Abstract
The human leukocyte antigen G (HLA-G) molecule exhibits limited tissue distribution and exerts multiple immunoregulatory functions. Recent studies indicate an ectopic up-regulation in tumor cells that may favor their escape from antitumor immune responses. The role of HLA-G in B-cell chronic lymphocytic leukemia (B-CLL) has not been defined. HLA-G expression was studied retrospectively in circulating B-CLL cells from 47 patients by flow cytometry using the anti-HLA-E specific monoclonal antibody MEM/G9. The proportion of leukemic cells expressing HLA-G varied from 1% to 54%. Patients with 23% or fewer HLA-G-positive cells (according to receiver operating characteristics [ROC] analysis; designated as HLA-G-negative group) had a significantly longer progression-free survival (PFS) time than patients with more than 23% positive cells (median PFS: 120 versus 23 months; P = .0001). In multivariate analysis, HLA-G expression (hazard ratio: 4.8; P = .002) was an even better independent prognostic factor than the zeta-associated protein 70 (ZAP-70) or CD38 status. Humoral and cellular immunosuppression were significantly more prominent in the HLA-G-positive compared with the HLA-G-negative patient group. In B-CLL, the level of HLA-G expression is correlated with the degree of immunosuppression and prognosis. HLA-G may contribute to the impairment of immune responses against tumor cells and infections. Thus, these findings need to be confirmed in a prospective study. (Blood. 2005;105:1694-1698)
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is the most common adult leukemia in Western countries. It is characterized by an accumulation of long-lived, functionally inactive, mature-appearing neoplastic B lymphocytes.1-3 B-CLL has a variable clinical course. Some patients have an excellent prognosis never requiring treatment, whereas in others survival is short despite early initiation of therapy. During the last few years, there has been a continuous effort to identify novel prognostic factors in B-CLL that may help define patient subgroups that may benefit from early therapeutic intervention.
Markers of poor prognosis in both early and advanced B-CLL include absence of mutations in the variable region of the immunoglobulin (Ig) heavy chain gene (IgVH gene), deletion of chromosome 11q23, loss or mutation of the p53 gene, as well as CD38 or zeta-associated protein 70 (ZAP-70) positivity. In multivariate analyses, IgVH gene mutational status has been identified as the best predictor of clinical outcome: Patients whose CLL cells have unmutated IgVH genes have a significantly worse prognosis than those whose IgVH genes have undergone somatic mutation.4-9
Classical major histocompatibility complex (MHC) class I antigens are highly polymorphic molecules that mediate the presentation of peptides to T lymphocytes. They are ubiquitously expressed and play an important role in the recognition of tumor cells and their products by the immune system.10 Human leukocyte antigen G (HLA-G) is a nonclassical MHC class I antigen with very little sequence variability. It is not expressed in normal tissues except in trophoblasts from early gestation placentas.11,12 HLA-G exerts multiple immunoregulatory functions such as inhibition of natural killer (NK) cell or T-cell-mediated cytolysis, induction of T-cell apoptosis, or inhibition of transendothelial NK cell migration.13,14 Since the net result of these effects is immunosuppression, HLA-G expression in tumor cells may favor their escape from antitumor immune responses, thus allowing tumor progression.15
Ectopic HLA-G expression has been demonstrated in various types of tumors including B-cell and T-cell lymphomas, melanoma, as well as lung, kidney, bladder, breast, malignant ascites, and colorectal carcinoma.16-22 In cutaneous B-cell lymphomas and lung cancer, HLA-G expression is paralleled by expression of interleukin-10 (IL-10), which has multiple inhibitory effects on the immune system.23 It has been hypothesized that IL-10 is also responsible for the up-regulation of HLA-G in tumor cells.22,24
In an effort to gain insight into the role of HLA-G in B-CLL, we measured its expression in circulating leukemic cells and correlated our findings with a variety of clinical and laboratory variables.
Materials and methods
Between June 2002 and January 2004, 47 white patients with B-CLL were enrolled into this retrospective study and analyzed for biologic and clinical characteristics including age, sex, Binet stage, treatment history, time from diagnosis to first treatment, white blood cell count, total T-cell count and T-cell subpopulations, hemoglobin concentration, platelet count, serum activities of lactate dehydrogenase and thymidine kinase, as well as serum concentrations of β2-microglobulin and immunoglobulins. In each patient, morphologic diagnosis of B-CLL was confirmed by flow cytometry revealing a typical CD19+ CD20+ CD5+ CD23+ Ig light chain (κ or λ) restricted immunophenotype. ZAP-70 and CD38 expression were determined by flow cytometry as described.8,25 Blood samples were obtained during routine follow-up visits to our institution, with all patients giving informed consent according to institutional guidelines. Indications for treatment were based on standard criteria.
Flow cytometry
Peripheral blood mononuclear cells were separated from freshly drawn anticoagulated blood by Ficoll-Paque (Pharmacia, Uppsala, Sweden) density gradient centrifugation. B cells were enriched by CD19 microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer's instructions. To block unspecific binding, the enriched B cells were incubated with 10% human serum in phosphate-buffer saline (PBS; pH 7.4) for one hour at 4°C. HLA-G expression on CD5+ B cells was evaluated by dual-color immunofluorescence staining. To this end, 1 × 106 B cells were incubated in 200 μL PBS at room temperature for 30 minutes in the dark with 15 μL (1 mg/mL) fluorescein isothiocyanate (FITC)-conjugated antihuman HLA-G-specific monoclonal antibody MEM/G9 (IgG1; Serotec, Oxford, United Kingdom) and 20 μL phycoerythrin (PE)-conjugated antihuman CD5-specific monoclonal antibody L17F12 (Becton Dickinson, San Jose, CA). After 2 washing steps with PBS, HLA-G expression was quantified by flow cytometry (FACS Calibur; Becton Dickinson, Heidelberg, Germany). Isotype-matched FITC- and PE-labeled mouse IgG served as a negative control. Data were acquired and analyzed using CellQuest software (Becton Dickinson, Heidelberg, Germany).
Soluble HLA-G (sHLA-G) and IL-10 levels
Levels of β2-microglobulin-associated sHLA-G molecules (G1 and G5 isoforms) in plasma samples were determined by enzyme-linked immunosorbent assay (ELISA) as described previously.26 IL-10 plasma levels were measured using a commercially available kit according to the manufacturer's instructions (OptEIA human IL-10 set; Pharmingen, San Diego, CA).
Statistical analysis
The impact of HLA-G expression on progression-free survival was assessed by performing receiver operating characteristics (ROC) curve analysis.27 Progression-free survival times were measured from the time of diagnosis to first therapy, plotted according to the Kaplan-Meier method,28 and compared using the log-rank test. Comparison of clinical or laboratory parameters between patient subgroups was performed using the nonparametric Mann-Whitney U test for continuous variables and the χ2 test for categoric data. Differences were regarded as significant at P less than .05. To identify prognostic factors for the duration of progression-free survival, multivariate Cox regression analysis was performed. The results were analyzed using GraphPad Prism 3.0 (GraphPad Software, San Diego, CA).
Results
Characteristics of patients with B-CLL
The cohort of patients analyzed was characteristic of B-CLL, with a median age of 61 years, male, and stage A predominance, and about half the patients displayed adverse prognostic factors such as high expression of CD38 or ZAP-70 (Table 1).
Clinical data and prognostic factors
Variable . | All patients . | Less than 23% HLA-G—positive cells . | 23% or more HLA-G—positive cells . | P* . |
|---|---|---|---|---|
| No. of patients (%) | 47 | 37 (79) | 10 (21) | |
| Age at diagnosis, y | 61 (30-88)† | 60.5 (30-80)† | 62.5 (41-88)† | NS |
| Male (%) | 37 (79)‡ | 29 (78)‡ | 8 (80)‡ | NS |
| Binet stage at diagnosis (%) | ||||
| A | 37 (79) | 30 (81) | 7 (70) | NS |
| B | 6 (13) | 5 (14) | 1 (10) | |
| C | 4 (9) | 2 (5) | 2 (20) | |
| Hemoglobin, g/dL, n = 47 | 13.05 ± 1.75§ | 13.35 ± 1.6§ | 11.94 ± 1.87§ | .023 |
| Platelets/nL, n = 47 | 147 ± 65§ | 153 ± 64§ | 124 ± 68§ | NS |
| Leukocytes/nL, n = 47 | 56.96 ± 64.5§ | 58.5 ± 69.1§ | 51.1 ± 45.6§ | NS |
| Neutrophils/nL, n = 46 | 4.8 ± 4.3§ | 5.3 ± 4.4§ | 3.0 ± 3.1§ | .06 |
| Lymphocytes/nL, n = 46 | 50.2 ± 61.0§ | 51.2 ± 65.7§ | 46.6 ± 42.4§ | NS |
| Monocytes/nL; n = 46 | 1.4 ± 2.6§ | 1.5 ± 2.9§ | 1.1 ± 1.1§ | NS |
| CD38+ leukemia (%), n = 47 | 24 (51) | 17 (46) | 7 (70) | NS |
| ZAP-70+ leukemia (%), n = 42 | 19 (45) | 15 (45) | 4 (44) | NS |
| β2-microglobulin, mg/L, n = 45 | 4.0 ± 2.2§ | 3.8 ± 2.0§ | 5.0 ± 2.5§ | NS |
| Thymidine kinase, U/L, n = 32 | 28.5 ± 34.5§ | 22.3 ± 29.3§ | 55.8 ± 44.8§ | .055 |
Variable . | All patients . | Less than 23% HLA-G—positive cells . | 23% or more HLA-G—positive cells . | P* . |
|---|---|---|---|---|
| No. of patients (%) | 47 | 37 (79) | 10 (21) | |
| Age at diagnosis, y | 61 (30-88)† | 60.5 (30-80)† | 62.5 (41-88)† | NS |
| Male (%) | 37 (79)‡ | 29 (78)‡ | 8 (80)‡ | NS |
| Binet stage at diagnosis (%) | ||||
| A | 37 (79) | 30 (81) | 7 (70) | NS |
| B | 6 (13) | 5 (14) | 1 (10) | |
| C | 4 (9) | 2 (5) | 2 (20) | |
| Hemoglobin, g/dL, n = 47 | 13.05 ± 1.75§ | 13.35 ± 1.6§ | 11.94 ± 1.87§ | .023 |
| Platelets/nL, n = 47 | 147 ± 65§ | 153 ± 64§ | 124 ± 68§ | NS |
| Leukocytes/nL, n = 47 | 56.96 ± 64.5§ | 58.5 ± 69.1§ | 51.1 ± 45.6§ | NS |
| Neutrophils/nL, n = 46 | 4.8 ± 4.3§ | 5.3 ± 4.4§ | 3.0 ± 3.1§ | .06 |
| Lymphocytes/nL, n = 46 | 50.2 ± 61.0§ | 51.2 ± 65.7§ | 46.6 ± 42.4§ | NS |
| Monocytes/nL; n = 46 | 1.4 ± 2.6§ | 1.5 ± 2.9§ | 1.1 ± 1.1§ | NS |
| CD38+ leukemia (%), n = 47 | 24 (51) | 17 (46) | 7 (70) | NS |
| ZAP-70+ leukemia (%), n = 42 | 19 (45) | 15 (45) | 4 (44) | NS |
| β2-microglobulin, mg/L, n = 45 | 4.0 ± 2.2§ | 3.8 ± 2.0§ | 5.0 ± 2.5§ | NS |
| Thymidine kinase, U/L, n = 32 | 28.5 ± 34.5§ | 22.3 ± 29.3§ | 55.8 ± 44.8§ | .055 |
In most patients, the laboratory parameters were not determined at diagnosis, but during the course of the disease. Normal ranges are as follows: hemoglobin, 14.0 to 18.0 g/dL (male), 12.0 to 14.0 g/dL (female); platelets, 140 to 440/nL; leukocytes, 4.0 to 10.0/nL; neutrophils, 1.5 to 7.0/nL; lymphocytes, 1.0 to 4.0/nL; monocytes, less than 1.0/nL; β2-microglobulin, less than 3.5 mg/L; and thymidine kinase less than 7 U/L. CD38+ or ZAP-70+ leukemias were diagnosed when more than 20% of the leukemic cells expressed the respective marker.
NS indicates not significant.
Comparison between the HLA-G-positive and HLA-G-negative subgroups using the chi-square test (Binet stage, CD38+ and ZAP-70+ leukemias) or the Mann-Whitney U test (all other variables).
Median (range).
Percent of all patients. Because of rounding, percentages do not always add up to 100.
Mean ± standard error of the mean.
HLA-G expression and prognosis
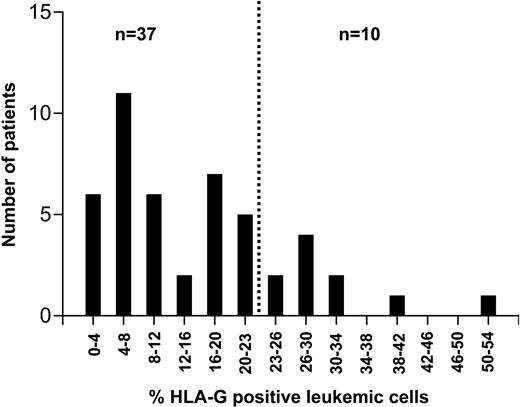
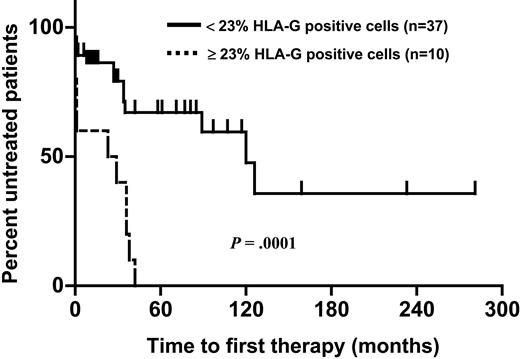
The proportion of CLL cells expressing HLA-G molecules varied from 1% to 54% (Figures 1-2). In an effort to assess the impact of HLA-G expression on the course of the disease, the percentage of cells expressing the molecule was correlated with the patients' progression-free survival time using ROC curve analysis. HLA-G expression was significantly correlated with survival, with the best separation of the curves achieved at a cut-off level of 23% positive leukemic cells. A total of 37 patients had leukemias with fewer than 23% HLA-G-positive cells (designated as the HLA-G-negative group). This group had a significantly longer progression-free survival time than the HLA-G-positive group (Figure 3). With a median follow-up of 34 months (range: 0-281 months), 23 patients (49%) required first-line chemotherapy. In the HLA-G-positive group, treatment became necessary in all patients, while in the HLA-G-negative group this was the case in only 13 patients (35%; P = .0003).
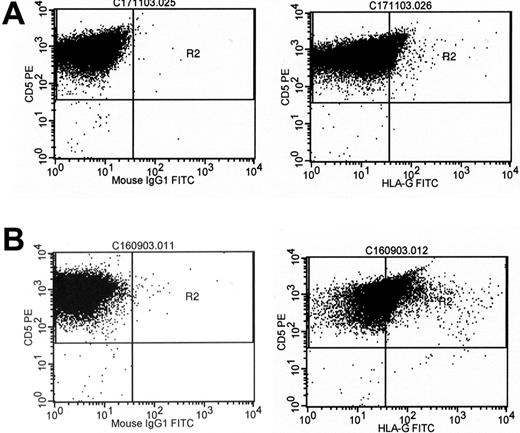
Cell surface expression of HLA-G in 2 cases of B-CLL. B-CLL cells were subjected to dual-color flow cytometry using a CD5-specific antibody and the HLA-G-specific antibody MEM/G9 (right column) or the respective isotype control (left column). (A) Low HLA-G expression. (B) High HLA-G expression.
Cell surface expression of HLA-G in 2 cases of B-CLL. B-CLL cells were subjected to dual-color flow cytometry using a CD5-specific antibody and the HLA-G-specific antibody MEM/G9 (right column) or the respective isotype control (left column). (A) Low HLA-G expression. (B) High HLA-G expression.
Proportion of HLA-G-positive leukemic cells in 47 patients with B-CLL. HLA-G expression was analyzed by flow cytometry. The dotted vertical line at 23% positive cells separates the patients into a prognostically favorable HLA-G-negative and an unfavorable HLA-G-positive group (cf Figure 3).
Proportion of HLA-G-positive leukemic cells in 47 patients with B-CLL. HLA-G expression was analyzed by flow cytometry. The dotted vertical line at 23% positive cells separates the patients into a prognostically favorable HLA-G-negative and an unfavorable HLA-G-positive group (cf Figure 3).
Probability of disease progression (interval from diagnosis to initiation of chemotherapy) in 10 patients with HLA-G-positive and 37 patients with HLA-G-negative B-CLL. Kaplan-Meier analysis. The median progression-free survival time was 23 months in HLA-G-positive and 120 months in HLA-G-negative B-CLL (P = .0001, log-rank test). All patients with HLA-G-positive B-CLL received chemotherapy within 42 months.
Probability of disease progression (interval from diagnosis to initiation of chemotherapy) in 10 patients with HLA-G-positive and 37 patients with HLA-G-negative B-CLL. Kaplan-Meier analysis. The median progression-free survival time was 23 months in HLA-G-positive and 120 months in HLA-G-negative B-CLL (P = .0001, log-rank test). All patients with HLA-G-positive B-CLL received chemotherapy within 42 months.
Of the known prognostic factors, only thymidine kinase was unevenly distributed between the HLA-G-negative and HLA-G-positive groups (Table 1). In order to test the prognostic factors for interdependency, HLA-G status, age (cut-off level: 60 years), Binet stage (A versus B versus C), β2-microglobulin (cut-off level: 3.5 mg/L), CD38 status, and ZAP-70 status (cut-off level: 20% positive cells) were subjected to multivariate Cox regression analysis. In addition to stage, and β2-microglubulin and ZAP-70 expression, HLA-G status was identified as an independent factor predicting disease progression (Table 2). The prognostic value of HLA-G expression in the B-CLL patients studied was even better than the value of the established markers such as ZAP-70 and CD38 status. The risk for HLA-G-positive patients receiving a first-line therapy is nearly 5 times higher compared with HLA-G-negative patients (hazard ratio: 4.8; P = .002). Only the initial staging according to Binet, known as one of the best prognostic markers, showed a higher hazard ratio (8.6; P = .0001).
Multivariate Cox regression analysis model
Variable . | n . | Hazard ratio . | CI, 95% . | P . |
|---|---|---|---|---|
| Age | 47 | 1.02 | 0.97-1.06 | .50 |
| Disease stage at diagnosis | 47 | 8.66 | 3.0-25.1 | .0001 |
| β2-microglobulin | 45 | 3.08 | 1.06-8.95 | .039 |
| ZAP-70 status | 42 | 3.58 | 1.14-11.27 | .029 |
| CD38 status | 47 | 1.83 | 0.48-6.94 | .37 |
| HLA-G status | 47 | 4.81 | 1.74-13.29 | .002 |
Variable . | n . | Hazard ratio . | CI, 95% . | P . |
|---|---|---|---|---|
| Age | 47 | 1.02 | 0.97-1.06 | .50 |
| Disease stage at diagnosis | 47 | 8.66 | 3.0-25.1 | .0001 |
| β2-microglobulin | 45 | 3.08 | 1.06-8.95 | .039 |
| ZAP-70 status | 42 | 3.58 | 1.14-11.27 | .029 |
| CD38 status | 47 | 1.83 | 0.48-6.94 | .37 |
| HLA-G status | 47 | 4.81 | 1.74-13.29 | .002 |
CI indicates confidence interval.
The cut-off levels used in the analysis were as follows: age, 60 years; disease stage at diagnosis, Binet stage A versus B versus C; β2-microglobulin, 3.5 mg/L; ZAP-70 or CD38 status, 20% positive cells; and HLA-G status, 23% positive cells. Because of missing data in a substantial proportion of patients, the prognostic marker thymidine kinase (cf Table 1) was not included in the analysis.
HLA-G expression and immunodeficiency
B-CLL is characterized by humoral immunodeficiency, and a substantial proportion of patients also develop abnormalities in the T-cell compartment. Table 3 shows that HLA-G status was correlated with both humoral and cellular immunosuppression. In comparison with the HLA-G-negative group, patients with HLA-G-positive leukemias had significantly lower levels of IgG, total T cells, and CD4+ T cells. In addition, plasma levels of IL-10 and soluble HLA-G tended to be higher in patients with HLA-G-positive leukemias than in patients with HLA-G-negative leukemias.
Immunologic parameters
Variable . | All patients . | Less than 23% HLA-G—positive cells . | 23% or more HLA-G—positive cells . | P* . |
|---|---|---|---|---|
| Immunglobulin G, g/L, n = 43 | 7.08 ± 3.03† | 7.51 ± 2.9† | 5.47 ± 3.1† | .042 |
| Immunglobulin A, g/L n = 43 | 0.88 ± 0.7† | 0.91 ± 0.7† | 0.77 ± 0.9† | NS |
| Immunglobulin M, g/L, n = 43 | 0.54 ± 0.6† | 0.54 ± 0.7† | 0.53 ± 0.6† | NS |
| Total T cells, per μL, n = 32 | 2780 ± 2475† | 3095 ± 2567† | 1076 ± 581† | .005 |
| CD4+ T cells, per μL, n = 32 | 1508 ± 1304† | 1685 ± 1344† | 556 ± 291† | .016 |
| CD8+ T cells, per μL, n = 32 | 1172 ± 1249† | 1299 ± 1317† | 586 ± 351† | .077 |
| CD4/CD8 ratio, n = 32 | 1.85 ± 1.6† | 1.93 ± 1.7† | 1.4 ± 0.7† | NS |
| IL-10, pg/mL, n = 47 | 5.1 (6.1-43.1)‡ | 4.9 (6.1-43.1)‡ | 10.2 (9.9-40.2)‡ | NS |
| sHLA-G, pg/mL, n = 47 | 17.3 (0.0-82.8)‡ | 16.7 (0.0-82.8)‡ | 26.9 (0.0-34.8)‡ | .043 |
Variable . | All patients . | Less than 23% HLA-G—positive cells . | 23% or more HLA-G—positive cells . | P* . |
|---|---|---|---|---|
| Immunglobulin G, g/L, n = 43 | 7.08 ± 3.03† | 7.51 ± 2.9† | 5.47 ± 3.1† | .042 |
| Immunglobulin A, g/L n = 43 | 0.88 ± 0.7† | 0.91 ± 0.7† | 0.77 ± 0.9† | NS |
| Immunglobulin M, g/L, n = 43 | 0.54 ± 0.6† | 0.54 ± 0.7† | 0.53 ± 0.6† | NS |
| Total T cells, per μL, n = 32 | 2780 ± 2475† | 3095 ± 2567† | 1076 ± 581† | .005 |
| CD4+ T cells, per μL, n = 32 | 1508 ± 1304† | 1685 ± 1344† | 556 ± 291† | .016 |
| CD8+ T cells, per μL, n = 32 | 1172 ± 1249† | 1299 ± 1317† | 586 ± 351† | .077 |
| CD4/CD8 ratio, n = 32 | 1.85 ± 1.6† | 1.93 ± 1.7† | 1.4 ± 0.7† | NS |
| IL-10, pg/mL, n = 47 | 5.1 (6.1-43.1)‡ | 4.9 (6.1-43.1)‡ | 10.2 (9.9-40.2)‡ | NS |
| sHLA-G, pg/mL, n = 47 | 17.3 (0.0-82.8)‡ | 16.7 (0.0-82.8)‡ | 26.9 (0.0-34.8)‡ | .043 |
NS indicates not significant.
In most patients, the laboratory parameters were not determined at diagnosis, but during the course of the disease. Normal ranges are as follows: IgG, 7 to 26 g/L; IgA, 0.7 to 4.0 g/L; IgM, 0.3 to 2 g/L; total T cells, 690 to 2540/μL; CD4+ T cells, 410 to 1590/μL; CD8+ T cells, 190 to 1140/μL; CD4/CD8 ratio, 1.0 to 3.0; IL-10, 20.1 ± 33.2 pg/mL; and sHLA-G, 22.9 ± 16.9 ng/mL.
Comparison between the HLA-G-positive and HLA-G-negative subgroups using the Mann-Whitney U test.
Mean ± standard error of the mean.
Median (range).
Discussion
Expression of HLA-G has been suggested to contribute to immune evasion of tumor cells. This view has recently been strengthened by data demonstrating cell surface expression of HLA-G in melanoma,18,29,30 renal cell carcinoma,17,31 cutaneous lymphoma,32 and glioblastoma33 to be associated with inhibitory effects on diverse types of immune effector cells.
Data regarding HLA-G expression in B-CLL are limited and controversial. While Amiot et al34 found transcription of the HLA-G gene in 3 of 6 cases by reverse transcriptase-polymerase chain reaction, Polakova et al35 and Mizuno et al36 were unable to detect the HLA-G antigen by flow cytometry in 8 or 2 samples of B-CLL, respectively. In our study, the HLA-G antigen was readily demonstrable in a variable proportion of tumor cells in all samples analyzed. Apart from problems related to small sample numbers, these differences are likely to be explained by differences in the properties of the monoclonal antibodies used to detect the HLA-G antigen. While Mizuno and colleagues36 used indirect immunofluorescence using the 87G antibody, our analysis involved direct immunofluorescence using the MEM/G9 antibody. This may be a more sensitive approach to visualize the HLA-G antigen. Furthermore, we measured the HLA-G expression in CD19-enriched B-CLL cells.
Using ROC curve analysis, we were able to show that the degree of HLA-G expression was correlated with the propensity of the disease to progress. Leukemias with a proportion of 23% HLA-G-positive cells or more had a more aggressive course than leukemias with fewer positive cells. Most other prognostic factors, such as the Binet stage, β2-microglobulin, or the CD38 or ZAP-70 expression status, were equally distributed between the 2 groups. Multivariate analysis revealed the HLA-G status to possess independent prognostic value even better than the prognostic value of the established prognostic markers such as ZAP-70 and CD38 status. The risk for HLA-G-positive patients receiving a first-line therapy was nearly 5 times higher in our study group compared with HLA-G-negative patients (Table 2). To our knowledge, this is the first report that describes an association of HLA-G antigen expression and the course of the disease in B-CLL. If our findings are confirmed in an unrelated cohort of patients, the expression of HLA-G will further refine our ability to identify prognostic subgroups in B-CLL.
In trophoblasts and monocytes, expression of HLA-G can be induced by IL-10, involving regulatory pathways not shared by other MHC class I genes.37 IL-10 may act as an autocrine growth factor for B-cell lymphomas. In primary cutaneous lymphomas, its expression is associated with up-regulation of HLA-G lending support to the speculation that IL-10 may control HLA-G expression.38 In our study, plasma levels of IL-10 tended to be higher in the group of patients with HLA-G-positive leukemias than in the group with HLA-G-negative leukemias. Thus, it appears possible that IL-10 may also be involved in the control of HLA-G expression in B-CLL.
The group of patients with HLA-G-positive B-CLL had more pronounced immunologic abnormalities than the group of patients with HLA-G-negative leukemias. HLA-G molecules possess potent immunosuppressive properties as exemplified in vitro by the induction of apoptosis in T cells and the inhibition of effector functions in NK and T cells. HLA-G-induced immunosuppression may result in inadequate responses of the immune system to tumor cells displaying the antigen.39 In our group of patients, HLA-G expression was associated with a more general state of immunosuppression involving both the humoral and the cellular immune systems. In accordance with LeMaoult et al,40 who demonstrated that HLA-G1-transfected antigen-presenting cells inhibit the proliferation of CD4+ T cells and induce CD4+ T-cell anergy, we could show a significantly decreased T-cell count, especially in CD4+ T cells, in the group of patients with HLA-G-positive leukemias. Such global immunodeficiency is a characteristic feature of B-CLL predisposing the patients to bacterial and viral infections. To what extent these observations are causally related to HLA-G, IL-10, or other regulators remains to be clarified.
To our knowledge, this is the first report demonstrating consistent expression of the HLA-G protein in B-CLL. Unexpectedly, its level of expression was correlated with the degree of immunosuppression accompanying the disease and the prognosis of the patients. The major limitation of our analysis is its retrospective nature. HLA-G and other immunologic parameters were determined at variable intervals from diagnosis, and chemotherapy was likely to have had an impact on the results obtained. Therefore, our findings need to be confirmed in a prospective study involving a larger cohort of patients.
Prepublished online as Blood First Edition Paper, October 5, 2004; DOI 10.1182/blood-2004-08-3335.
H.N. and V.R. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Brigitte Fischer for generously contributing information on the clinical course and the treatment histories of the study patients. We thank Babette Pohle and Annika Dolar for expert technical assistance with flow cytometry and measuring sHLA-G and IL-10.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal