonies. The depletion factor was determined by fluorescence-activated cell sorting (FACS) measurements of EP frequencies before and after sorting. Percentages and total numbers of various populations (Figures 1 and 4; Tables 1, 2) were determined by counting nucleated BM cells and by flow cytometry.
Phenotypic isolation, morphology, and potential of 6 distinct hematopoietic progenitor populations. (A) Lin–IL-7Rα–Sca-1– BM cells were analyzed for expression of c-Kit versus IL-3Rα (i) or c-Kit versus antibody isotype control (ii). Cells shown in i correspond to 2% of all nucleated BM cells. Lin–Sca-+–IL-7Rα– cells were sorted into c-Kit+IL-3Rα– (green gate in i) and c-Kit+IL-3Rα+ (red gate in i) populations. Cells shown in iii and xi correspond to 0.52% and 0.68% of all nucleated BM cells, respectively. Purified c-Kit+IL-3Rα– cells (iii) were restained for expression CD71 versus CD41 (iv), and sorted into CD71+CD41– (v), CD71–CD41– (vii), or CD71–CD41+ (ix) cells. Sorted c-Kit+IL-3Rα+ cells (xi) were stained for expression of FcγR (CD16/CD32) versus CD41 (xii), and further separated into FcγRhiCD41– (xiii), FcγRloCD41– (xv), or FcγRloCD41+ (xvii) cells. Numbers in i, iv, and xii are relative percentages of the populations in the indicated regions. Numbers shown in v, vii, ix, xiii, xv, and xvii are percentages of each population per total nucleated BM cells. Cytospins from each population were stained with May-Grünwald-Giemsa stain (vi,viii,x,xiv,xvi,xviii). The scale bar (shown in xviii) corresponds to 15 μM and applies to panels vi, viii, x, xiv, xvi, and xviii. Based on their potential, the 6 progenitors were designated as erythrocyte progenitors (EPs; v), IL-3Rα– common myeloid progenitors (CMP IL-3Rα–; vii), IL-3Rα – megakaryocyte progenitors (MP1; ix), granulocyte-monocyte progenitors (GMPs; xiii), IL-3Rα+ common myeloid progenitors (CMP IL-3Rα+; xv), and IL-3Rα+ megakaryocyte progenitors (MP2; xvii). (B) Purified BM progenitors (as shown in panel A) were plated at 2000 cells/culture into methylcellulose assays promoting the generation of BFU-E (i), CFU-Meg (ii), or CFU-GM (iii) colonies. Colonies were counted after 8 days. BFU-E colonies were typed by benzidine staining for globin. CFU-Meg and CFU-GM colonies were identified by morphology. EPs lack potential for BFU-E, CFU-Meg, or CFU-GM colonies. Data shown are representative for one of 3 experiments. Formation of more mature CFU-Ss day 8 (iv), and more immature CFU-Ss day 12 (v) was analyzed by transfer of HSCs (2500 cells/mouse; number of recipients [n] = 2), CMP IL-3Rα– (2500 cells/mouse; n = 2), CMP IL-3Rα+ (2500 cells/mouse; n = 2), GMP (2500 cells/mouse; n = 2), EP (2500 cells/mouse; n = 2), MP1 (2500 cells/mouse; n = 2), or MP2 (2500 cells/mouse; n = 2). Injection of total BM (2 × 105 cells/mouse; n = 2) or PBS (n = 2) served as positive and negative controls, respectively. After 8 or 12 days macroscopic colonies were counted. CFU-Ss are restricted to HSC and CMP subpopulations (iv-v). Data shown are representative for one of 2 experiments.
Phenotypic isolation, morphology, and potential of 6 distinct hematopoietic progenitor populations. (A) Lin–IL-7Rα–Sca-1– BM cells were analyzed for expression of c-Kit versus IL-3Rα (i) or c-Kit versus antibody isotype control (ii). Cells shown in i correspond to 2% of all nucleated BM cells. Lin–Sca-+–IL-7Rα– cells were sorted into c-Kit+IL-3Rα– (green gate in i) and c-Kit+IL-3Rα+ (red gate in i) populations. Cells shown in iii and xi correspond to 0.52% and 0.68% of all nucleated BM cells, respectively. Purified c-Kit+IL-3Rα– cells (iii) were restained for expression CD71 versus CD41 (iv), and sorted into CD71+CD41– (v), CD71–CD41– (vii), or CD71–CD41+ (ix) cells. Sorted c-Kit+IL-3Rα+ cells (xi) were stained for expression of FcγR (CD16/CD32) versus CD41 (xii), and further separated into FcγRhiCD41– (xiii), FcγRloCD41– (xv), or FcγRloCD41+ (xvii) cells. Numbers in i, iv, and xii are relative percentages of the populations in the indicated regions. Numbers shown in v, vii, ix, xiii, xv, and xvii are percentages of each population per total nucleated BM cells. Cytospins from each population were stained with May-Grünwald-Giemsa stain (vi,viii,x,xiv,xvi,xviii). The scale bar (shown in xviii) corresponds to 15 μM and applies to panels vi, viii, x, xiv, xvi, and xviii. Based on their potential, the 6 progenitors were designated as erythrocyte progenitors (EPs; v), IL-3Rα– common myeloid progenitors (CMP IL-3Rα–; vii), IL-3Rα – megakaryocyte progenitors (MP1; ix), granulocyte-monocyte progenitors (GMPs; xiii), IL-3Rα+ common myeloid progenitors (CMP IL-3Rα+; xv), and IL-3Rα+ megakaryocyte progenitors (MP2; xvii). (B) Purified BM progenitors (as shown in panel A) were plated at 2000 cells/culture into methylcellulose assays promoting the generation of BFU-E (i), CFU-Meg (ii), or CFU-GM (iii) colonies. Colonies were counted after 8 days. BFU-E colonies were typed by benzidine staining for globin. CFU-Meg and CFU-GM colonies were identified by morphology. EPs lack potential for BFU-E, CFU-Meg, or CFU-GM colonies. Data shown are representative for one of 3 experiments. Formation of more mature CFU-Ss day 8 (iv), and more immature CFU-Ss day 12 (v) was analyzed by transfer of HSCs (2500 cells/mouse; number of recipients [n] = 2), CMP IL-3Rα– (2500 cells/mouse; n = 2), CMP IL-3Rα+ (2500 cells/mouse; n = 2), GMP (2500 cells/mouse; n = 2), EP (2500 cells/mouse; n = 2), MP1 (2500 cells/mouse; n = 2), or MP2 (2500 cells/mouse; n = 2). Injection of total BM (2 × 105 cells/mouse; n = 2) or PBS (n = 2) served as positive and negative controls, respectively. After 8 or 12 days macroscopic colonies were counted. CFU-Ss are restricted to HSC and CMP subpopulations (iv-v). Data shown are representative for one of 2 experiments.
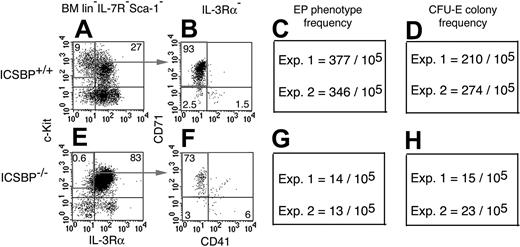
Parallel reduction in the frequencies of CFU-E colonies and EPs in mice lacking the transcription factor ICSBP. Lin–Sca-1–IL-7Rα– cells from ICSBP+/+ (A) and from ICSBP–/– (E) mice were analyzed for expression of c-Kit and IL-3Rα. C-Kit+IL-3Rα– cells were further separated by staining for CD71 versus CD41 (B,F). Frequencies of EPs were strongly reduced in ICSBP–/– (G) compared to ICSBP+/+ (C) BM. This decline in the EP frequencies parallels the reduction in the CFU-E frequencies in total BM from ICSBP–/– (H) compared to ICSBP+/+ mice (D). Numbers in panels A, B, E, and F are relative percentages of the populations in the indicated regions. Data in shown panels C, D, G, and H are mean numbers from duplicate CFU-E cultures for each experiment (Exp.).
Parallel reduction in the frequencies of CFU-E colonies and EPs in mice lacking the transcription factor ICSBP. Lin–Sca-1–IL-7Rα– cells from ICSBP+/+ (A) and from ICSBP–/– (E) mice were analyzed for expression of c-Kit and IL-3Rα. C-Kit+IL-3Rα– cells were further separated by staining for CD71 versus CD41 (B,F). Frequencies of EPs were strongly reduced in ICSBP–/– (G) compared to ICSBP+/+ (C) BM. This decline in the EP frequencies parallels the reduction in the CFU-E frequencies in total BM from ICSBP–/– (H) compared to ICSBP+/+ mice (D). Numbers in panels A, B, E, and F are relative percentages of the populations in the indicated regions. Data in shown panels C, D, G, and H are mean numbers from duplicate CFU-E cultures for each experiment (Exp.).
CFU-E colonies are highly enriched in the EP phenotype in BM
Experiment no. . | CFU-E colonies/105 total BM cells . | CFU-E colonies/105 EP cells . | CFU-E colonies/105 BM cells depleted of EP cells . |
|---|---|---|---|
| 1 | Not done | 76 000 | Not done |
| 2 | 235 | 69 000 | 30 |
| 3 | 260 | 81 000 | 17 |
Experiment no. . | CFU-E colonies/105 total BM cells . | CFU-E colonies/105 EP cells . | CFU-E colonies/105 BM cells depleted of EP cells . |
|---|---|---|---|
| 1 | Not done | 76 000 | Not done |
| 2 | 235 | 69 000 | 30 |
| 3 | 260 | 81 000 | 17 |
CFU-E colony frequencies were determined in methylcellulose assays for total BM cells, for sorted EPs, and for BM cells depleted of EPs by cell sorting. CFU-E colony numbers were measured per 105 total BM cells and per 105 BM cells depleted of EPs. For EPs, CFU-E colony numbers were calculated per 105 cells. We obtained 648 CFU-E colonies per 850 EPs plated (experiment 1), 345 CFU-E colonies per 500 EPs plated (experiment 2), and 405 CFU-E colonies per 500 EPs plated (experiment 3). Thus, EPs gave rise to CFU-E colonies with about 70% efficiency. An approximate 8-fold depletion of EPs from BM reduced the CFU-E frequencies in BM by about 10-fold (see “Materials and methods”). Hence, the vast majority of CFU-E activity in BM was quantitatively isolated with the EP phenotype.
Parallel increase in the frequencies of CFU-E colonies and the EP phenotype in BM before and after PHZ-induced anemia
. | CFU-E colonies per 105 total BM cells . | . | EP phenotype per 105 total BM cells . | . | ||
|---|---|---|---|---|---|---|
. | PBS control . | PHZ-treated* . | PBS control . | PHZ-treated* . | ||
| Exp. 1 | 245 | 520 | 400 | 912 | ||
| Exp. 2 | 255 | 610 | 378 | 887 | ||
. | CFU-E colonies per 105 total BM cells . | . | EP phenotype per 105 total BM cells . | . | ||
|---|---|---|---|---|---|---|
. | PBS control . | PHZ-treated* . | PBS control . | PHZ-treated* . | ||
| Exp. 1 | 245 | 520 | 400 | 912 | ||
| Exp. 2 | 255 | 610 | 378 | 887 | ||
Frequencies of CFU-E colonies (left) and the EP population (right) were determined in methylcellulose assays and by flow cytometry 3 days after a single injection of PHZ (60 mg/kg body weight). Control mice were injected with PBS only. Both CFU-E colony and EP frequencies increased more than 2-fold.
Measured 3 days after a single injection of PHZ (60 mg/kg body weight).
In vitro colony assays
Cell sorter-purified progenitors were placed in methylcellulose cultures as described.12,13 Briefly, cells were cultured in MethoCult M3231 (StemCell Technologies, Vancouver, BC, Canada) for BFU-E, CFU-GM, CFU-megakaryocyte (CFU-Meg), and mixed CFU (CFU-Mix) colonies, or in MethoCult M3334 with erythropoietin (Epo) for CFU-E colonies. Added growth factors were granulocyte-macrophage colony-stimulating factor (GM-CSF; 1 ng/mL), macrophage colony-stimulating factor (M-CSF; 10 U/mL), thrombopoietin (Tpo; 10 ng/mL), stem cell factor (SCF; 50 ng/mL); all of these were murine (R&D Systems, Minneapolis, MN); IL-3 (1% supernatant, expressed as reported14 ), and human Epo (3 U/mL; Erypo, FS2000, Janssen-Cilag, Neuss, Germany). CFU-E and BFU-E colonies were stained with 0.4% benzidine (Sigma, Taufkirchen, Germany) in 12% glacial acetic acid and 0.3% H2O2. Cytospins were stained by Diff-Quik (Dade Behring, Marburg, Germany) according to the manufacturer's recommendation. Cells were inspected with an Axioskop microscope (Zeiss, Oberkochen, Germany) using an objective with a 100 × magnification and a 1.3 aperture (Zeiss) with oil, and photomicrographs (Figure 1) were taken using the OM-11 color camera (Olympus, Hamburg, Germany).
Adoptive cell transfers
Mice were irradiated with 850 rad (split dose) and given intravenous injections of HSCs (2500 cells/mouse), total BM (2 × 105), CMP IL-3Rα–, CMP IL-3Rα+, GMP, EP, MP1, or MP2 (each at 5000 cells/mouse). Percentages of reticulocytes in peripheral blood were determined on an ADVIA 120 Hematology System (Bayer, Leverkusen, Germany). For spleen colony-forming units (CFU-Ss), 900-rad irradiated mice were given intravenous injections of total BM (2 × 105), HSC, CMP IL-3Rα–, CMP IL-3Rα+, GMP, EP, MP1, or MP2 (each at 2500 cells/mouse).
Phenylhydrazine-induced anemia
Mice were treated with a single intraperitoneal injection of phenylhydrazine (PHZ; 60 mg/kg body weight) as described.15 Frequencies of CFU-E colonies and EPs were determined in BM 3 days after the injection.
DNA microarray gene expression analyses and bioinformatics
Total RNA was isolated from purified populations from 10- to 16-week-old mice using the RNeasy kit including DNase digestion (Qiagen, Hilden, Germany). Total RNA (5 μg) was used to generate cDNA according to the technical manual (Affymetrix, Santa Clara, CA). cRNA was generated with the BioArray high-yield transcript labeling kit (ENZO, Farmingdale, NY), and 15 μg cRNA was hybridized to Affymetrix mouse expression 430A arrays at 45°C for 16 hours. DNA chips were stained, washed, and scanned according to the manufacturer's protocol. Scanned GeneChip DAT files were analyzed by the GeneChip Analysis Suite Software (Affymetrix) with global scaling to 500. Further analysis of data output was carried out using Data Mining Tool and NetAffx Web-Based Database (Affymetrix) as well as Microsoft Excel. The data have been submitted to the GEO Web site (http://www.ncbi.nlm.nih.gov/geo; submission no. GSE1584).
Results
Division of BM populations based on IL-3Rα chain expression identifies 6 distinct progenitor populations
Downstream from HSCs (lin–c-Kit+Sca-1+), progenitor potential for erythrocyte, megakaryocyte, monocyte, and granulocyte lineages is contained in the lin–c-Kit+Sca-1–IL-7Rα– population.6 Because IL-3 is a potent growth factor for stem/progenitor cells (for reviews, see Ihle,7 Ogawa,8 McKinstry et al,16 and Leonard17 ), we reasoned that IL-3Rα chain expression might correlate with lineage commitment within this compartment. Whereas staining with an anti-IL-3Rα monoclonal antibody18,19 was very weak or undetectable on whole BM, the lin–c-Kit+Sca-1–IL-7Rα– fraction could be separated into IL-3Rα+ and IL-3Rα– subpopulations (Figure 1Ai). Antibody isotype control staining (Figure 1Aii), and analysis for expression of IL-3Rα mRNA by reverse transcription-polymerase chain reaction (RT-PCR) on sorted cells (not shown) confirmed the specificity of the anti-IL-3Rα antibody staining. As a first guideline, we searched by RT-PCR for lineage-specific gene expression in lin–c-Kit+Sca-1–IL-7Rα–IL-3Rα+ (termed c-Kit+IL-3Rα+) and lin–c-Kit+Sca-1–IL-7Rα–IL-3Rα– (termed c-Kit+IL-3Rα–). The “myeloid” gene Pu1 and the mast cell-specific protease Mc-cpa were strongly expressed in c-Kit+IL-3Rα+ (not shown). In contrast, c-Kit+IL-3Rα– highly expressed the erythropoietin (Epo) receptor (EpoR) gene, suggesting that IL-3Rα expression marks a GM versus erythrocyte branch.
Based on the suspected erythrocyte potential within the c-Kit+IL-3Rα– population, these cells were further stained (Figure 1Aiv) for molecules expressed on erythrocyte progenitors (transferrin receptor [CD71]),20 and on megakaryocytes (integrin αIIb chain [CD41]).21 This staining resolved c-Kit+IL-3Rα– cells into CD71+CD41– (Figure 1Av), CD71–CD41– (Figure 1Avii), and CD71–CD41+ (Figure 1Aix) subpopulations. Because of their presumed GM potential, c-Kit+IL-3Rα+ cells were stained for expression of FcγR, a marker previously used to define hematopoietic progenitors.6,22 Expression of FcγR, in combination with CD41, resolved c-Kit+IL-3Rα+ cells (Figure 1Axi) into 3 subsets (Figure 1Axii) characterized by their FcγRhiCD41– (Figure 1Axiii), FcγRloCD41– (Figure 1Axv), or FcγRloCD41+ (Figure 1Axvii) phenotypes.
All 6 populations were isolated at high purity (> 98%) by FACS. Based on their developmental potential (Figure 1, Table 1), we designated the 3 c-Kit+IL-3Rα– subsets as (1) erythrocyte progenitors (EPs; CD71+CD41–), (2) CMP IL-3Rα– (CD71–CD41–), and (3) IL-3Rα– MP1 (CD71–CD41+). The c-Kit+IL-3Rα+ subsets were termed (4) GMP (FcγRhiCD41–), (5) CMP IL-3Rα+ (FcγRloCD41–), and (6) IL-3Rα+ MP2 (FcγRloCD41+). The frequencies of these populations among nucleated BM cells ranged from 0.02% to 0.4% (Figure 1A). By morphology (Figure 1Avi,viii,x,xiv,xvi,xviii), EPs and GMPs were the largest cells. CMP subsets and both MP subsets were of similar size but the CMP populations were more basophilic when compared to the MPs. CMP IL-3Rα– and CMP IL-3Rα+, on the one hand, and MP1 and MP2, on the other hand, were morphologically similar. Hence, we observed little heterogeneity within each population but clear differences between the populations.
Phenotype of c-Kit+IL-3Rα+ and c-Kit+IL-3Rα– subsets according to FcγR and CD34 expression
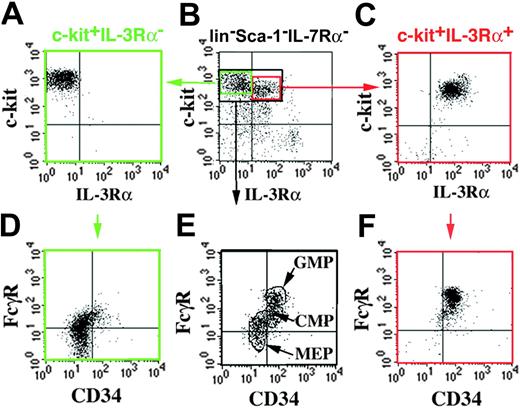
Lin–c-Kit+Sca-1–IL-7Rα– cells have previously been subdivided based on expression of FcγR and CD34 into CMPs (CD34+FcγRlo), GMPs (CD34+FcγRhi), and MEPs (CD34–FcγRlo).6 The 6 populations that we isolated according to expression of IL-3Rα, CD41, CD71, and FcγR (Figure 1A) may partially overlap with the model proposed by Akashi et al.6 Therefore, we “cross-phenotyped” BM cells by both types of analyses (Figure 2). Resolving lin–c-Kit+Sca-+–cells according to CD34 and FcγR expression confirmed the reported pattern of distinct but, at least in our hands, partially overlapping populations (Figure 2E). When lin–c-Kit+Sca-1– cells were first separated into IL-3Rα– and IL-3Rα+ populations, and then restained for CD34 and FcγR expression, the IL-3Rα– cells and IL-3Rα+ cells represented the “lower” (Figure 2D) and “upper” (Figure 2F) parts, respectively, of the nonseparated pattern shown in Figure 2E. This distribution suggested that the IL-3Rα– population includes MEPs and a subset of CMPs, whereas the IL-3Rα+ population includes GMPs and another subset of CMP phenotypes.
“Cross-phenotyping” of IL-3Rα+ and IL-3Rα– BM progenitors for CD34 and FcγR expression. Lin–c-Kit+Sca-1–IL-7Rα– cells (black gate in panel B) were stained for CD34 versus FcγR expression and resolved (E) into CD34+FcγRlo (CMP), CD34+FcγRhi (GMP), and CD34–FcγRlo (MEP).6 For comparison, c-Kit+lin–Sca-1–IL-7Rα– cells were sorted into IL-3Rα– (A) and IL-3Rα+ (C) subsets, and then restained for CD34 and FcγR expression. c-Kit+IL-3Rα– cells include MEP and CMP phenotypes (D), and c-Kit+IL-3Rα+ cells include GMP and CMP phenotypes (F).
“Cross-phenotyping” of IL-3Rα+ and IL-3Rα– BM progenitors for CD34 and FcγR expression. Lin–c-Kit+Sca-1–IL-7Rα– cells (black gate in panel B) were stained for CD34 versus FcγR expression and resolved (E) into CD34+FcγRlo (CMP), CD34+FcγRhi (GMP), and CD34–FcγRlo (MEP).6 For comparison, c-Kit+lin–Sca-1–IL-7Rα– cells were sorted into IL-3Rα– (A) and IL-3Rα+ (C) subsets, and then restained for CD34 and FcγR expression. c-Kit+IL-3Rα– cells include MEP and CMP phenotypes (D), and c-Kit+IL-3Rα+ cells include GMP and CMP phenotypes (F).
EPs generate CFU-E colonies at about 70% cloning efficiency
CFU-E colonies are small aggregates of approximately 16 to 32 globin-positive cells scored after 48 hours of culture in Epo.23-25 A clonogenic, phenotypically defined CFU-E has not been purified from BM. In 2 experiments, 105 total BM cells contained about 235 and 260 CFU-E (Table 1) colonies, a frequency similar to published data.23-25 Interestingly, 105 purified EPs gave rise to 76 000 (experiment 1), 69 000 (experiment 2), and 81 000 (experiment 3) CFU-E colonies. This corresponds to a cloning efficiency of at least 70% (Table 1). To estimate whether or not most CFU-E activity in BM was quantitatively retrieved within the EP fraction, the EP population was removed from BM by cell sorter depletion. In BM depleted of EPs by a factor of 8 (see “Materials and methods”), the CFU-E colony frequency dropped 10-fold (Table 1). Moreover, none of the other isolated progenitors (Figure 1A) had CFU-E potential (not shown). These data show that the vast majority of CFU-Es were quantitatively recovered from BM by isolating EPs.
EPs are CFU-E committed
To analyze whether EPs were CFU-E committed, EPs and all other progenitors depicted in Figure 1A were analyzed for BFU-E, megakaryocyte, and GM colony formation (Figure 1B). BFU-E colonies were measured as large colonies of globin-positive cells, visualized by dark blue benzidine staining, scored after 8 days of culture in SCF (or Kit ligand), Epo, and IL-3. BFU-E activity was present in both CMP IL-3Rα– and CMP IL-3Rα+ subsets but absent from all other progenitors including EPs (Figure 1Bi).
The potential to form CFU-Megs (Figure 1Bii) was examined by culture in Tpo and SCF. CFU-Meg potential was highly enriched in one of the c-Kit+IL-3Rα– subsets (CD71– CD41+; MP1), and in one of the c-Kit+IL-3Rα+ subsets (FcγRloCD41+; MP2). With much lower frequencies, CFU-Meg potential was also found in both CMP subsets. CFU-Meg potential was absent from EPs.
CFU-GM potential was analyzed in the presence of IL-3, SCF, GM-CSF, and M-CSF (Figure 1Biii). High frequencies of CFU-GMs were detected in the c-Kit+IL-3Rα+FcγRhiCD41– population. These cells are probably highly overlapping, if not identical with the previously described GMPs.6 In addition, both CMP populations contained CFU-GMs. EPs lacked CFU-GM potential. Finally, EPs also lacked mixed-lineage potential when analyzed under conditions permissive for the parallel development of erythrocyte, megakaryocyte, and GM colonies. The only populations with mixed-lineage potential were the CMP subsets (data not shown).
Next, all progenitor populations were assayed for their potential to form day 8 (CFU-S day 8) and day 12 (CFU-S day 12) spleen colonies in lethally irradiated mice.26 As positive controls, 2500 HSCs, or 2 × 105 total BM cells, were injected. Both for day 8 (Figure 1Biv) and for day 12 (Figure 1Bv), CFU-S potential was restricted to CMP subsets. In marked contrast, EPs as well as MP1, MP2, and GMPs lacked CFU-S potential.
Erythrocyte potential in vivo
In vitro, CMP subsets and EPs revealed erythrocyte potential indicative of early (BFU-E+CFU-E–) and late (BFU-E–CFU-E+) stages of erythropoiesis, respectively. We next asked whether progenitors with this in vitro potential could also contribute to erythropoiesis in vivo (Figure 3). Mice were myeloablated by irradiation with 850 rad, which led to the absence of reticulocytes in the circulation due to interruption of their de novo production. In PBS-injected mice, reticulocytes did not reappear in the peripheral blood for at least 2 weeks following irradiation (Figure 3A). Transfer of total BM caused an increase in the percentage of reticulocytes, which appeared as early as on day 10. Percentages increased to approximately 30% by day 14 (Figure 3B). HSCs and both CMP progenitors also reconstituted reticulocytes in vivo (Figure 3C-D,G). Reconstitution kinetics suggested that reticulocytes appeared earlier from CMP IL-3Rα+ (Figure 3G) than from CMP IL-3Rα– (Figure 3D) cells.
Erythrocyte reconstitution in vivo. Host erythropoiesis was transiently suppressed by irradiation (850 rad; split dose). After irradiation, mice were given intravenous injections of PBS (number of recipients [n] = 2; A), total BM (2 × 105 cells/mouse; n = 2; B), HSC (2500 cells/mouse; n = 2; C), CMP IL-3Rα– cells (5000 cells/mouse; n = 2; D), EPs (5000 cells/mouse; n = 2; E), MP1 (5000 cells/mouse; n = 1; F), CMP IL-3Rα+ (5000 cells/mouse; n = 2; G), GMP (5000 cells/mouse; n = 2; H), or MP2 (5000 cells/mouse; n = 1; I). Percentages of reticulocytes in peripheral blood were determined at the indicated time points. Data shown are representative for one of 2 experiments.
Erythrocyte reconstitution in vivo. Host erythropoiesis was transiently suppressed by irradiation (850 rad; split dose). After irradiation, mice were given intravenous injections of PBS (number of recipients [n] = 2; A), total BM (2 × 105 cells/mouse; n = 2; B), HSC (2500 cells/mouse; n = 2; C), CMP IL-3Rα– cells (5000 cells/mouse; n = 2; D), EPs (5000 cells/mouse; n = 2; E), MP1 (5000 cells/mouse; n = 1; F), CMP IL-3Rα+ (5000 cells/mouse; n = 2; G), GMP (5000 cells/mouse; n = 2; H), or MP2 (5000 cells/mouse; n = 1; I). Percentages of reticulocytes in peripheral blood were determined at the indicated time points. Data shown are representative for one of 2 experiments.
Interestingly, injection of EPs led to a marked increase in reticulocytes (Figure 3E). Unexpectedly, reticulocytes appeared later from EPs compared to CMPs. None of the remaining populations, that is, MP1 (Figure 3F), MP2 (Figure 3I), and GMP (Figure 3H), had reticulocyte potential. These experiments demonstrate that BFU-E and CFU-E assays correlated perfectly with the in vivo function of the respective progenitors.
Frequencies of EPs and CFU-Es following PHZ-induced anemia
If the EP phenotype represents the majority of the CFU-Es in BM, one should expect to measure parallel changes in both the frequencies of EP and the frequencies of CFU-E colonies. We tested this idea directly in a model of experimentally induced erythropoietic stress. Treatment of mice with PHZ (60 mg/kg body weight; single injection) leads to a drop in the hematocrit from about 45% to about 30%. This hemolytic anemia is accompanied by an increase in the frequency of CFU-E colonies in the BM15 (Table 2). We measured 245 (experiment 1) and 255 (experiment 2) CFU-E colonies/105 BM cells in untreated mice. Numbers rose to 520 (experiment 1) and 610 (experiment 2) CFU-E colonies/105 BM cells in mice 3 days after PHZ treatment. Parallel measurements of the abundance of cells with the EP phenotype revealed an increase from 400 (experiment 1) and 378 (experiment 2) EP/105 BM cells in untreated mice to 912 (experiment 1) and 887 (experiment 2) EP cells/105 BM cells in treated mice (Table 2).
The major stress erythropoiesis response occurs usually in the spleen.15 To analyze whether the EP phenotype also reflects stress erythropoiesis in this organ, we analyzed the spleen for the EP phenotype and followed this phenotype after PHZ treatment. Spleen EPs had CFU-E colony activity in vitro, and 5 days after a single injection of PHZ, the EP phenotype in spleen was increased from 0.03% in untreated mice to 0.53% in treated mice (not shown).
Collectively, an increase in the frequency of the EP phenotype faithfully reflects the changes of CFU-E colonies in this model of experimentally induced anemia.
Reduced frequencies of both CFU-E colonies and EPs in mice lacking the transcription factor ICSBP
We next asked whether numbers of EPs also correlated with numbers of CFU-E colonies in a chronic, genetically based erythropoietic defect. To this end, we determined the frequencies of EP and CFU-E colonies in mice lacking ICSBP. ICSBP is an important regulator of hematopoiesis. In vivo, ICSBP–/– BM is characterized by expansion of myeloid progenitors and concomitant suppression of erythropoiesis.10 With age, ICSBP–/– mice develop a myeloproliferative syndrome reminiscent of human chronic myelogenous leukemia (CML). We analyzed ICSBP+/+ and ICSBP–/– BM at 3 to 6 months of age, a time when the suppression of erythropoiesis was evident.
Numbers of CFU-E colonies and EPs were compared between ICSBP+/+ and ICSBP–/– BM. In wild-type mice, 27% and 9% of lin–IL-7R–Sca-1–c-Kit+ cells were IL-3Rα+ and IL-3Rα–, respectively, a ratio of 3:1 (Figure 4A). In ICSBP–/– mice, 83% and 0.6% of lin–IL-7R–Sca-1–c-Kit+ cells were IL-3Rα+ and IL-3Rα–, respectively, a ratio of 138:1 (Figure 4E). Hence, the c-Kit+IL-3Rα– population was dramatically reduced in ICSBP–/– BM. c-Kit+IL-3Rα– cells included 93% and 73% EPs (CD71+CD41–) in ICSBP+/+ and ICSBP–/–, respectively (Figure 4B,F). Based on absolute cell numbers and these phenotypic analyses, ICSBP+/+ BM harbored 377 (experiment 1) and 346 (experiment 2) EPs/105 cells (Figure 4C). Parallel measurements revealed 210 (experiment 1) and 274 (experiment 2) CFU-E colonies/105 ICSBP+/+ BM cells (Figure 4D).
Frequencies of both EPs (Figure 4G) and CFU-E colonies (Figure 4H) were 10- to 20-fold reduced comparing ICSBP+/+ (Figure 4C-D) and ICSBP–/– (Figure 4G-H) BM cells. These experiments demonstrate that the frequency of EPs is a very good indicator of CFU-E frequencies in this anemic BM.
Global gene expression profiles in EPs and GMPs reveal EP-specific gene expression
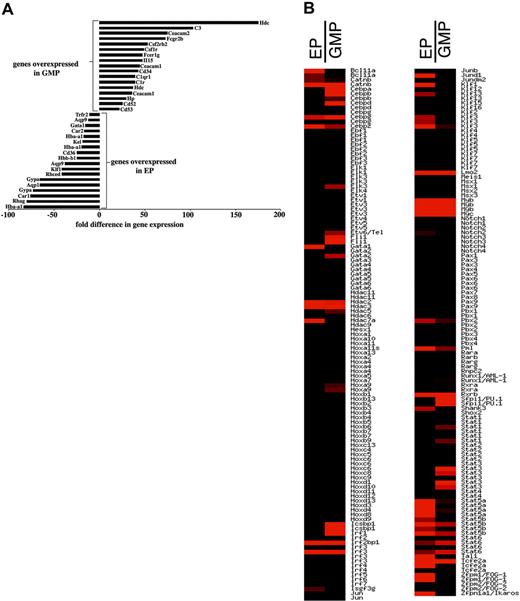
To characterize the RNA expression profile of the CFU-Es, we compared RNA expression in EPs and GMPs by array analysis. We chose this comparison because EPs and GMPs are both hematopoietic progenitors but located “closely” downstream from the erythroid versus GM branch. We identified 14 080 transcripts that were present in EPs or GMPs or both populations; 7795 of those were not differentially expressed. Using a signal ratio cutoff of more than 2.5, we identified 627 genes that were expressed higher in EPs than in GMPs; 136 genes were expressed at least 10-fold stronger in EPs compared to GMPs. Among those were hemoglobin A (Hba-a1), Rhesus blood group-associated A glycoprotein (Rhag), carboanhydrase 1 (Car1), glycophorin A (Gypa), aquaphorin (Aqp) 1 and 9, Rhesus blood group CE and D (Rhced), erythroid Kruppel-like factor (Klf1), CD36, Kell blood group (Kel), Gata-1, and transferrin receptor 2 (Trfr2) as shown in Figure 5A.
Differential gene expression profile comparing EPs and GMPs. RNAs from cell sorter-purified EPs and GMPs from normal mice were analyzed by microarray analyses for differences in global gene expression. Panel A depicts a selection of genes that are highly and differentially expressed in either GMPs (top) or EPs (bottom). Depicted is the fold difference in GMP over EP (positive numbers) and EP over GMP (negative numbers) hybridization signals. Panel B shows the relative expression pattern of transcription factors in alphabetical order comparing EPs and GMPs. Red indicates increased expression; black indicates lack of expression. Repeats of the same gene name reflect distinct probes for that gene. The full list of expression data has been deposited at http://www.ncbi.nlm.nih.gov/geo.
Differential gene expression profile comparing EPs and GMPs. RNAs from cell sorter-purified EPs and GMPs from normal mice were analyzed by microarray analyses for differences in global gene expression. Panel A depicts a selection of genes that are highly and differentially expressed in either GMPs (top) or EPs (bottom). Depicted is the fold difference in GMP over EP (positive numbers) and EP over GMP (negative numbers) hybridization signals. Panel B shows the relative expression pattern of transcription factors in alphabetical order comparing EPs and GMPs. Red indicates increased expression; black indicates lack of expression. Repeats of the same gene name reflect distinct probes for that gene. The full list of expression data has been deposited at http://www.ncbi.nlm.nih.gov/geo.
Using the same cutoff, we found 1255 genes whose expression was higher in GMPs than EPs; 330 genes were expressed at least 10-fold higher. Among those were histidine decarboxylase (Hdc), complement component 3 (C3), GM-CSFR (CSF2rb2), M-CSFR (Csf1r), high-affinity IgE receptor γ chain (Fcer1g), IL-15, CD34, CD52, and CD53 (Figure 5A). These data indicate a perfect correlation between each isolated progenitor population and its lineage-specific gene expression. In particular, the “erythroid gene expression pattern” of EPs is a strong indicator of the lineage assignment of this progenitor.
Next, we analyzed our array data for transcription factors expression. A total of 1317 transcripts involved in gene regulation were identified that were expressed in one or both of these populations. Among those, 48 and 103 were differentially expressed (2.5-fold or higher) in EPs and GMPs, respectively. Among the CCAAT/enhancer-binding proteins, C/ebpa, C/ebpb, and C/ebpd were expressed in GMPs but not in EPs. The 430A chip lacks C/ebpe probe sets. From the Ets family Elk3, Etv6/Tel, Fli1, and Pu.1 were expressed higher in GMPs than in EPs. Within the Gata family, Gata1 was expressed exclusively in EPs and Gata-2 specifically but weakly in GMPs. Friend of Gata (Fog)1 (also termed Zfpm1) and Tal1 expressions were very strong in EPs and absent (Fog1) and low (Tal1) in GMPs. Within the homeobox domain gene family, only HoxA9 was expressed, with lower expression in EPs and higher expression in GMPs. Among the interferon-regulated factors, Irf1 was expressed in both populations but was much stronger in GMPs. Irf3 was strongly expressed in EPs and GMPs. Icsbp was exclusively and strongly expressed in GMPs. Within the Kruppel-like factors, Klf1 and Klf3 were predominantly expressed in EPs. In contrast, Klf4 expression was exclusive for GMPs. Finally, within the STAT family, Stat3 and Stat4 expressions were stronger in GMPs, whereas Stat5a and Stat5b were predominantly expressed in EPs. The relative expression of these genes is shown in Figure 5B, and the full list of array data are available at http://www.ncbi.nlm.nih.gov/geo. This analysis should be taken as a systematic starting point to study and dissect the physiologic network of genes regulating and determining the developmental potential of undifferentiated, yet lineage-committed erythroid progenitors.27
Discussion
Prospective isolation of a clonogenic EP
Distinct stages of erythrocyte development have been measured for a long time by BFU-E and CFU-E assays.23-25 Despite the wide usage of CFU-E assays to characterize erythropoiesis (for examples, see Wu et al,28 Zang et al,29 and Jegalian et al30 ), the colony-forming “unit” had not been purified to homogeneity by cell surface phenotype. We have now prospectively identified CFU-Es as lin–c-Kit+Sca-1–IL-7Rα–IL-3Rα–CD41–CD71+ cells. CD71, the transferrin receptor, has been used in combination with the erythroid-restricted marker TER11931 to dissect distinct stages of erythroblast maturation.20 However, CD71 is expressed on many cycling cells and is not specific for the erythrocyte lineage.32 In fact, CD71 alone is not sufficient to isolate EPs because we also detected CD71+IL-3Rα+ cells, which are not erythrocyte committed (not shown). Thus, BM separation based on IL-3Rα expression was crucial for the identification of EPs.
In transgenic mice expressing a Gata-1 promoter-driven GFP transgene, GFP+CD71+ progenitors had only CFU-E potential32 indicating that Gata-1, together with CD71, can be a useful intracellular marker of CFU-Es. However, GFP+CD71+ cells gave rise to CFU-E colonies with considerably lower frequencies (1/10) compared to EPs described here. Zhang and colleagues,43 using day 14.5 fetal liver (FL) cells, enriched CFU-Es prospectively by phenotype from 7400 CFU-Es/105 total FL cells to 41 200/105 total FL cells by isolation of CD71lowTER119– FL cells. Thus, CD71lowTER119– FL cells are at least 41% pure CFU-Es. Taking the overall frequency of CD71lowTER119– cells in FL into consideration, the CD71lowTER119– phenotype can quantitatively account for about 30% (2000 of 7400 CFU-Es/105 total FL cells) of all CFU-Es in FL. Very recently, introduction of GFP as a reporter into the EpoR locus showed that CFU-Es are included in the lin–Sca-1– c-Kithi population.33
We have now identified CFU-Es among lin–Sca-1–c-Kithi BM cells as IL-3Rα–CD41–CD71+ cells. EPs were at least 70% pure CFU-Es (Table 1). Given that about 0.4% of all nucleated BM cells are EPs (Figure 1), and that at least 70% of EPs can generate CFU-E colonies (Table 1), we estimate that EPs can account for about 300 CFU-Es/105 total BM cells. This is in the range of the total CFU-E frequency (∼200-300 CFU-Es/105 total BM cells) reported by others23-25 and measured by us (Table 1).13 Our conclusion that EPs are quantitatively identical with CFU-Es is further supported by the 10-fold reduction of CFU-Es in BM depleted of EPs by a factor of 8 (Table 2). To our knowledge, this is the first report of the near-complete and prospective isolation of CFU-Es from BM by cell surface phenotype.
Global gene expression pattern in EPs
The isolation of EPs has opened access for global gene expression profiling of the CFU-E stage. Many genes specifically expressed in EPs, when compared to GMPs, are known to be erythroid, prototypically Gata-1, Klf1, and blood group-related genes (Figure 5). It was not known, however, which genes are expressed in a committed erythroid progenitor. The earliest erythroid commitment may take place prior to the CFU-E stage, at the BFU-E stage; however, up to now, BFU-E potential has not been prospectively separated from nonerythroid potential because, at the population level, all progenitors that contain BFU-Es also contain non-BFU-E potential. Hence, EPs are the only pure erythroid-committed progenitors available for analysis at present. This study now offers information on a large fraction of those genes that are expressed specifically at the CFU-E stage. Future experiments could determine which of these genes are crucial for the development of erythroid cells from EPs.
Relationship of BFU-Es and CFU-Es
It has been established a long time ago that BFU-Es and CFU-Es represent erythroid precursors at sequential stages of differentiation.24 BFU-Es and CFU-Es differ in cell size, sensitivity to cycle-active agents, response to plethora, and effects of the W/Wv24 and W/W13 genotypes. Based on these data, BFU-E and CFU-E colonies were thought to arise from committed progenitors. We now demonstrate the erythroid commitment of CFU-Es directly by showing prospectively that these cells give rise to erythroid colonies but not to any other lineage assayed for. Our data also show that BFU-Es and CFU-Es can be prospectively separated because EPs lack BFU-E potential (Figure 1B). BFU-Es arise from the CMP compartments. The generation of mixed BFU-E/CFU-GM colonies demonstrates that a single CMP can express “true” common myeloid potential. However, CMPs also generate pure BFU-E colonies. Until the prospective isolation of a phenotypically defined BFU-E will be achieved, it remains open whether a committed BFU-E exists, or whether single BFU-E colonies arise from CMPs stochastically.
The identification of EPs as the CFU-Es, as well as the assignment of BFU-E activity to the multipotent CMP stage, places EPs downstream from HSCs and BFU-Es (Figure 6). Transplanted EPs also have significant potential in vivo as shown by a burst of reticulocytes following injection of EPs into myelosuppressed mice (Figure 3). Selective substitution of erythropoiesis may be clinically relevant under myelosuppressive conditions. We noted that reticulocytes did not arise earlier from EPs when compared to CMPs. Although the basis for this kinetic difference is unknown, it is possible that the irradiated host does not immediately support reticulocyte development from EPs. An example of an irradiation-induced developmental delay has been noted when pro–T cells were injected into an irradiated but not into a nonirradiated thymus.34 In this regard it is noteworthy that, on day 3, reticulocytes were absent even in mice injected with total BM, which should contain all CFU-E colony activity, regardless of any purification method or progenitor subset. Yet, at the typical time when the CFU-E colony arises in vitro, that is 2 to 3 days, we see no reticulocytes in vivo (Figure 3). We conclude that, in irradiated mice in vivo, it takes up to 10 days before reticulocytes are detectable from any cell type in the BM, including EPs.
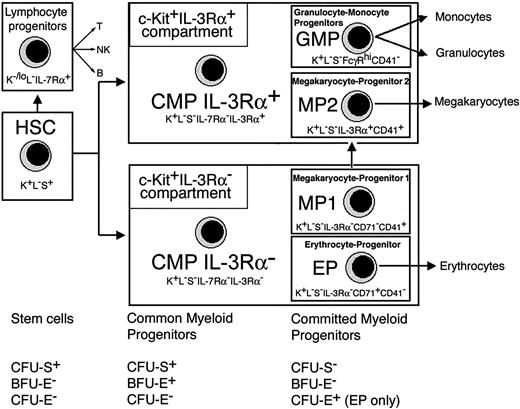
Phenotypic and functional map of myeloid pathways in mouse BM. IL-3Rα expression defines subsets of CMPs and progenitors with prospective potential for erythrocytes (EP), megakaryocytes (MP1 and MP2), or granulocytes and monocytes (GMP). The c-Kit+IL-3Rα+ population includes the IL-3Rα+ CMP subpopulation, GMPs, and the IL-3Rα+ megakaryocyte progenitor termed MP2. The c-Kit+IL-3Rα– population includes the IL-3Rα– CMP subsets, the IL-3Rα– megakaryocyte progenitor termed MP1, and EPs. The relationship of stem cells and common myeloid and committed myeloid stages to CFU-S, BFU-E, and CFU-E is indicated for each compartment. Despite their major myeloid potential, we also detected a minor level of B-cell progenitor activity in mice given transplants of CMP 3Rα– or CMP 3Rα+ cells. The level of B-cell reconstitution from CMPs was about 10-fold lower when compared to CLPs and about 40-fold lower when compared to HSCs or BM. Phenotypic abbreviations are c-Kit (K), lin (L), and Sca-1 (S).
Phenotypic and functional map of myeloid pathways in mouse BM. IL-3Rα expression defines subsets of CMPs and progenitors with prospective potential for erythrocytes (EP), megakaryocytes (MP1 and MP2), or granulocytes and monocytes (GMP). The c-Kit+IL-3Rα+ population includes the IL-3Rα+ CMP subpopulation, GMPs, and the IL-3Rα+ megakaryocyte progenitor termed MP2. The c-Kit+IL-3Rα– population includes the IL-3Rα– CMP subsets, the IL-3Rα– megakaryocyte progenitor termed MP1, and EPs. The relationship of stem cells and common myeloid and committed myeloid stages to CFU-S, BFU-E, and CFU-E is indicated for each compartment. Despite their major myeloid potential, we also detected a minor level of B-cell progenitor activity in mice given transplants of CMP 3Rα– or CMP 3Rα+ cells. The level of B-cell reconstitution from CMPs was about 10-fold lower when compared to CLPs and about 40-fold lower when compared to HSCs or BM. Phenotypic abbreviations are c-Kit (K), lin (L), and Sca-1 (S).
Identification of 2 late MPs
Our phenotypic BM dissection has uncovered 2 megakaryocyte progenitors, termed MP1 and MP2 (Figure 6). Both progenitors are committed to the megakaryocyte lineage, but they cannot be recognized by their morphology as megakaryocytes when isolated ex vivo. We propose that MP1 and MP2 represent subsequent, late stages of megakaryocyte development. MP1 are probably earlier than MP2. It remains to be determined whether MP1 and MP2 share a direct precursor-product relationship. IL-3 is a growth factor for the megakaryocyte lineage,35 and our data suggest that the effect of IL-3 on this lineage may occur at the level of the IL-3Rα+ MP2 rather than the IL-3Rα– MP1. It remains to be shown directly that IL-3Rα expression fluctuates during megakaryocyte development.
Relationship of CFU-Es and MPs
The CFU-E colony is composed of 16 to 32 erythroid cells. Thus, EPs undergo 4 to 5 cell divisions to form a CFU-E colony. Interestingly, MPs have a very similar limited expansion potential. One MP often differentiates into only one megakaryocyte, and this megakaryocyte gives rise to 16 to 32 platelets. Therefore, we propose that EP and MP populations represent correspondingly late stages in erythrocyte and megakaryocyte development, respectively. The expansion potential of both types of progenitors is similar, the difference being “extracellular” colony formation from CFU-E versus “intracellular” colony formation from MPs. Collectively, MPs represent a later stage in megakaryocyte development when compared to CMPs, or the megakaryocyte-committed progenitors defined by Nakorn et al (lin–c-Kit+Sca-1–IL7Rα–CD9+CD41+FcγRloCD34+CD38+).5 CD41 is megakaryocytic marker but CD41 can also be expressed on erythroid progenitors.36 A CD34+CD41+ population had bipotent erythroid/megakaryocytic rather than single erythroid potential. This progenitor appears, therefore, to be earlier than EPs. CD41+ progenitors that we tested in our model were megakaryocyte committed (Figure 1B), and EPs lacked CD41 expression (Figure 1A). Thus, CD41 may mark stages prior to EPs or may be expressed on a very small subset of CFU-Es not included in the EP phenotype.
Assignment of key hematopoietic properties to common versus committed myeloid progenitors
Analyses of isolated BM fractions revealed a clear correlation between key hematopoietic properties (CFU-S, BFU-E, CFU-E) and phenotypically defined stages of commitment from stem cells to myeloid lineages (Figure 6). BFU-E and CFU-S potential is restricted to CMPs. HSCs and CMPs both have CFU-S potential. Benzidine-positive BFU-E colonies originate from CMPs but not from HSCs within 8 days. Thus, from a practical viewpoint, CFU-Ss measure HSCs and CMPs, whereas benzidine-positive BFU-E colonies reflect CMP activity. CFU-E assays measure EPs because CFU-E potential is absent from HSCs, CMPs, and committed progenitors other than EPs. Because CFU-S activity is present in a common MEP37 but absent from MPs or EPs (Figure 6), it is likely that CFU-S potential is lost at the MEP-to-EP transition.
IL-3R α chain expression marks way-points in myelopoiesis
Cytokine receptor expression can be associated with lineage commitment,2,38 but cytokine receptor expression has not been widely used to purify progenitors. We have now divided myeloid progenitors into functionally distinct IL-3Rα+ and IL-3Rα– subpopulations. The analysis of IL-3Rα expression was crucial for the identification of EPs. It is, however, unlikely that IL-3 plays a major role for the generation or function of EPs because mice lacking IL-3, or components of the IL-3R, show normal steady-state hematopoiesis.39,40 Functional roles for IL-3 have been noted under conditions of limited signaling mediated by c-Kit,41 EpoR,30 or GM-CSF40 in vivo. At the CFU-E stage, c-Kit ligand (references in Waskow et al13 ) and Epo28 are crucial growth factor. Overexpression of Epo can partially substitute for lack of c-Kit at this stage.13 Despite this lack of a definitive functional assignment of IL-3 to certain stages of hematopoiesis, IL-3Rα expression marks important way-points in the normal BM (Figure 6) and in certain leukemias.42 It may be interesting to readdress the roles of IL-3 by applying the proposed IL-3Rα–based high-resolution map to steady state as well as pathologic hematopoiesis and for the precise characterization of hematopoietic mutants.
Prepublished online as Blood First Edition Paper, November 2, 2004; DOI 10.1182/blood-2004-09-3459.
Supported by the DFG-RO754/2-1 and the IZKF Ulm (H.-R.R.), by the Landesstiftung Baden-Württemberg P-LS-AL/11 (C.W.), by DFG-HO-493/12.1 (I.H.), and DFG-CA-306/1 (D.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs P. Besmer (New York, NY), H. Lodish (Boston, MA), A. Potocnik (London, United Kingdom), T. Borggrefe (Ulm, Germany), and H. J. Fehling (Ulm, Germany) for discussions; O. Hummel (Berlin, Germany) for help with data analysis, and G. Heller (Department for Internal Medicine III, Ulm, Germany) for blood parameter analyses.

![Figure 1. Phenotypic isolation, morphology, and potential of 6 distinct hematopoietic progenitor populations. (A) Lin–IL-7Rα–Sca-1– BM cells were analyzed for expression of c-Kit versus IL-3Rα (i) or c-Kit versus antibody isotype control (ii). Cells shown in i correspond to 2% of all nucleated BM cells. Lin–Sca-+–IL-7Rα– cells were sorted into c-Kit+IL-3Rα– (green gate in i) and c-Kit+IL-3Rα+ (red gate in i) populations. Cells shown in iii and xi correspond to 0.52% and 0.68% of all nucleated BM cells, respectively. Purified c-Kit+IL-3Rα– cells (iii) were restained for expression CD71 versus CD41 (iv), and sorted into CD71+CD41– (v), CD71–CD41– (vii), or CD71–CD41+ (ix) cells. Sorted c-Kit+IL-3Rα+ cells (xi) were stained for expression of FcγR (CD16/CD32) versus CD41 (xii), and further separated into FcγRhiCD41– (xiii), FcγRloCD41– (xv), or FcγRloCD41+ (xvii) cells. Numbers in i, iv, and xii are relative percentages of the populations in the indicated regions. Numbers shown in v, vii, ix, xiii, xv, and xvii are percentages of each population per total nucleated BM cells. Cytospins from each population were stained with May-Grünwald-Giemsa stain (vi,viii,x,xiv,xvi,xviii). The scale bar (shown in xviii) corresponds to 15 μM and applies to panels vi, viii, x, xiv, xvi, and xviii. Based on their potential, the 6 progenitors were designated as erythrocyte progenitors (EPs; v), IL-3Rα– common myeloid progenitors (CMP IL-3Rα–; vii), IL-3Rα – megakaryocyte progenitors (MP1; ix), granulocyte-monocyte progenitors (GMPs; xiii), IL-3Rα+ common myeloid progenitors (CMP IL-3Rα+; xv), and IL-3Rα+ megakaryocyte progenitors (MP2; xvii). (B) Purified BM progenitors (as shown in panel A) were plated at 2000 cells/culture into methylcellulose assays promoting the generation of BFU-E (i), CFU-Meg (ii), or CFU-GM (iii) colonies. Colonies were counted after 8 days. BFU-E colonies were typed by benzidine staining for globin. CFU-Meg and CFU-GM colonies were identified by morphology. EPs lack potential for BFU-E, CFU-Meg, or CFU-GM colonies. Data shown are representative for one of 3 experiments. Formation of more mature CFU-Ss day 8 (iv), and more immature CFU-Ss day 12 (v) was analyzed by transfer of HSCs (2500 cells/mouse; number of recipients [n] = 2), CMP IL-3Rα– (2500 cells/mouse; n = 2), CMP IL-3Rα+ (2500 cells/mouse; n = 2), GMP (2500 cells/mouse; n = 2), EP (2500 cells/mouse; n = 2), MP1 (2500 cells/mouse; n = 2), or MP2 (2500 cells/mouse; n = 2). Injection of total BM (2 × 105 cells/mouse; n = 2) or PBS (n = 2) served as positive and negative controls, respectively. After 8 or 12 days macroscopic colonies were counted. CFU-Ss are restricted to HSC and CMP subpopulations (iv-v). Data shown are representative for one of 2 experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/5/10.1182_blood-2004-09-3459/6/m_zh80050574850001.jpeg?Expires=1769090960&Signature=Aev8t~Fc8n8WipbplhFwAG5Lz5HQ2NFGKMe2YikexD~J3DTdQd701o7q0caa8l3o-6T27vwWbh-BtQoA3CAGU77MqaE97h7IXt4xfHuadojZcQZ8lC8WHR2jzf24~Nss1A5FOvckp3PXD0Z9PqjQdGjz10RakJnFk7M286XlBch29tfH01lUM5o-94Rg4CqaqOenEyIu8QSht0x0cg5DcyIAQ-Gy-p2LxgZXFD4NFqJ2-P-kt0DsAnsEU4B-x6ytrbSoVB9OhXi6GURNUvuiqnamCkxC7~Z6Wl3lQPv2Lv-HqV3tx5-mHHf-elzuNfXbWmPFmO~jGyUwZitlgsSvVg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Erythrocyte reconstitution in vivo. Host erythropoiesis was transiently suppressed by irradiation (850 rad; split dose). After irradiation, mice were given intravenous injections of PBS (number of recipients [n] = 2; A), total BM (2 × 105 cells/mouse; n = 2; B), HSC (2500 cells/mouse; n = 2; C), CMP IL-3Rα– cells (5000 cells/mouse; n = 2; D), EPs (5000 cells/mouse; n = 2; E), MP1 (5000 cells/mouse; n = 1; F), CMP IL-3Rα+ (5000 cells/mouse; n = 2; G), GMP (5000 cells/mouse; n = 2; H), or MP2 (5000 cells/mouse; n = 1; I). Percentages of reticulocytes in peripheral blood were determined at the indicated time points. Data shown are representative for one of 2 experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/5/10.1182_blood-2004-09-3459/6/m_zh80050574850003.jpeg?Expires=1769090960&Signature=bSWX6Z4Q0rpzOyq0PgIrKCoLaqjcqP00eFe0555lhXQGLabcFOuPlaiaUYOqYrJ8m1tykbdee57TnmPotsm35-aYMFjcip8w7mu4v1FW8sMp0xQeUAbuvoC1esP9R1n~1jYnA00LMu328ciyt3zVW-t6oOaVOb7jvP5x1ItCGz3QBmzzeG18u8F4syp-jCp3-5FSwbbQQbrsaR10hqUYJq41FUmR9CFTjjUbL2Uw1s6THBiJz0bIdaHynYfx4ZBOUe2cXNiGqdZx1j9SYlewXkeJmwuK8kGZq3z7JEs7nDvPeNqrYyvoLkEyYbel~JLdKpughwi-OoV1O2S2sQBH5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal