Abstract
The hyper immunoglobulin M (IgM) syndrome (HIGM), characterized by recurrent infections, low serum IgG and IgA, normal or elevated IgM, and defective class switch recombination and somatic hypermutation, is a heterogenous disorder with at least 5 distinct molecular defects, including mutations of the genes coding for the CD40 ligand (CD40L) and IKK-gamma (NEMO) genes, both X-linked; and mutations of CD40, activation-induced cytidine deaminase (AICDA), and uracil-DNA glycosylase (UNG), associated with autosomal recessive HIGM syndromes. To investigate the molecular basis of HIGM, we determined the prevalence of mutations affecting these 5 genes in a cohort of 140 patients (130 males and 10 females). Those patients without a molecular diagnosis were subsequently evaluated for mutations of the following genes: inducible CO-stimulator molecule (ICOS), ICOS ligand (ICOSL), and if male, Bruton tyrosine kinase (Btk) and SLAM-associated protein (SAP/SH2D1A). We found mutations of CD40L in 98 males; AICDA in 4 patients (3 males, 1 female); UNG in one adult male; and Btk in 3 boys. Of the remaining 25 males, one infant with hypohidrotic ectodermal dysplasia had a mutation of NEMO. None of the remaining 33 patients (24 males/9 females) had mutations affecting CD40, ICOS, ICOSL, or SH2D1, and are best classified as common variable immune deficiency (CVID), although other genes, including some not yet identified, may be responsible.
Introduction
Immunoglobulin (Ig) deficiency with “elevated” IgM (HIGM) is characterized by recurrent infections associated with low serum levels of IgG, IgA, normal or increased IgM, and grossly intact T-cell function. At least 5 genes, directly or indirectly involved in B-cell signaling via CD40 and required for class switch recombination (CSR) and somatic hypermutation (SHM), have been identified to cause HIGM if mutated. The most common form of HIGM is thought to be X-linked and the consequence of mutations of the CD40 ligand gene (CD40L, HIGM1, Xq26).1-5 Most CD40L-deficient patients present in infancy with severe recurrent sinopulmonary infections, Pneumocystis carinii pneumonia (PCP), chronic diarrhea due to Cryptosporidium infection that may lead to sclerosing cholangitis and intermittent or persistent neutropenia.2-7 Mutations of CD40 (HIGM3, 20q12-q13.2), the receptor of CD40L, is a rare autosomal recessive disorder, presenting with a clinical phenotype similar to that observed in males with CD40L deficiency.8,9 A more frequent genetically defined HIGM syndrome with autosomal recessive inheritance is caused by mutations of the activation-induced cytidine deaminase gene (AICDA or AID, HIGM2, 12q13) expressed in activated B cells. Patients with AID deficiency often have lymphoid hyperplasia, lack opportunistic infections, and generally have a better prognosis.10,11 Another gene expressed in activated B cells, uracil-DNA glycosylase (UNG, 12q24.1), is known to generate abasic sites within the variable Ig regions, a process required for efficient CSR and SHM.12 UNG-deficient mice characteristically have defective CSR, SHM, and decreased deoxyguanine (dG) and deoxycytidine (dC) transversion.13 Recently, 3 unrelated patients with autosomal recessive HIGM due to a mutation of UNG have been reported.14 A second X-linked form of HIGM is caused by mutations of the nuclear factor κB (NF-κB) essential modulator gene (NEMO, Xq28), also known as IKK gamma.15 NEMO serves as a scaffold protein, forming a complex with IKKα and IKKβ, 2 phosphokinases. Following CD40 crosslinking, NF-κB inhibiting protein (I-κB) is phosphorylated, resulting in its degradation and the release of NF-κB, allowing its transfer from the cytoplasma to the nucleus. Mutations of NEMO block NF-κB release and thus interfere with NF-κB signaling. Male patients with mutations of NEMO may present with hypohidrotic (anhidrotic) ectodermal dysplasia (EDA) and immunodeficiency (ID)16-18 ; some, but not all, have hypogammaglobulinemia with elevated IgM.18
Inducible costimulator molecule (ICOS, 2q33), a third member of the CD28/CTLA4 family, is expressed on activated T cells19 and interacts with its ligands, human GL50 and B7-H2 (or B7-related protein-1, B7RP-1), 2 splice variants constitutively expressed by human B cells.20,21 Marked defects in CSR and germinal center formation were observed in ICOS and ICOSL (B7-H2) knockout mice.22,23 These findings in mice and the recent discovery of homozygous mutations of ICOS in adult patients with common variable immunodeficiency (CVID) phenotype24 make this receptor/ligand pair candidate genes for patients with the HIGM phenotype. An HIGM/CVID phenotype has also been observed in subsets of male patients with mutations of Btk or SAP/SH2D1A,25,26 the genes responsible for X-linked agammaglobulinemia (XLA) and X-linked lymphoproliferative disease (XLP), respectively. Other defined primary immune deficiency disorders may present with an HIGM phenotype. Approximately 20% of patients with hypogammaglobulinemia and major histocompatibility complex class II (MHCII) deficiency and a subset of patients with ataxia telangiectasia (AT) have elevated serum IgM levels. These disorders, however, can be recognized by experienced clinicians and confirmed by simple laboratory tests; for example, lack of MHCII molecules on the surface of B cells or elevated α-fetoprotein in the serum of patients with AT. However, in a considerable proportion of patients with the HIGM phenotype, the molecular defect remains unknown.27
In this study, we investigated the molecular basis of HIGM in a large, mainly North American cohort of patients with the HIGM phenotype. Initially, we focused on genes known to cause HIGM if mutated, including CD40L, CD40, AICDA, UNG, and NEMO. If their genes were not mutated, male patients were evaluated for mutations of Btk and SAP. The remaining patients, both male and female, were studied for possible mutations of ICOS and ICOSL. With this approach we successfully identified a genetic defect in all but 33 individuals from the original cohort of 140 patients with HIGM belonging to 115 unrelated families.
Patients, materials, and methods
Patients
The patients included in this study were followed in our Immunodeficiency Clinic or were cared for by US, Canadian, Japanese, or Thai physicians between 1993 and 2003. All expressed MHCII and none had clinical findings suggesting AT. Patients with persistent lymphopenia, abnormal mitogen responses, human immunodeficiency virus or congenital rubella infections, antiepileptic or immunosuppressive drug exposure, or neoplastic disease were excluded. To be accepted in the study, patients had to have low serum IgG and IgA (≤ 2 standard deviation [SD] below normal values for age), normal or elevated IgM, and recurrent or severe infections. After informed consent was obtained, 10 mL to 20 mL venous blood was collected from each patient into heparin-containing syringes and delivered to our laboratory within 24 to 72 hours. When possible, blood samples from other family members and a healthy adult control were obtained simultaneously.
Cell preparations and culture
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque (Pharmacia Biotech, Piscataway, NJ) centrifugation. RPMI 1640 supplemented with 10% fetal calf serum (FCS; Hyclone, Logan, UT), 2 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (complete medium) was used for culture. Interleukin-2 (IL-2)–dependent T-cell lines were established from PBMCs using a standard method.28 B-cell lines were established as previously described using Epstein Barr virus (EBV) prepared from B95-8 cells.29
Detection of CD40L and ICOS expression by flow cytometry
The expression of CD40L and ICOS by activated T cells was evaluated by immunostaining as described previously.24,30,31 Briefly, fresh PBMCs were stained with phycoerythrin (PE)–labeled anti-CD4 monoclonal antibody (mAb) (IgG1; Ancell, Bayport, MN) at 4°C for 20 minutes, washed and incubated overnight in the presence of phorbol 12-myristate 13-acetate (PMA; GIBCO-BRL, Gaithersburg, MD) at 10 ng/mL and ionomycin (Sigma, St Louis, MO) at 1 μg/mL. We used 2 or more anti–human CD40L mAbs including mAb 106 (mouse IgG1; provided by Dr Tony Siadak, formerly at Bristol-Myers Squibb, Pharmaceutical Research Institute, Seattle, WA), mAb 5c8 (mouse IgG2a; provided by Biogen, Cambridge, MA), and a commercially available anti–human CD40L mAb (mouse IgG1; Pharmingen, San Diego, CA). In addition, we identified CD40L expression with a biotinylated-CD40 human Ig fusion protein (provided by Dr Stephen J. Klaus, formerly at the Department of Microbiology, University of Washington, Seattle) or with a human CD40 mouse Ig fusion protein (Ancell); fluorescein isothiocyanate (FITC)–conjugated streptavidin (Biosource International, Camarillo, CA) or FITC-labeled goat anti–mouse IgG1 (SouthernBiotech, Birmingham, AL) were used as secondary antibody. Immunostaining for ICOS was performed by incubating the activated cells with the ICOS ligand fusion protein B7RP-1 Fc (a gift from Amgen, Thousand Oaks, CA) at a concentration of 20 μg/mL for one hour at room temperature; the cells were then washed and stained with FITC-conjugated anti-human Fc antibody (Chemicon, Temecula, CA) at a final dilution of 1:200. FITC-labeled anti-human CD69 (mouse IgG1; Pharmingen) served as activation control. The immunostained cells underwent flow cytometric analysis (FACScan; Becton Dickinson, San Jose, CA); a minimum of 10 000 events were evaluated and analyzed by Lysis II software (Becton Dickinson).
Sequence analysis of the CD40L, CD40, AICDA, UNG, NEMO, ICOS, ICOSL, Btk, and SAP/SH2D1A genes
Total RNA was isolated from activated PBMCs (4-hour incubation with PMA at 10 ng/mL and ionomycin at 1 μg/mL) with TRIzol (Invitrogen, Carlsbad, CA). Reverse transcription of mRNA followed by polymerase chain reaction (RT-PCR) was performed as follows: 1 μgto2 μg total RNA in a volume of 20 μL was reverse-transcribed into cDNA using oligo-dT primers and SuperScript II reverse transcriptase (Invitrogen). Amplification was performed using 1 μL cDNA in a volume of 20 μL containing 0.5 U High Fidelity Taq DNA polymerase, 1.875 mM MgSO4, 200 μM deoxynucleoside triphosphate (dNTP), 500 nM of each pair of oligonucleotide primers and 10× buffer (Invitrogen). One or 2 pairs of oligonucleotide primers (Table 1) were selected for each gene to cover the entire coding region. The mutations identified from cDNA were confirmed by sequence analysis of genomic DNA. The individual exons, including exon-intron boundaries, were amplified by previously designed primers.8,10,14,23,26,31-34 Sequencing of the products was performed using the Big Dye Terminator kit (Applied Biosystems, Foster City, CA) and an ABI PRISM 3100 DNA Sequencer (Applied Biosystems).
Primers used for RT-PCR
. | Complementary DNA . | . | Reference . | |
|---|---|---|---|---|
. | Forward primer (5′→3′) . | Reverse primer (5′→3′) . | Genebank . | |
| CD40L | GCC AGA AGA TAC CAT TTC AAC | CCG CTG TGC TGT ATT ATG AA | NM 000074 | |
| CD40 | GTG GTC CTG CCG CCT GGT CTC | GCT CCA GGG TGA AGT GAG AG | X 60592 | |
| AICDA | CAG GGA GGC AAG AAG ACA CT | TGA CAT TCC TGG AAG TTG CT | AB 040431 | |
| UNG | CAG CTC CAG GAT GAT CGG CCA | ATT CCT GGA CAG CCT GGT TC | NM 080911 | |
| NEMO2-4 | CCC TTG CCC TGT TGG ATG | ACC CTC CAG AGC CTG GCA TTC | NM 003639 | |
| NEMO4-10 | GAC AAG GCC TCT GTG AAG C | TCA GAC GCG AAA GGA GA GG | XM 003639 | |
| ICOS | GAA CAC TGA ACG CGA GGA CT | GTT GAG GAA AAC TGG CCA AC | AJ 277832 | |
| ICOSL | GTT GCT CCT CTC CGA GGT CT | GCC TCA TTC CAG GAT CAC AG | NM 015259 | |
| Btk1-13 | CAG TGT CTG CTG CGA TCG AG | CAG TGG AAG GTG CAT TCT TG | NM 000061 | |
| Btk11-19 | TCA TTG TCA GAG ACT CCA GC | TTG CTC AGA AGC CAC TAT CC | NM 000061 | |
| SAP | GTT GAC TTG TGC CTG GCT GC | ATT TGT AGC TCA CCG AAC TG | NM 002351 | |
. | Complementary DNA . | . | Reference . | |
|---|---|---|---|---|
. | Forward primer (5′→3′) . | Reverse primer (5′→3′) . | Genebank . | |
| CD40L | GCC AGA AGA TAC CAT TTC AAC | CCG CTG TGC TGT ATT ATG AA | NM 000074 | |
| CD40 | GTG GTC CTG CCG CCT GGT CTC | GCT CCA GGG TGA AGT GAG AG | X 60592 | |
| AICDA | CAG GGA GGC AAG AAG ACA CT | TGA CAT TCC TGG AAG TTG CT | AB 040431 | |
| UNG | CAG CTC CAG GAT GAT CGG CCA | ATT CCT GGA CAG CCT GGT TC | NM 080911 | |
| NEMO2-4 | CCC TTG CCC TGT TGG ATG | ACC CTC CAG AGC CTG GCA TTC | NM 003639 | |
| NEMO4-10 | GAC AAG GCC TCT GTG AAG C | TCA GAC GCG AAA GGA GA GG | XM 003639 | |
| ICOS | GAA CAC TGA ACG CGA GGA CT | GTT GAG GAA AAC TGG CCA AC | AJ 277832 | |
| ICOSL | GTT GCT CCT CTC CGA GGT CT | GCC TCA TTC CAG GAT CAC AG | NM 015259 | |
| Btk1-13 | CAG TGT CTG CTG CGA TCG AG | CAG TGG AAG GTG CAT TCT TG | NM 000061 | |
| Btk11-19 | TCA TTG TCA GAG ACT CCA GC | TTG CTC AGA AGC CAC TAT CC | NM 000061 | |
| SAP | GTT GAC TTG TGC CTG GCT GC | ATT TGT AGC TCA CCG AAC TG | NM 002351 | |
Results
X-linked HIGM and CD40L mutations
Because mutations of CD40L are thought to be the most common cause of HIGM, we began our investigation by testing activated lymphocytes from every male patient with HIGM (n = 130) for the expression of CD40L. The exclusive use of mAbs for the identification of CD40L would have missed 32% of CD40L mutations, falsely suggesting a normal staining pattern (Table 2). In contrast, binding of the CD40-Ig fusion protein by activated CD4+ T cells was observed in only 5 of 61 unique mutations (8%) and staining always was less intense than in healthy controls. One of these 5 patients (patient 1 in Table 2) with residual CD40L activity, reported previously,31,35 had a nonsense mutation affecting Arg11 located within the intracellular domain. This mutation resulted in the use of Met21 as initiation codon and expression of a truncated protein that lacked the first 20 amino acids, affecting efficiency of production or stability. The remaining 4 patients had splice site mutations (IVS2 + 2t>a; 367G>A; IVS3 + 2t>c; IVS3 + 5g>c) that allowed the generation of small amounts of normally spliced CD40L. In addition, we sequenced CD40L cDNA (and genomic DNA if a mutation was identified) from every male patient with HIGM and found that 98 individuals from 77 unrelated families (including brothers and cousins from 15 families) had mutations of CD40L (Table 2).
Mutations of the CD40L gene and CD40L expression in patients with X-linked HIGM
. | . | . | . | . | Protein expression by flow cytometry . | . | . | |
|---|---|---|---|---|---|---|---|---|
| Family/Patient . | Genomic DNA mutation (nucleotide no.)* . | cDNA mutation (nucleotide no.)*if different from gDNA . | Predicted effect on protein . | Affected domain . | Anti-CD40L mAb . | CD40-Ig . | Reference‡ . | |
| Exon 1/Intron 1 | ||||||||
| Nonsense and missense mutations | ||||||||
| 1 | 52C > T | — | Arg 11 stop | IC | + | (+) | 31,35 | |
| 2 | 99T > G† | — | Tyr 26 stop | TM | D | D | ||
| 3 | 128T > G | — | Met 36 Arg | TM | NA | NA | 36 | |
| 4 | 175A > T | — | Lys 52 stop | EC | D | D | ||
| Splice site mutations | ||||||||
| 5 | IVS1 + 1 g > a† | Del 82 bp (nt 96-177) | fs, stop at 26 | TM | D | D | ||
| 6 | IVS1 + 1 g > t | Del 82 bp (nt 96-177) | fs, stop at 26 | TM | D | D | ||
| Del 10 bp (nt 168-177) | fs, stop at 50 | EC | ||||||
| Wild type cDNA | ||||||||
| 7 a,b,c | IVS1 + 1 g > t | Del 82 bp (nt 96-177) | fs, stop at 26 | EC | D | D | 31 | |
| Del 43 bp (nt 135-177) | fs, stop at 39 | EC | ||||||
| Del 10 bp (nt 168-177) | fs, stop at 50 | TM | ||||||
| Del 14 bp (nt 168-181) | fs, stop at 51 | TM | ||||||
| Wild type cDNA | ||||||||
| 8 | IVS1 + 1 g > t | Del 82 bp (nt 96-177) | fs, stop at 26 | TM | D | D | ||
| Wild type cDNA | ||||||||
| Exon2/Intron 2 | ||||||||
| Nonsense mutations | ||||||||
| 9 | 187G > T | — | Glu 56 stop | EC | D | D | 31 | |
| 10 | 229C > T | — | Gln 70 stop | EC | D | D | ||
| Deletions | ||||||||
| 11 | Del 9 bp aatagATAG | 178-81 Del ATAG | fs, stop at 65 | EC | NA | NA | ||
| 12 | Del 8 bp atagATAG | 178-81 Del ATAG | fs, stop at 65 | EC | NA | NA | 31,36 | |
| 13 | Del 8 bp atagATAG | 178-81 Del ATAG | fs, stop at 65 | EC | D | D | ||
| 14 | Del 4 bp ATAG | 178-81 Del ATAG | fs, stop at 65 | EC | NA | NA | ||
| 15 | Del 2 bp CA (229-30) | 229-30 Del CA | fs, stop at 84 | EC | NA | NA | ||
| Splice site mutations | ||||||||
| 16 a,b | IVS2 + 1 g > a | E2 skip | Inframe del 44 aa | EC | + | D | 31,36 | |
| Ins 19 bp (5′ part of 12) | fs, stop at 118 | |||||||
| 17 | IVS2 + 2 t > a | E2 skip | Inframe del 44 aa | EC | + | (+) | 31,35 | |
| Ins 19 bp (5′ part of 12) | fs, stop at 118 | |||||||
| Wild type cDNA | ||||||||
| Exon 3/Intron 3 | ||||||||
| Deletions | ||||||||
| 18 | 344-7 Del A† | A344-7 Del A | fs, stop at 127 | EC | D | D | ||
| Splice site mutations | ||||||||
| 19 | 367G > A | E3 skip (del 310-67) | fs, stop at 108 | EC | + | (+) | 31 | |
| Wild type cDNA | ||||||||
| 20 | 367G > C† | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| Lys 96 Asn | ||||||||
| 21 | IVS3 + 1 g > a | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 22 | IVS3 + 1 g > a | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 23 | IVS3 + 1 g > a | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 24 | IVS3 + 1 g Del | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 25 | IVS3 + 2 t > c | E3 skip (del 310-67) | fs, stop at 108 | EC | + | (+) | 31 | |
| Wild type cDNA | ||||||||
| 26 | IVS3 + 5 g > c | E3 skip (del 310-67) | fs, stop at 108 | EC | + | (+) | 31 | |
| Wild type cDNA | ||||||||
| 27 | IVS3-915a > t† | New splice site Ins 59 bp between E3&E4 | fs, stop at 127 | EC(TNF) | D | D | ||
| 28 a,b | IVS3-1 g > a | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | NA | NA | ||
| 29 | IVS3-1 g > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | D | NA | ||
| Exon 4/Intron 4 | ||||||||
| Missense mutations | ||||||||
| 30 | 405T > A | — | Ser 128 Arg | EC(TNF) | + | D | 31 | |
| 407A > G | — | Glu 129 Gly | ||||||
| Insertions | ||||||||
| 31 | 403-405 ins G | — | fs, stop at 129 | EC(TNF) | D | D | 31 | |
| 32 | 403-405 ins G | — | fs, stop at 129 | EC(TNF) | D | D | 31 | |
| 33 a,b,c | 411-413 ins A | — | fs, stop at 133 | EC(TNF) | NA | NA | 37 | |
| 34 | 417-422 ins A† | — | fs, stop at 142 | EC(TNF) | NA | NA | ||
| Splice site mutations | ||||||||
| 35 a,b,c,d,e | IVS4 + 1 g > c | E4 skip (del 368-430) | Inframe del 21 aa fs | EC(TNF) | + | D | 31 | |
| Ins 5′ part of IVS4 (23, 73, 152 bp) | ||||||||
| 36 | IVS4 + 1 g > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | + | D | 31 | |
| 37 | IVS4 + 1 g > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | NA | NA | ||
| 38 | IVS4 + 2 i > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | D | NA | ||
| Exon 5/Intron 5 | ||||||||
| Nonsense and missense mutations | ||||||||
| 39 | 436C > T† | — | Gln 139 stop | EC(TNF) | D | D | ||
| 40 | 440G > A | — | Trp 140 stop | EC(TNF) | D | D | 36 | |
| 41 | 456C > A | — | Thy 145 stop | EC(TNF) | D | NA | ||
| 42 | 461C > A | — | Thr 147 Asn | EC(TNF) | + | D | 31 | |
| 43 | 517C > T | — | Gln 166 stop | EC(TNF) | D | D | ||
| 44 a,b | 517C > T | — | Gln 166 stop | EC(TNF) | D | D | ||
| 45 a,b | 526T > G | — | Tyr 169 Asp | EC(TNF) | D | NA | ||
| 46 a,b | 530C > A | — | Tyr 170 Cys | EC(TNF) | D | D | 31 | |
| 47 | 541C > T† | — | Gln 174 stop | EC(TNF) | D | D | ||
| 48 | 577C > T | — | Gln 186 stop | EC(TNF) | D | D | 31 | |
| 49 | 577C > T | — | Gln 186 stop | EC(TNF) | NA | NA | ||
| 50 | 619A > T | — | Arg 200 stop | EC(TNF) | NA | NA | ||
| 51 | 619A > T | — | Arg 200 stop | EC(TNF) | D | D | ||
| 52 | 629G > T† | — | Arg 203 Ile | EC(TNF) | + | D | ||
| 53 | 675C > A | — | Cys 218 stop | EC(TNF) | NA | NA | ||
| 54 | 675C > A | — | Cys 218 stop | EC(TNF) | D | NA | ||
| 55 | 679C > T | — | Gln 220 stop | EC(TNF) | D | NA | ||
| 56 | 679C > T | — | Gln 220 stop | EC(TNF) | D | D | ||
| 57 | 682C > T | — | Gln 221 stop | EC(TNF) | D | D | 31 | |
| 58 | 701G > T | — | Gly 227 Val | EC(TNF) | + | D | 31 | |
| 59 a,b | 713G > T | — | Leu 231 Ser | EC(TNF) | NA | NA | 31 | |
| 60 a,b | 724G > C | — | Ala 235 Pro | EC(TNF) | D | D | 31 | |
| 61 | 782C > T | — | Thr 254 Met | EC(TNF) | + | D | ||
| 62 | 782C > T | — | Thr 254 Met | EC(TNF) | + | D | 31,35 | |
| 63 | 782C > T | — | Thr 254 Met | EC(TNF) | + | D | 31 | |
| 64 a,b,c | 794T > C | — | Leu 258 Ser | EC(TNF) | + | D | 31 | |
| 65 a,b | 794T > C | — | Leu 258 Ser | EC(TNF) | + | D | 31 | |
| Deletions | ||||||||
| 66 | IVS4-1 g Del | Del 8 bp (431-8) | fs, stop 139 | EC(TNF) | NA | NA | 31 | |
| 67 | 445-50 Del 6 bp† | DelGAA AAA | Del 142 Glu, 143 Lys | EC(TNF) | + | D | ||
| 68 | 454-6 Del 3 bp† | Del TAC | Del 145 Tyr | EC(TNF) | + | D | ||
| 69 | 580 Del G† | — | fs, stop 190 | EC(TNF) | D | NA | ||
| 70 | 610-616 Del C† | — | fs, stop 241 | EC(TNF) | NA | NA | ||
| 71 | 699-701 Del 3 bp | Del AGG | Del 227 Gly | EC(TNF) | NA | NA | 36 | |
| Insertions | ||||||||
| 72 a,b | Ins 12 bp between IVS4-1 & nt 431 | Ins 12 bp between E4&E5 | Ins 4 aa between E4&E5 | EC(TNF) | D | D | ||
| 73 | Ins 9 bp between IVS4-1 & nt 431 | Ins 9 bp between E4&E5 | Ins 3 aa between E4&E5 | EC(TNF) | + | D | ||
| 74 | (445-451) insert A | — | fs, stop 164 | EC(TNF) | D | D | 31 | |
| 75 a,b | Duplicate CAGCC between 593C&599T | — | fs, stop 197 | EC(TNF) | D | D | 31 | |
| Deletions involving more than 2 exons | ||||||||
| 76 a,b | Del 33Kb including E1, E2, E3 | Del E1-E3 | Unstable protein | IC,TM,EC | D | D | 31 | |
| 77 | Del 56 Kb including E1-5 | Nondetectable cDNA | Unstable protein | IC,TM,EC | D | D | ||
. | . | . | . | . | Protein expression by flow cytometry . | . | . | |
|---|---|---|---|---|---|---|---|---|
| Family/Patient . | Genomic DNA mutation (nucleotide no.)* . | cDNA mutation (nucleotide no.)*if different from gDNA . | Predicted effect on protein . | Affected domain . | Anti-CD40L mAb . | CD40-Ig . | Reference‡ . | |
| Exon 1/Intron 1 | ||||||||
| Nonsense and missense mutations | ||||||||
| 1 | 52C > T | — | Arg 11 stop | IC | + | (+) | 31,35 | |
| 2 | 99T > G† | — | Tyr 26 stop | TM | D | D | ||
| 3 | 128T > G | — | Met 36 Arg | TM | NA | NA | 36 | |
| 4 | 175A > T | — | Lys 52 stop | EC | D | D | ||
| Splice site mutations | ||||||||
| 5 | IVS1 + 1 g > a† | Del 82 bp (nt 96-177) | fs, stop at 26 | TM | D | D | ||
| 6 | IVS1 + 1 g > t | Del 82 bp (nt 96-177) | fs, stop at 26 | TM | D | D | ||
| Del 10 bp (nt 168-177) | fs, stop at 50 | EC | ||||||
| Wild type cDNA | ||||||||
| 7 a,b,c | IVS1 + 1 g > t | Del 82 bp (nt 96-177) | fs, stop at 26 | EC | D | D | 31 | |
| Del 43 bp (nt 135-177) | fs, stop at 39 | EC | ||||||
| Del 10 bp (nt 168-177) | fs, stop at 50 | TM | ||||||
| Del 14 bp (nt 168-181) | fs, stop at 51 | TM | ||||||
| Wild type cDNA | ||||||||
| 8 | IVS1 + 1 g > t | Del 82 bp (nt 96-177) | fs, stop at 26 | TM | D | D | ||
| Wild type cDNA | ||||||||
| Exon2/Intron 2 | ||||||||
| Nonsense mutations | ||||||||
| 9 | 187G > T | — | Glu 56 stop | EC | D | D | 31 | |
| 10 | 229C > T | — | Gln 70 stop | EC | D | D | ||
| Deletions | ||||||||
| 11 | Del 9 bp aatagATAG | 178-81 Del ATAG | fs, stop at 65 | EC | NA | NA | ||
| 12 | Del 8 bp atagATAG | 178-81 Del ATAG | fs, stop at 65 | EC | NA | NA | 31,36 | |
| 13 | Del 8 bp atagATAG | 178-81 Del ATAG | fs, stop at 65 | EC | D | D | ||
| 14 | Del 4 bp ATAG | 178-81 Del ATAG | fs, stop at 65 | EC | NA | NA | ||
| 15 | Del 2 bp CA (229-30) | 229-30 Del CA | fs, stop at 84 | EC | NA | NA | ||
| Splice site mutations | ||||||||
| 16 a,b | IVS2 + 1 g > a | E2 skip | Inframe del 44 aa | EC | + | D | 31,36 | |
| Ins 19 bp (5′ part of 12) | fs, stop at 118 | |||||||
| 17 | IVS2 + 2 t > a | E2 skip | Inframe del 44 aa | EC | + | (+) | 31,35 | |
| Ins 19 bp (5′ part of 12) | fs, stop at 118 | |||||||
| Wild type cDNA | ||||||||
| Exon 3/Intron 3 | ||||||||
| Deletions | ||||||||
| 18 | 344-7 Del A† | A344-7 Del A | fs, stop at 127 | EC | D | D | ||
| Splice site mutations | ||||||||
| 19 | 367G > A | E3 skip (del 310-67) | fs, stop at 108 | EC | + | (+) | 31 | |
| Wild type cDNA | ||||||||
| 20 | 367G > C† | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| Lys 96 Asn | ||||||||
| 21 | IVS3 + 1 g > a | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 22 | IVS3 + 1 g > a | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 23 | IVS3 + 1 g > a | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 24 | IVS3 + 1 g Del | E3 skip (del 310-67) | fs, stop at 108 | EC | D | D | ||
| 25 | IVS3 + 2 t > c | E3 skip (del 310-67) | fs, stop at 108 | EC | + | (+) | 31 | |
| Wild type cDNA | ||||||||
| 26 | IVS3 + 5 g > c | E3 skip (del 310-67) | fs, stop at 108 | EC | + | (+) | 31 | |
| Wild type cDNA | ||||||||
| 27 | IVS3-915a > t† | New splice site Ins 59 bp between E3&E4 | fs, stop at 127 | EC(TNF) | D | D | ||
| 28 a,b | IVS3-1 g > a | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | NA | NA | ||
| 29 | IVS3-1 g > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | D | NA | ||
| Exon 4/Intron 4 | ||||||||
| Missense mutations | ||||||||
| 30 | 405T > A | — | Ser 128 Arg | EC(TNF) | + | D | 31 | |
| 407A > G | — | Glu 129 Gly | ||||||
| Insertions | ||||||||
| 31 | 403-405 ins G | — | fs, stop at 129 | EC(TNF) | D | D | 31 | |
| 32 | 403-405 ins G | — | fs, stop at 129 | EC(TNF) | D | D | 31 | |
| 33 a,b,c | 411-413 ins A | — | fs, stop at 133 | EC(TNF) | NA | NA | 37 | |
| 34 | 417-422 ins A† | — | fs, stop at 142 | EC(TNF) | NA | NA | ||
| Splice site mutations | ||||||||
| 35 a,b,c,d,e | IVS4 + 1 g > c | E4 skip (del 368-430) | Inframe del 21 aa fs | EC(TNF) | + | D | 31 | |
| Ins 5′ part of IVS4 (23, 73, 152 bp) | ||||||||
| 36 | IVS4 + 1 g > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | + | D | 31 | |
| 37 | IVS4 + 1 g > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | NA | NA | ||
| 38 | IVS4 + 2 i > c | E4 skip (del 368-430) | Inframe del 21 aa | EC(TNF) | D | NA | ||
| Exon 5/Intron 5 | ||||||||
| Nonsense and missense mutations | ||||||||
| 39 | 436C > T† | — | Gln 139 stop | EC(TNF) | D | D | ||
| 40 | 440G > A | — | Trp 140 stop | EC(TNF) | D | D | 36 | |
| 41 | 456C > A | — | Thy 145 stop | EC(TNF) | D | NA | ||
| 42 | 461C > A | — | Thr 147 Asn | EC(TNF) | + | D | 31 | |
| 43 | 517C > T | — | Gln 166 stop | EC(TNF) | D | D | ||
| 44 a,b | 517C > T | — | Gln 166 stop | EC(TNF) | D | D | ||
| 45 a,b | 526T > G | — | Tyr 169 Asp | EC(TNF) | D | NA | ||
| 46 a,b | 530C > A | — | Tyr 170 Cys | EC(TNF) | D | D | 31 | |
| 47 | 541C > T† | — | Gln 174 stop | EC(TNF) | D | D | ||
| 48 | 577C > T | — | Gln 186 stop | EC(TNF) | D | D | 31 | |
| 49 | 577C > T | — | Gln 186 stop | EC(TNF) | NA | NA | ||
| 50 | 619A > T | — | Arg 200 stop | EC(TNF) | NA | NA | ||
| 51 | 619A > T | — | Arg 200 stop | EC(TNF) | D | D | ||
| 52 | 629G > T† | — | Arg 203 Ile | EC(TNF) | + | D | ||
| 53 | 675C > A | — | Cys 218 stop | EC(TNF) | NA | NA | ||
| 54 | 675C > A | — | Cys 218 stop | EC(TNF) | D | NA | ||
| 55 | 679C > T | — | Gln 220 stop | EC(TNF) | D | NA | ||
| 56 | 679C > T | — | Gln 220 stop | EC(TNF) | D | D | ||
| 57 | 682C > T | — | Gln 221 stop | EC(TNF) | D | D | 31 | |
| 58 | 701G > T | — | Gly 227 Val | EC(TNF) | + | D | 31 | |
| 59 a,b | 713G > T | — | Leu 231 Ser | EC(TNF) | NA | NA | 31 | |
| 60 a,b | 724G > C | — | Ala 235 Pro | EC(TNF) | D | D | 31 | |
| 61 | 782C > T | — | Thr 254 Met | EC(TNF) | + | D | ||
| 62 | 782C > T | — | Thr 254 Met | EC(TNF) | + | D | 31,35 | |
| 63 | 782C > T | — | Thr 254 Met | EC(TNF) | + | D | 31 | |
| 64 a,b,c | 794T > C | — | Leu 258 Ser | EC(TNF) | + | D | 31 | |
| 65 a,b | 794T > C | — | Leu 258 Ser | EC(TNF) | + | D | 31 | |
| Deletions | ||||||||
| 66 | IVS4-1 g Del | Del 8 bp (431-8) | fs, stop 139 | EC(TNF) | NA | NA | 31 | |
| 67 | 445-50 Del 6 bp† | DelGAA AAA | Del 142 Glu, 143 Lys | EC(TNF) | + | D | ||
| 68 | 454-6 Del 3 bp† | Del TAC | Del 145 Tyr | EC(TNF) | + | D | ||
| 69 | 580 Del G† | — | fs, stop 190 | EC(TNF) | D | NA | ||
| 70 | 610-616 Del C† | — | fs, stop 241 | EC(TNF) | NA | NA | ||
| 71 | 699-701 Del 3 bp | Del AGG | Del 227 Gly | EC(TNF) | NA | NA | 36 | |
| Insertions | ||||||||
| 72 a,b | Ins 12 bp between IVS4-1 & nt 431 | Ins 12 bp between E4&E5 | Ins 4 aa between E4&E5 | EC(TNF) | D | D | ||
| 73 | Ins 9 bp between IVS4-1 & nt 431 | Ins 9 bp between E4&E5 | Ins 3 aa between E4&E5 | EC(TNF) | + | D | ||
| 74 | (445-451) insert A | — | fs, stop 164 | EC(TNF) | D | D | 31 | |
| 75 a,b | Duplicate CAGCC between 593C&599T | — | fs, stop 197 | EC(TNF) | D | D | 31 | |
| Deletions involving more than 2 exons | ||||||||
| 76 a,b | Del 33Kb including E1, E2, E3 | Del E1-E3 | Unstable protein | IC,TM,EC | D | D | 31 | |
| 77 | Del 56 Kb including E1-5 | Nondetectable cDNA | Unstable protein | IC,TM,EC | D | D | ||
The small letters (a,b,c,d,e) indicate members from the same family.
Del indicates deletion; ins, insertion; fs, frameshift; E, exon; I, intron; IC, intracytoplasmic; TM, transmembrane; EC, extracellular; TNF, tumor necrosis factor homology domain; +, CD40L expression is normal; (+), CD40L detected, but decreased compared with normal; D, dramatically decreased or absent CD40L expression; NA, not available because of limited blood samples; —, no difference from gDNA.
Nucleotide number is based on the sequence data described by Hollenbaugh et al.57
Novel mutation, not yet reported.
References in which a specific patient was reported previously (see “References”).
Of those, 61 mutations were unique and consisted of 11 missense (observed in 14 unrelated families) and 15 nonsense mutations (20 families); 13 deletions (3 in frame) were observed in 14 unrelated families, including 2 large deletions: patient 76 had a 33 996 bp deletion from upstream 21 669 to downstream 55 634, resulting in deletion of exons 1-3, and patient 77 had a 56 294 bp deletion from upstream 22 297 to downstream 78 590, resulting in deletion of the entire coding region (based on Genebank: AL135 783). Seven unique insertions (2 in frame) were identified in 8 families, and 15 distinct splice site mutations were found in 21 families. Only one mutation (Arg11stop) affected the intracellular domain. Mutations within the transmembrane domain were identified in 5 families; the rest of the mutations affected exons 2-5, which code for the extracellular domain. Four possible hotspot mutations were identified; each one was observed in 3 unrelated families. These include a splice site mutation in intron 1 (IVS1 + 1g>t); a splice site mutation in intron 3 (IVS3 + 1g>a); a splice site mutation in intron 4 (IVS4 + 1g>c), and a missense mutation (782C>T [Thr254Met]) in exon 5. Based on published data, 13 of the 61 unique mutations observed were novel and not yet published (Table 2), including one missense mutation (Arg203Ile), 3 nonsense mutations (Tyr26stop; Gln139stop; Gln174stop), one insertion (417-422insA), 5 deletions ([344-347] delA; del 6bp resulting in the inframe deletion of 142Glu and 143Leu; del 3bp resulting in deletion of 145Tyr; 580delG; [610-616] delC), and 3 splice site mutations (IVS1 + 1g>a, resulting in a deletion of 85 bp and stop at codon 26; 367G>C, resulting in skipping of exon 3, frameshift and stop at codon 108; and IVS3-915a>t, creating a new splice site and activating a cryptic splice site at –1019, resulting in the insertion of 59 bp between exons 3 and 4).
More than half of the patients with CD40L mutations were diagnosed before one year of age and generally had a more severe clinical phenotype than patients with HIGM with normal CD40L. Lymphoid hyperplasia was not reported, and serum IgM levels were only moderately elevated and not infrequently normal. PCP was reported in 42% and neutropenia in 62%. Persistent cholangiolitis and liver cirrhosis occurred in 5 patients (patients 7a and 7b; patient 18; patient 35a; and patient 75a). Five patients developed tumors of the gastrointestinal tract, including hepatic/pancreatic carcinoid (patient 7b and patient 74), bile duct carcinoma (patient 35c), and adenocarcinoma of unknown origin (both patients of family 75).
Six unrelated patients had a mild clinical phenotype; 3 of those patients were 14 years or older at the time symptoms appeared (Table 3). Of the other 3 unrelated patients, one was found to have Arg11stop (resulting in the generation of a 19–amino acid shorter protein), one had IVS2 + 2t>a, and one had 367G>A (both mutations generating small amounts of wild-type CD40L). Three of these 6 patients presented with parvovirus B19–induced pure red cell aplasia.
Clinical features of 6 CD40L-deficient patients with mild disease
. | . | Atypical mild phenotype . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Typical phenotype . | Thr 254 Met Patient 61 . | Thr 254 Met Patient 62 . | Thr 254 Met Patient 63 . | Arg 11 stop Patient 1 . | IVS2 + 2 t > a Patient 17 . | 367G > A Patient 19 . | |||||
| Age at onset of symptoms, y | 1.2 (range, 0.18-5) | 23 | 14 | 5 | 8 | 17 | 5 | |||||
| Age when analyzed, y | 63 | 22 | 28 | 22 | 24 | 17 | ||||||
| PCP | 42% | - | - | - | - | - | - | |||||
| Neutropenia | 62% | - | - | Intermittent | - | - | - | |||||
| B19-induced red blood cell aplasia | No | No | Yes | No | Yes | Yes | No | |||||
| Anti-CD40L mAb binding | D/+ | + | + | + | + | + | + | |||||
| CD40-Ig binding | D | D | D | D | (+) | (+) | (+) | |||||
. | . | Atypical mild phenotype . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Typical phenotype . | Thr 254 Met Patient 61 . | Thr 254 Met Patient 62 . | Thr 254 Met Patient 63 . | Arg 11 stop Patient 1 . | IVS2 + 2 t > a Patient 17 . | 367G > A Patient 19 . | |||||
| Age at onset of symptoms, y | 1.2 (range, 0.18-5) | 23 | 14 | 5 | 8 | 17 | 5 | |||||
| Age when analyzed, y | 63 | 22 | 28 | 22 | 24 | 17 | ||||||
| PCP | 42% | - | - | - | - | - | - | |||||
| Neutropenia | 62% | - | - | Intermittent | - | - | - | |||||
| B19-induced red blood cell aplasia | No | No | Yes | No | Yes | Yes | No | |||||
| Anti-CD40L mAb binding | D/+ | + | + | + | + | + | + | |||||
| CD40-Ig binding | D | D | D | D | (+) | (+) | (+) | |||||
PCP indicates pneumocystis carinii pneumonia; +, CD40L expressed normally; (+), CD40L detected, but decreased compared with normal; D, dramatically decreased or absent CD40L expression; -, absent.
Analysis of the CD40 gene
All patients with normal CD40L (32 males and 10 females) were studied for mutations of the CD40 gene. Flow cytometric analysis demonstrated that CD40 was expressed normally by CD19+ peripheral blood B cells and/or EBV-induced B lymphoblasts in all patients (data not shown). Two patients were heterozygous for an amino acid substitution: a female patient had a Pro227Ala substitution (CCC/GCC) in exon 8 and a male patient had a Ser124Leu substitution (TCA/TTA) in exon 4. Each of the 2 patients had a healthy parent who was heterozygous for the same allele, suggesting that these changes are not responsible for the HIGM phenotype in their offspring.
Autosomal recessive HIGM due to mutations of AID
The same group (32 males and 10 females) was evaluated for mutations of AID. Three unrelated males and one female were homozygous for 3 unique missense mutations (Table 4), including 2 individuals from Quebec, Canada, who had the previously reported Arg112Cys mutation characteristic for French-Canadians.11 All 4 patients reported relatively mild bacterial but not opportunistic infections; 3 had lymphoid hyperplasia as reported by others10,11 ; serum IgG and IgA were consistently low, IgM levels markedly elevated (437 mg/dL-1820 mg/dL); neutrophil and lymphocyte counts and subsets were normal.
Sequence analysis, immune status, and clinical findings in patients with HIGM due to mutations of AID, NEMO, UNG, and Btk
. | . | . | . | Age, y . | . | Immunoglobulins (mg/dL) . | . | . | Absolute counts, per mm3 . | . | Lymphocyte subsets, per mm3 . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no./sex . | gDNA mutation, nucleotide . | Predicted effect on protein . | Exon . | Onset . | Now . | IgM . | IgG . | IgA . | Neutrophil . | Lymphocyte . | CD4 . | CD8 . | CD19 . | Lymphocyte proliferation mitogens/antigens* . | Clinical findings . | ||||||
| AID | |||||||||||||||||||||
| 78/M | 259T > C | Cys B7 Arg | 3 | 1.6 | 4 | 437 (43-207) | <33 | <7 | 5582 | 5832 | 1982 | 1341 | 175 | N/D | Recurrent sinopulmonary infections, chronic diarrhea, cervical lymph node hyperplasia | ||||||
| 79/F | 292T > G† | Leu 98 Arg | 3 | 4 | 25 | 640 (43-207) | 40 | 24 | 3124 | 3110 | 1555 | 212 | 196 | N/N | Recurrent sinopulmonary infections, cervical lymph node hyperplasia, adenitis with abscess formation | ||||||
| 80/M | 334C > T | Arg 112 Cys | 3 | 1.3 | 12 | 1820 (43-207) | <33 | 29 | 6875 | 1860 | 668 | 428 | 353 | N/D | Staphylococcal arthritis and osteomyelitis, consanguinity | ||||||
| 81/M | 334C > T | Arg 112 Cys | 3 | 2 | 7 | 1220 (43-207) | <33 | <7 | 6386 | 2141 | 771 | 299 | 792 | N/D | Cervical lymph node hyperplasia, atypical mycobacteria, bronchioctasis, consanguinity | ||||||
| UNG | |||||||||||||||||||||
| 82/M | 497-498 del AT | fs, stop at 159 | 2 | 15 | 39 | 785 (56-352) | 209 | <6.7 | 5687 | 3568 | 1268 | 687 | 549 | NA/D | Recurrent sinopulmonary infections, chronic epididymitis, pulmonary lymphoid hyperplasia, Sjógren syndrome, consanguinity | ||||||
| NEMO | |||||||||||||||||||||
| 83/M | 1426 ins C | fs, stop at 393 | 10 | 0.03 | 1.2 | 41 (27-101) | 109 | <6 | 6245 | 10 414 | 8022 | 2392 | 2533 | N/N | EDA, recurrent sinopulmonary infections, abdominal wall abscess, gram-negative sepsis, meningitis, chronic diarrhea, persistent molluseum contagiosum, failure to thrive | ||||||
| Btk | |||||||||||||||||||||
| 84/M | 338A > T | Lys 199 stop | 8 | 3 | 4 | 42 (43-207) | <7 | <7 | 1840 | 3950 | 2805 | 909 | 40 (122-632) | N/N | Neutropenia, impetigo, osteomyelitis, mandibular abscess due to atypical mycobacteria | ||||||
| 85a/M | 1984A > G | Arg 618 Gly | 18 | 9 | 11 | 73 (43-207) | 130 | <7 | 6897 | NA | NA | NA | NA | N/N | Recurrent sinopulmonary infections, chronic diarrhea, pocumococcal meningitis | ||||||
| 85b/M | 1984A > G | Arg 618 Gly | 18 | 10 | 13 | 58 (43-207) | 199 | <7 | 5894 | 5264 | 3368 | 1579 | 526 (122-632) | N/N | Recurrent sinopulmonary infections | ||||||
. | . | . | . | Age, y . | . | Immunoglobulins (mg/dL) . | . | . | Absolute counts, per mm3 . | . | Lymphocyte subsets, per mm3 . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no./sex . | gDNA mutation, nucleotide . | Predicted effect on protein . | Exon . | Onset . | Now . | IgM . | IgG . | IgA . | Neutrophil . | Lymphocyte . | CD4 . | CD8 . | CD19 . | Lymphocyte proliferation mitogens/antigens* . | Clinical findings . | ||||||
| AID | |||||||||||||||||||||
| 78/M | 259T > C | Cys B7 Arg | 3 | 1.6 | 4 | 437 (43-207) | <33 | <7 | 5582 | 5832 | 1982 | 1341 | 175 | N/D | Recurrent sinopulmonary infections, chronic diarrhea, cervical lymph node hyperplasia | ||||||
| 79/F | 292T > G† | Leu 98 Arg | 3 | 4 | 25 | 640 (43-207) | 40 | 24 | 3124 | 3110 | 1555 | 212 | 196 | N/N | Recurrent sinopulmonary infections, cervical lymph node hyperplasia, adenitis with abscess formation | ||||||
| 80/M | 334C > T | Arg 112 Cys | 3 | 1.3 | 12 | 1820 (43-207) | <33 | 29 | 6875 | 1860 | 668 | 428 | 353 | N/D | Staphylococcal arthritis and osteomyelitis, consanguinity | ||||||
| 81/M | 334C > T | Arg 112 Cys | 3 | 2 | 7 | 1220 (43-207) | <33 | <7 | 6386 | 2141 | 771 | 299 | 792 | N/D | Cervical lymph node hyperplasia, atypical mycobacteria, bronchioctasis, consanguinity | ||||||
| UNG | |||||||||||||||||||||
| 82/M | 497-498 del AT | fs, stop at 159 | 2 | 15 | 39 | 785 (56-352) | 209 | <6.7 | 5687 | 3568 | 1268 | 687 | 549 | NA/D | Recurrent sinopulmonary infections, chronic epididymitis, pulmonary lymphoid hyperplasia, Sjógren syndrome, consanguinity | ||||||
| NEMO | |||||||||||||||||||||
| 83/M | 1426 ins C | fs, stop at 393 | 10 | 0.03 | 1.2 | 41 (27-101) | 109 | <6 | 6245 | 10 414 | 8022 | 2392 | 2533 | N/N | EDA, recurrent sinopulmonary infections, abdominal wall abscess, gram-negative sepsis, meningitis, chronic diarrhea, persistent molluseum contagiosum, failure to thrive | ||||||
| Btk | |||||||||||||||||||||
| 84/M | 338A > T | Lys 199 stop | 8 | 3 | 4 | 42 (43-207) | <7 | <7 | 1840 | 3950 | 2805 | 909 | 40 (122-632) | N/N | Neutropenia, impetigo, osteomyelitis, mandibular abscess due to atypical mycobacteria | ||||||
| 85a/M | 1984A > G | Arg 618 Gly | 18 | 9 | 11 | 73 (43-207) | 130 | <7 | 6897 | NA | NA | NA | NA | N/N | Recurrent sinopulmonary infections, chronic diarrhea, pocumococcal meningitis | ||||||
| 85b/M | 1984A > G | Arg 618 Gly | 18 | 10 | 13 | 58 (43-207) | 199 | <7 | 5894 | 5264 | 3368 | 1579 | 526 (122-632) | N/N | Recurrent sinopulmonary infections | ||||||
Values in parentheses indicate normal values for age, according to individual laboratories.
N indicates normal; D, decrease; NA, not available; EDA, ectodermal dysplasia; fs, frameshift.
Mitogens are PHA, PMA, or ConA: antigens are tetanus candida, diphtheria.
Novel mutation, not yet reported.
While sequencing the AICDA gene, we found a polymorphism (T544C) that resulted in a silent mutation (CAT/CAC) at His155. CAT was seen in 21 individuals, CAT/CAC in 18, and CAC in 3. This silent mutation does neither affect nor create new splice sites and therefore cannot be responsible for the HIGM phenotype in these patients.
A normal variant, the deletion of 30 bp in exon 4 of AID due to alternative splicing, previously reported by Noguchi et al,38 was found in 6 of the 42 patients analyzed. Two individuals were homozygous and 4 were heterozygous for this variant.
Autosomal recessive HIGM due to mutations of uracil-DNA glycosylase (UNG)
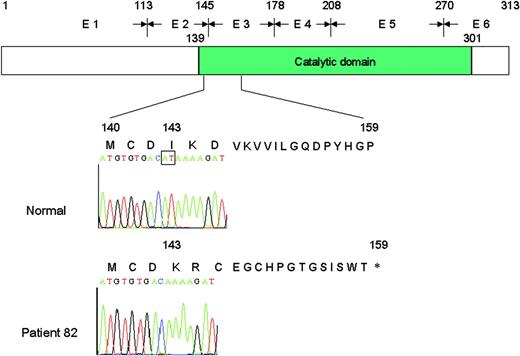
The remaining patients (29 males and 9 females) were studied for mutations of UNG.39 A 39-year-old male (patient 82 in Table 4) whose parents were first cousins, was found to be homozygous for a 2-bp deletion in exon 2 (497, 498delAT) affecting codon 143 of the UNG gene. This deletion resulted in frameshift and premature termination at codon 159 (Figure 1).14
Representation of the normal UNG protein consisting of 313 amino acids and the putative truncated protein of 158 amino acids identified in patient 82. The deletion of 2 nucleotides (AT) in exon 2 (E2) causes frameshift at amino acid 143, resulting in a stop codon at position 159.
Representation of the normal UNG protein consisting of 313 amino acids and the putative truncated protein of 158 amino acids identified in patient 82. The deletion of 2 nucleotides (AT) in exon 2 (E2) causes frameshift at amino acid 143, resulting in a stop codon at position 159.
The UNG-deficient patient was asymptomatic until the age of 15 years, when he presented with recurrent sinopulmonary infections. He subsequently developed chronic epididymitis and Sjögren syndrome, and was found to have pulmonary lymphoid hyperplasia. He never had neutropenia or opportunistic infections. At the time the diagnosis of HIGM was established, he had low serum IgG (209 mg/dL), IgA (< 7 mg/dL), clearly elevated IgM (785 mg/dL), and abnormal antibody responses to pneumococcal antigens. Intravenous Ig therapy reduced the number of infections but had no effect on the progression of the Sjögren syndrome and pulmonary lymphoid hyperplasia.
X-linked HIGM and NEMO mutations
Because a subset of male patients with mutations of NEMO have the HIGM phenotype18 and because not all patients with mutations of NEMO present with EDA, we evaluated all remaining male patients with HIGM for NEMO mutations.
One of 28 males, a 15-month-old infant (patient 83 in Table 4) with EDA and onset of frequent infections at age 10 days, presented with hypogammaglobulinemia (IgG 109 mg/dL, IgA<6 mg/dL) and normal IgM (41 mg/dL). He was found to have a C insertion at position 1426 (Genebank: NM 003 639) in exon 10, resulting in frameshift and premature termination with loss of the Zinc finger domain.
Other genes associated with antibody deficiency (ICOS, ICOSL, Btk, and SAP/SH2D1A)
Because a mutation of the ICOS gene has recently been observed in several patients with the diagnosis of CVID,24 we evaluated all patients with HIGM without a molecular diagnosis for ICOS expression by activated T cells and sequenced both ICOS and ICOSL; no mutations were identified.34
To rule out the possibility of atypical XLA or XLP, all male patients without a molecular diagnosis (n = 27) were examined for mutations of Btk and SAP/SH2D1A. Three patients from 2 unrelated families were found to have mutations of Btk (Table 4). A 4-year-old boy (patient 84) with lymph node hypoplasia presented with recurrent infections (cellulitis, impetigo, osteomylitis) at 3 years of age; he was found to have a nonsense mutation in the Tec homology domain (338A>T, resulting in Lys199stop). As expected, this patient had hypogammaglobulinemia (IgG of < 7 mg/dL, IgA < 7 mg/dL), normal serum IgM (42 mg/dL), and decreased CD19+ B cells (40/mm3; normal range: 122-632). The 2 brothers (family 85) were well until the ages of 8 and 9 years, respectively, when they started to develop recurrent infections (pneumonia, sinusitis, meningitis due to Strep pneumoniae). IgG serum levels were 130/190 mg/dL, IgA < 7 mg/dL, and IgM 73/58 mg/dL, respectively. One brother had a B-cell count of 526/mm3. Sequence analysis revealed a missense mutation in exon 18 affecting the kinase domain (1984A>G, resulting in Arg618Gly). Three male relatives of the 2 brothers reportedly died of chronic lung disease or meningitis during childhood.
None of the male patients studied had mutations of the SAP/SH2D1A gene.
Discussion
The production of high-affinity antibody of a specific isotype is orchestrated by the CD40L-driven CD40 signaling pathway. Mutations of several genes known to participate in this signaling circuit (CD40L, CD40, NEMO, AICDA, and UNG) disrupt CSR, SHM, and germinal center formation. A direct consequence of defective CD40 signaling is an antibody deficiency syndrome characterized by frequent infections associated with hypogammaglobulinemia and normal or elevated serum IgM. Taking advantage of new insights into the CD40 signaling cascade, we studied the genetic basis of HIGM in a large cohort of symptomatic patients. Included in this analysis were genes known to cause HIGM; genes that had been associated with other genetically defined antibody deficiencies (often with atypical presentations) resembling HIGM, for example, mutations of Btk and SAP/SH2D1A; and genes known to have functions that potentially make them candidates for molecularly undefined HIGM syndromes.
The patient population included in this analysis was predominantly male, which is not unexpected because the largest group by far (70% in our series of 140 patients) of molecularly defined HIGM syndromes have mutations of the X-chromosome–associated CD40L gene. Because of this bias, based on historic data, it is possible that female patients with hypogammaglobulinemia and normal IgM are classified as CVID and, as a consequence, not examined for molecular defects involving autosomal recessive genes known to cause an HIGM phenotype.
Using flow cytometry to screen for defective CD40L expression followed by sequence analysis, we found CD40L mutations in 98 males. This represents 75% of all male patients with HIGM enrolled in the study. Anti-CD40L monoclonal antibodies, by not recognizing mutated CD40L due to nonsense mutations and out-of-frame deletions and insertions, successfully identified patients with these mutations, but often failed to discriminate between wild-type CD40L and nonfunctional CD40L due to missense mutations or in-frame deletions/insertions. The soluble CD40-Ig fusion protein, on the other hand, identified all CD40 mutations except for the deletion of most of the intracellular tail (patient 1), and the splice site mutations affecting IVS 2 and IVS 3 that allowed the production of small amounts of wild-type CD40L (patients 17, 19, 25, and 26 in Table 2).
The majority of patients with mutations of CD40L had the typical phenotype of X-linked HIGM characterized by early onset of severe recurrent sinopulmonary infections, including PCP, and the development of cholangitis associated with persistent cryptosporidium infection, with or without neutropenia. Response to intravenous immunoglobulin therapy was less than optimal in some patients. Exceptions were 6 male patients with a mild phenotype, late onset of symptoms, and no history of PCP, cryptosporidium infection, or neutropenia (Table 3). Three of the 6 atypical cases (patients 61-63) had the same amino acid substitution (Thr254Met) affecting the carboxy terminal of exon 5. One patient had a nonsense mutation (Arg11stop) that resulted in truncation of the intracellular domain. Patient 17 had an in-frame deletion of exon 2 (44 amino acids) due to a splice site mutation (IVS2 + 2t>a) resulting in the production of a small population of wild-type mRNA, presumably enough to generate a low number of functionally normal CD40L homotrimeric molecules.31,35 The last of the atypical patients (no. 19) had a “silent” point mutation (367G>A) affecting the last nucleotide of exon 3, causing a splicing defect that results in skipping of exon 3 and frameshift, but also in the production of a small amount of normally spliced wild-type mRNA. Activated T cells from 3 of these 6 individuals with a mild phenotype were able to bind CD40-Ig fusion protein although at reduced quantity. Patients 1, 17, and 19 were asymptomatic until they presented with pure red cell aplasia caused by chronic parvovirus B19 infection.35 Interestingly, patient 20, with the 367G>C mutation (resulting in skipping of exon 3 and production of a small amount of mutated [Lys96Asn] nonfunctional CD40L) developed PCP at 4 months of age and subsequently suffered from recurrent sinopulmonary infections. Thus, a detailed analysis of the CD40L mutation and its effect on CD40L expression and function provides useful information that may affect prognostic and therapeutic decisions.
None of our cohort had a mutation of CD40, a rare autosomal recessive form of HIGM observed to date in only 4 unrelated infants born to consanguineous parents living in the Mediterranean region.8,9
Two enzymes, AID and UNG, are of crucial importance for CSR and SHM.10-14,40-42 Both genes are expressed by activated B cells, including EBV-transformed B-cell lines. AID and UNG knockout mice have a severe defect in CSR and SHM.13,41 Based on its homology to APOPEC-1, which deaminases cytidine residues of the ApoB mRNA transcript to uracil residues, AID was originally considered a member of the RNA editing deaminase family.40,41 However, recent data provide strong evidence that AID acts directly on DNA43 and deaminases cytidine to uracil within immunoglobulin V-genes,13 indicating a DNA editing function. DNA polymerase, an enzyme that copies DNA by reading uracil as thymidine during replication, is responsible for the transition of cytidine → thymidine, a UNG-independent SHM mechanism. The removal of uracil by UNG leaves a single nucleotide gap, which results in double strand breaks, thus initiating the process of CSR. If the gap is filled, at random, with purine or pyrimidine nucleotides by low fidelity polymerase, both transitions and transversions are generated, a prerequisite for effective SHM. In UNG-deficient mice, an SHM pattern occurs in which mutations at dG and dC residues are biased toward transition, whereas at dA and dT residues, the ratio of transitions to transversions are similar to control values.13 Taking advantage of this new insight into the molecular basis of CSR and SHM, selected patients with HIGM and normal CD40L were evaluated for mutations of AID and UNG and defects were identified in both genes.10,11,14,44 Based on these reports, we sequenced AID and UNG in 32 males with normal CD40L and 10 females with the HIGM phenotype. Four patients, 3 males and one female, with recurrent sinopulmonary but not opportunistic infections, and with characteristic hypogammaglobulinemia and elevated IgM, were found to have mutations of AID (Table 4). In addition to the common Arg112Cys mutation of AID previously reported in French-Canadians11 and present in 2 of our AID-deficient patients, we found 2 missense mutations of AID (Cys87Arg and Leu98Arg), one affecting the cytidine deaminase motif and the other the link between the active and pseudoactive sites of AID. Using an in vitro system, 2 of these mutations were studied by Ta et al45 and were found to lack CSR and SHM activities. A fifth patient (patient no. 82), a 33-year-old male with late-onset HIGM and relatively mild symptoms, was found to have a homozygous deletion of 2 bp in exon 2 of UNG, which resulted in frameshift and premature stop of transcription. As reported previously, a B-lymphoblastoid cell line established from this patient lacked UNG protein and his B cells demonstrated profound impairment of CSR and partial disturbance of SHM.14
NEMO, also known as IKKγ, is a component of IκB kinase, which is essential for the phosphorylation of IκB, a step required for activation of NF-κB, which, after nuclear transfer, regulates the expression of multiple genes involved in immune responses. Mutations of NEMO that result in loss of function cause incontinentia pigmenti, which is a rare X-linked dominant geno-dermatosis that is ante-natally lethal in males; heterozygous females, in addition to incontinentia pigmenti, develop anomalies of teeth, eyes, hair, and the central nervous system.46 Those mutations that result in residual biologic function of NEMO, usually missense mutations in the Zinc finger domain, cause the phenotype of X-linked recessive hypohidrotic ectodermal dysplasia and immunodeficiency (EDA-ID).16-18,46 Mutations of NEMO that generate a stop codon, usually nonsense mutations, insertions, or deletions, if not lethal, cause a more severe syndrome in male patients and is often associated with osteopetrosis or lymphedema, as well as EDA-ID.16 Of 25 male patients with HIGM without a molecular diagnosis, one, a 12-month-old boy, had a single nucleotide insertion in exon 10 of NEMO, which resulted in frameshift and premature termination that led to a truncated, probably unstable protein that was no longer demonstrable by monoclonal or polyclonal anti-NEMO antibodies. The infant presented with severe bacterial infections, persistent molluscum contagiosum, and failure to thrive. However, not all male patients with NEMO mutations have the HIGM phenotype.16,17 We have investigated a 10-year-old boy with EDA who presented with normal or elevated serum immunoglobulin levels (and therefore was excluded from this report) and recurrent sinopulmonary infections that were well controlled with antibiotics, who had a missense mutation of NEMO (Arg217Gly). In this latter patient, the NEMO protein could be readily detected with monoclonal and polyclonal antibodies and phosphorylation of IκB-α was demonstrable when EBV-transformed B lymphoblasts were stimulated with soluble CD40L-Ig (W.-IL., unpublished observation, January 2003).
Finally, because of some overlap in clinical presentation of patients with mutations of Btk or SAP/SH2D1A and CVID25,26 we sequenced both genes in every male patient without a molecular diagnosis. Three patients, including 2 brothers, all with normal levels of IgM, were found to have mutations of Btk. One of the 2 brothers had normal numbers of B cells. None of our patients with HIGM had mutations of ICOS or ICOS ligand, suggesting that in humans, unlike in mice, mutations of these genes do not result in the HIGM syndrome.19-24
All patients included in this study had lymphocytes that expressed MHCII and none had AT. Both these molecularly defined immunodeficiency disorders with characteristic clinical and laboratory findings can present with hypogammaglobulinemia, elevated IgM, and frequent infections.47-49
The intent of this review has been to demonstrate that the clinically defined HIGM syndromes consist of patients with a range of clinical findings and are caused by multiple genetic defects. The molecular analyses of a large cohort of patients with HIGM referred to a single center for molecular analysis supports the impression that CD40L mutations predominate, although a selection bias may exist.
Overall, 33 individuals (23%) in the entire group had no identifiable molecular defect (Table 5). The sex distribution (24 male, 9 female) within this group reflects the bias in favor of referring male patients suspected to have an X-linked form of HIGM. Based on the clinical presentation (recurrent sinopulmonary infections, low serum IgG and IgA with normal IgM [16 patients] or elevated IgM [17 patients]), we propose to consider this molecularly undefined group of patients as CVID. We expect, however, that subsets of this group of patients may have well-defined single gene defect(s). Of our own CVID cohort, 5 individuals (10%) met the inclusion criteria and were entered into the study; one of these patients (no. 29) was found to have a mutation of CD40L. Durandy and coworkers27 have recently identified a group of patients with HIGM with a selective deficiency of CSR caused by unknown molecular defect(s). Candidate genes suggested from knockout mice to be directly involved in CSR and/or SHM include nonhomologous end-joining enzymes (the Ku70/Ku80 recombination complex, DNA-PK),50,51 mismatch repair enzymes (Msh2, Mlh1, Pms2),52-55 exonuclease 1,56 nucleotide-excision-repair enzymes, and error-prone polymerases.42
Molecular defects identified in a cohort of 140 patients with HIGM
. | . | Number of patients . | . | . | |
|---|---|---|---|---|---|
| Gene . | Percentage* . | Male . | Female . | Number of families . | |
| CD40L | 70.0 | 98 | 0 | 77 | |
| AID | 2.9 | 3 | 1 | 4 | |
| CD40 | 0 | 0 | 0 | 0 | |
| NEMO | 0.7 | 1 | 0 | 1 | |
| ICOS | 0 | 0 | 0 | 0 | |
| ICOSL | 0 | 0 | 0 | 0 | |
| UNG2 | 0.7 | 1 | 0 | 1 | |
| Btk | 2.1 | 3 | 0 | 2 | |
| SAP | 0 | 0 | 0 | 0 | |
| Unknown | 23.6 | 24 | 9 | 30 | |
| Total | 100 | 130 | 10 | 115 | |
. | . | Number of patients . | . | . | |
|---|---|---|---|---|---|
| Gene . | Percentage* . | Male . | Female . | Number of families . | |
| CD40L | 70.0 | 98 | 0 | 77 | |
| AID | 2.9 | 3 | 1 | 4 | |
| CD40 | 0 | 0 | 0 | 0 | |
| NEMO | 0.7 | 1 | 0 | 1 | |
| ICOS | 0 | 0 | 0 | 0 | |
| ICOSL | 0 | 0 | 0 | 0 | |
| UNG2 | 0.7 | 1 | 0 | 1 | |
| Btk | 2.1 | 3 | 0 | 2 | |
| SAP | 0 | 0 | 0 | 0 | |
| Unknown | 23.6 | 24 | 9 | 30 | |
| Total | 100 | 130 | 10 | 115 | |
Percentage of all patients.
Prepublished online as Blood First Edition Paper, September 9, 2004; DOI 10.1182/blood-2003-12-4420.
Supported by grants from the National Institutes of Health (HD 17427-33), the March of Dimes Birth Defects Foundation (6-FY96-0330), the Immune Deficiency Foundation, and the Jeffrey Modell Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank the following physicians who provided samples from their patients: Javeed Akhter, Julian Ambrus, Andrea Apter, Prescott Atkinson, Scott Baker, Conrad Belnap, Jonathan Bernstein, John Bohnsack, Ellen Bolotin, Tony Bonilla, Rebecca Buckley, Tom Cleary, Charlotte Cunningham-Rundles, Noorbibi Day, Alexandro Dorenbaum, Melissa Elder, Allen Ettinger, Mary Fasano, Ronald Ferdman, Senih Fikrig, David Freyer, Erwin Gelfand, Deborah Gentile, Barry Gray, Jeff Harris, Henrik Hasle, Harry Hill, Robert Hostoffer, Anna Huttenlocher, Anne-Marie Irani, Anne Junker, Rhoda Kagan, Steven Klein, Gary Kleiner, Michelle Klinek, Alan Knutsen, Roger Kobayashi, Lisa Kobrynski, Mark Kosinski, Lawrence Larson, Howard Ledermann, Sok Lee, Bruce Mazer, Doug McDonald, Marian Melish, Alton Melton, Hilaire Meuwissen, Robert Nelson, Shigeaki Nonoyama, Henry Pabst, Elena Perez, Daniel Pietryga, Wendell Richmond, Robert Roberts, Howard Rosenblatt, Frank Saulsbury, Christine Seroogy, Ralph Shapiro, John Sleasman, Monica Smogorzewska, Richard Stiehm, Daniel Suez, Katharine Sullivan, Dale Umetsu, Pakit Vichyanond, Mary Wakim, Diane Wara, Debora Williams-Herman, Charles Wilson, and Jerry Winkelstein.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal