Abstract
Epstein-Barr virus (EBV) latently infects and immortalizes B lymphocytes and causes lymphoproliferative malignancies. We show here that the EBV nuclear antigen EBNA2 induces expression of the 2 chains of the interleukin-18 receptor (IL-18R) in Burkitt lymphoma (BL) cell lines and in nontransformed B cells. Activation of IL-18R expression by EBNA2 is independent of its interaction with the transcriptional repressor RBPJκ. It occurs in the absence of any other viral protein but requires de novo synthesis of cellular proteins. IL-18R induction is a highly specific function of EBNA2, because neither other EBV latent proteins nor the cellular proteins c-myc or Notch can exert this effect. Using cDNA microarray expression profiling, we find that the IL-18 receptor expressed in EBV-infected BL cells has signaling capacity, because IL-18 significantly modified gene expression. We report that EBNA2 expression is associated with IL-18R expression in vivo in EBV-positive B-lymphomas from AIDS patients. (Blood. 2005;105:1632-1639)
Introduction
Epstein-Barr virus (EBV) is a ubiquitous human lymphotropic γ herpesvirus that latently infects and immortalizes B cells and persists lifelong in resting memory B cells. Ultimately, latent EBV infection can lead to the development of Burkitt lymphoma (BL), nasopharyngeal carcinoma, Hodgkin disease, posttransplantation lymphoproliferative disease, and immunoblastic lymphoma in immunocompromised patients (reviewed by Young and Murray1 ).
B cells latently infected with EBV express variable subsets of the EBV latency genes, which code for the nuclear antigens EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNA-LP and the membrane proteins LMP1 (latent membrane protein-1), LMP2A (or TP1), and LMP2B (or TP2). Freshly infected naive B cells and in vitro-transformed lymphoblastoid cell lines express the full set of latency genes, while their expression may be restricted to EBNA1 alone in BL (reviewed by Young et al2 ). Uncontrolled proliferation of EBV-immortalized B cells in vivo is prevented by strong primary and memory CD8+ cytotoxic T lymphocyte (CTL) responses directed essentially toward HLA allele-specific epitopes from the latent proteins EBNA3A, EBNA3B, and EBNA3C.3,4 Virus-bearing memory B cells or B-cell tumors escape immune surveillance by down-regulation of immunogenic latent proteins, particularly EBNA2, which activates transcription of the EBNA3 genes.
EBV latent proteins induce the synthesis of surface and soluble proteins, which can interfere with the host's immune response. Thus, EBV infection activates expression of the receptors CD21,5 CD23,6 CD25,7 Fas ligand,8 and BLR2/CCR79 and the cytokines tumor necrosis factor-α (TNF-α),10 lymphotoxin α (LT-α),11 granulocyte colony-stimulating factor (G-CSF),10 interferon-β (IFN-β),12 interleukin-6 (IL-6),13 IL-8,14 IL-10,15 and IL-12.16 The cytokine IL-18 is mainly produced by activated macrophages and dendritic cells; it has multiple functions in the regulation of innate and adaptive immune responses17 and has antitumoral18,19 and antiviral20 properties.
IL-18 binds to the cell through a specific receptor, IL-18R, belonging to the Toll-like receptor family.21 It is composed of IL-18Rα,22 which binds IL-18 with low affinity,23 and IL-18Rβ, which does not bind IL-18 but together with IL-18Rα forms the high-affinity IL-18 receptor and mediates signaling24 through pathways shared with the IL-1 receptor. IL-18R is expressed on a variety of cell types, including T cells, natural killer (NK) cells, and peripheral CD19+ B cells.25 Freshly isolated human tonsillar B cells, however, do not express IL-18R.26,27
IL-18 was found to participate in host reactions to EBV-infected cells. Significant amounts of IL-18 are detected in lymphoid tissues during EBV-induced infectious mononucleosis.28 In athymic mice, murine IL-18 accumulates in experimental tumors of EBV-positive BL cells and may contribute to their rejection.29 The availability of IL-18 in the environment of EBV-infected cells raised the question of whether these cells express the IL-18 receptor and might thus respond to IL-18. It has indeed been found recently that IL-18Rα was strongly expressed in some EBV-transformed cell lines but absent in a series of EBV-negative B-cell tumors.25 These observations prompted us to investigate whether EBV infection could lead to expression of a functional IL-18R in B cells. We report that the latent EBV protein, EBNA2, drives IL-18R expression, allowing the infected cell to respond to IL-18.
Materials and methods
Cell lines
The EBV-negative BL cell lines DG75, BL2, BL30, BL31, BL41, and BL70 and their in vitro-infected counterparts with the EBV strains P3HR-1 (P3) or B95-8 (B95), BL2-P3, BL41-P3, BL2-B95, BL30-B95, BL31-B95, BL41-B95, and BL70-B95 were a gift from G. Lenoir and have been described previously.30-33 Conditionally immortalized EREB2-5 cells, which proliferate as a lymphoblastoid cell line in the presence of estrogen, were obtained from B. Kempkes.34 The cell lines P493-6,33 LMPtet EREB,35 and 141436 were established by stable transfection of EREB2-5 cells with tetracycline-controlled expression plasmids for c-myc, LMP1, and the intracellular domain of Notch1, respectively. The myelomonocytic cell line KG-1 was purchased from the American Type Culture Collection (Manassas, VA).
Cells were cultured in RPMI 1640 medium (Gibco, Gaithersburg, MD) supplemented with 10% fetal calf serum (Gibco), 2 mM l-glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin. For induction of estrogen receptor (ER) chimeras, β-estradiol (estrogen) was added to the culture medium at 1 μM. For repression of tetracycline-controlled transgenes, the medium was supplemented with 0.1 μg/mL tetracycline.
Transfection and plasmids
Cells were transfected by electroporation (240 to 280 V, 975 μF). Expression plasmids coding for the EBV latent proteins EBNA2 and LMP1 were obtained from A. Sergeant and W. Hammerschmidt, respectively. pPDL152, an expression plasmid coding for the RBPJκ-binding deficient mutant WW323SR of EBNA2,37 was obtained from S. D. Hayward.
Luciferase assays were performed as described.38 The nuclear factor-κB (NF-κB) reporter plasmids Igκ3-ConA-Luc39 and p(IL6κB)3-50hu.luc40 were obtained from A. Israel and G. Haegeman, respectively. A luciferase reporter construct for the human IFN-γ promoter41 was obtained from K. Barbulescu. The previously described reporter plasmid Ga981-16 contains 12 tandemly repeated RBPJκ binding sites from the viral TP1 promoter.42 Complementary DNA (cDNA) coding for the human IL-18 receptor alpha chain (IL-18Rα) was obtained from J. E. Sims.22
Immunofluorescence and flow cytometry
For detection of IL-18Rα, cells were reacted with mouse monoclonal antibody MAB840 (R&D Systems, Minneapolis, MN), followed by a secondary fluorescein isothiocyanate (FITC)-labeled antibody or by the phycoerythrin (PE)-labeled mouse monoclonal antibody FAB840P (R&D Systems), according to the manufacturer's recommendations. IL-18Rβ was detected by incubation with goat polyclonal antibody AF118 (R&D Systems), followed by biotinylated antibodies to goat immunoglobulin G (IgG) and streptavidin coupled to FITC. Staining was visualized by fluorescence microscopy using a Zeiss Axiovert 405 microscope equipped with an Achroplan 20 ×/0.45 objective lens (Carl Zeiss, Oberkochen, Germany) and a Hamamatsu C5810 camera (Hamamatsu, Hamamatsu City, Japan). Images were processed with Adobe Photoshop 4.0 software (Adobe, San Jose, CA). For Figure 4 acquisition, we used a Leica DMR microscope equipped with HCPL Fluotar 20 ×/0.5 and Apo oil 63 ×/1.32 objective lenses (Leica, Heidelberg, Germany) and a KAPPA PS30C camera; images were processed with KAPPA ImageBase Control 2.5 software (KAPPA, Gleichen, Germany). Flow cytometry was performed using a FACScan instrument (Becton Dickinson, San Jose, CA) with CellQuest acquisition software.
Northern blots and polymerase chain reaction
Northern blotting was performed according to standard procedures. The radiolabeled cDNA for human IL-18Rα was used as a probe.
RT-PCR for IL-18Rα, IL-18Rβ, and HPRT was as follows: RNA was isolated using the RNeasy Kit (Qiagen, Valencia, CA) and treated with RNAse-free DNase I. Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed using 1 μg total cellular RNA incubated with avian myeloblastosis virus (AMV) reverse transcriptase, primer oligo(dT), deoxyribonucleoside triphosphate (dNTP), and RNase inhibitor for 1 hour at 42°C (cDNA synthesis kit; Boehringer Mannheim Biochemica, Indianapolis, IN). PCR amplification was performed as previously described.19 The sense and antisense primers, respectively, and the size of the PCR products were as follows: IL-18Rα, 5′-TTGGAGTGATGACAGGAACAC-3′, 5′-CATCAGATAGGTCGTTACTACTACC-3′, 223 bp; IL-18Rβ, 5′-GGTTATTACTCCTGCGTGC-3′, 5′-CCATTTTCTTCCCCGAACATCC-3′, 273 bp; and hypoxanthine phosphoribosyltransferase (HPRT), 5′-TTCAAATCCAACAAAGTCTG-3′, 5′-AGCACTGAATAGAAATAGTGATAGA-3′, 278 bp.
Immunohistochemistry: case selection
Tissue specimens from patients with cerebral diffuse large B-cell lymphomas (n = 6) were obtained from J. J. Hauw, Department of Neuropathology, CHU Pitie-Salpetrière, Paris, France, and 7 cases of systemic large B-cell lymphoma from the Department of Pathology, Hôtel-Dieu, Paris. Patient samples were obtained from the pathologists after completion of the diagnostic procedures. Large B-cell lymphomas were diagnosed on the basis of clinical and characteristic histologic features and were classified according to the Revised European-American Lymphoma classification.43 Pertinent selected information on each tissue sample is given in Figure 4.
The avidin-biotin-peroxidase method was performed on frozen sections using the LSAB+ peroxidase kit (Dako, Glostrup, Denmark). Briefly, tissue sections were deparaffinized, subjected to microwave treatment (3 times for 5 minutes), and then incubated in hydrogen peroxide for 15 minutes and blocked with bovine serum albumin for 30 minutes before incubation with the primary antibodies. The complex was visualized with diaminobenzidine, and the nuclei were counterstained with hematoxylin. Anti-IL-18R monoclonal IgG1 (R&D Systems) was used at a final concentration of 0.25 μg/mL, and anti-EBNA2 monoclonal IgG1 (Dako) was used at a final concentration of 32 μg/mL. Irrelevant murine IgG1 was used at the same final concentrations as negative controls. EBV was detected by in situ hybridization of paraffin sections using an FITC-labeled oligonucleotide complementary to EBV-encoded RNA1 (EBER1) (Dakopatts, Glostrup, Denmark).44
Complementary DNA microarrays
BL2-B95 cells (1 × 107) were kept in serum-free medium for 24 hours. Incubation was continued for 24 hours in the presence of 100 ng/mL IL-18 or bovine serum albumin (BSA). RNA was extracted using RNAgents (Promega, Madison, WI). Complemenatary DNA made from 10 μg RNA from each sample and from reference RNA (R) (Clontech, Palo Alto, CA) was labeled with deoxycytidine triphosphate (dCTP) dyes cyanine-3 and cyanine-5 (Perkin Elmer, Norwalk, CT), respectively, according to the instructions of Agilent Technologies (Palo Alto, CA). Sample cDNAs were mixed with reference cDNA and hybridized for 16 hours to the Human 1 cDNA Microarray (Agilent Technologies) containing 12814 unique clones from the clone sets Incyte Unigen 1 and Human Drug Target DNA (Incyte Genomics, St Louis, MO). The reproducibility has been evaluated for these microarrays by the manufacturer (interarray standard deviation of log ratios less than 0.1 for 24 arrays). All microarray hybridizations are assessed for labeling, hybridization efficiency, and sensitivity using specially designed control elements that are added to each labeling reaction (for details, see Galon et al45 ). The detection limit (sensitivity) of the microarrays is such that control targets added to the hybridization reaction are easily detected over background at a concentration of 3.0 copies per cell per million cells. The microarrays were scanned on a dynamic autofocus microarray scanner (Agilent Technologies). Feature Extraction Software (Agilent Technologies) was used to extract data information and to perform statistical analysis of signals. The intensity of the fluorescence at each array element was proportional to the expression level of that gene in the sample. The ratio of the 2 fluorescence intensities provided a quantitative measurement of the relative gene expression level in the 2 samples. The statistical significance of the correlation between 2 DNA microarray experiments was determined using correlation matrix algorithm, Kendall correlation algorithm, and Spearman correlation algorithm. Similar results were found with all algorithms. When no fluorescent signal was detected (signal intensity = intensity of local background + SD local background) in one or both experiments, the corresponding gene was removed from the statistical analysis. Correlation analyses were performed by plotting differential gene expression of one experiment against the value from the other experiment. The correlation coefficient (r) and the statistical significance (P) were determined. Levels of expression after IL-18 treatment were compared with those of BSA-treated cells. Data are presented as mean ± SD and were analyzed by analysis of variance (ANOVA) t test for each gene.
Results
EBNA2 induces expression of IL-18Rα in the absence of other EBV genes
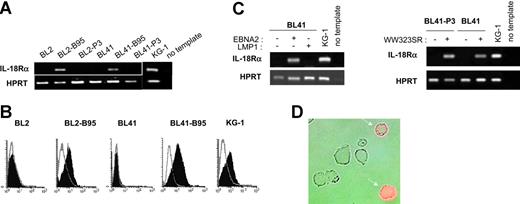
To study whether latent infection of BL cells by EBV could lead to IL-18Rα expression, we analyzed a series of EBV-negative BL cell lines and their in vitro EBV-converted counterparts. RT-PCR using total RNA from the EBV-negative BL cell lines DG75, BL2, BL30, BL31, BL41, and BL70 demonstrated the absence of transcripts specific for IL-18Rα. In contrast, the cell lines BL2, BL30, BL31, BL41, and BL70 accumulated IL-18Rα RNA after conversion with the wild-type EBV-strain B95-8 (Figure 1A and data not shown). Surface expression of the receptor was readily detected in these cells by flow cytometry (Figure 1B). The mean fluorescence intensity was comparable to that observed with the KG-1 cell line derived from an acute myeloblastic leukemia, which has been shown to strongly express IL-18Rα.46 Conversion with the EBNA2-deficient, nontransforming EBV strain P3HR1, however, did not induce IL-18Rα expression (Figure 1A). These results indicate that in vitro infection of BL cells with wild-type EBV leads to expression of IL-18Rα and suggest that this process depends on EBNA2 expression. To identify the EBV latent proteins involved in induction of IL-18Rα expression, we transfected the EBV-negative BL cell lines BL2 and BL41 and their P3HR1 convertants with expression plasmids for EBNA2 or LMP1 or an empty control vector. Proper expression from the EBNA2 and LMP1 coding plasmids was monitored by transactivation of the cotransfected reporter genes Ga981-16 and Igκ3-ConA-Luc, respectively (“Materials and methods” and data not shown). Forty-eight hours after transfection, we analyzed the cells for the presence of IL-18Rα mRNA (Figure 1C) and protein (Figure 1D and data not shown). While EBNA2 transfectants expressed IL-18Rα, IL-18Rα expression was undetectable in LMP1 and control transfectants. These observations imply that EBNA2 can induce IL-18Rα in the absence of other viral proteins and that overexpression of LMP1, even in combination with the latent proteins EBNA1, EBNA3A, EBNA3B, and EBNA3C, which are expressed in P3HR1 convertants,47 cannot substitute for this function of EBNA2. EBNA2 can transactivate viral and cellular genes by binding to and converting the site-specific transcriptional repressor RBPJκ (or CBF1) into a transcriptional activator.48 The expression plasmid pPDL152 codes for the point mutant WW323SR in conserved region 6 (CR6) of EBNA2. The mutation specifically abrogates binding of RBPJκ and derepression of RBPJκ-repressed genes.37 We cotransfected EBV-negative BL cells with plasmid pPDL152 and the Ga981-16 reporter gene. As shown in Figure 1C, IL-18Rα mRNA is efficiently induced by this mutant, which only marginally enhanced transcription of the reporter (data not shown).
EBNA2 induces IL-18Rα expression in BL cells. (A) RT-PCR for IL-18Rα and the housekeeping gene, HPRT, in BL2, BL2-B95, BL2-P3, BL41, BL41-B95, BL41-P3, and KG-1 cells. (B) Flow cytometric analysis of IL-18Rα expression in BL2, BL2-B95, BL41, BL41-B95, and KG-1 cells. Cells were reacted with a mouse monoclonal antibody to IL-18Rα (filled histogram) or an isotype-matched control antibody (open histogram), followed by an FITC-labeled secondary antibody. (C) RT-PCR for IL-18Rα and HPRT in BL41 and BL41-P3 cells transfected with expression plasmids for EBNA2, LMP1, or the EBNA2-mutant WW323SR, as indicated. (D) BL2 cells, transiently transfected with an expression plasmid for EBNA2, were reacted 48 hours after transfection with a phycoerythrin-labeled mouse monoclonal antibody to IL-18Rα. Micrograph shows phase contrast and red fluorescence.
EBNA2 induces IL-18Rα expression in BL cells. (A) RT-PCR for IL-18Rα and the housekeeping gene, HPRT, in BL2, BL2-B95, BL2-P3, BL41, BL41-B95, BL41-P3, and KG-1 cells. (B) Flow cytometric analysis of IL-18Rα expression in BL2, BL2-B95, BL41, BL41-B95, and KG-1 cells. Cells were reacted with a mouse monoclonal antibody to IL-18Rα (filled histogram) or an isotype-matched control antibody (open histogram), followed by an FITC-labeled secondary antibody. (C) RT-PCR for IL-18Rα and HPRT in BL41 and BL41-P3 cells transfected with expression plasmids for EBNA2, LMP1, or the EBNA2-mutant WW323SR, as indicated. (D) BL2 cells, transiently transfected with an expression plasmid for EBNA2, were reacted 48 hours after transfection with a phycoerythrin-labeled mouse monoclonal antibody to IL-18Rα. Micrograph shows phase contrast and red fluorescence.
We conclude that IL-18Rα induction by EBNA2 is independent of its capacity to bind RBPJκ and does not require other EBV proteins.
Induction of IL-18Rα is highly specific to EBNA2
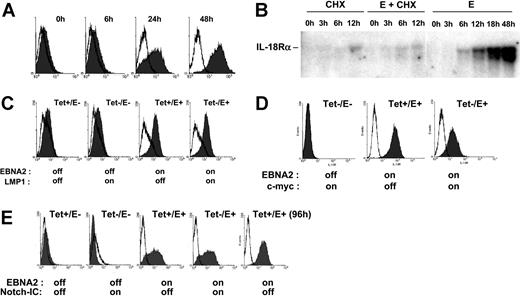
To further examine the role of EBNA2 in IL-18Rα expression, we used conditionally immortalized EREB2-5 cells. These cells harbor the EBNA2-deficient nontransforming EBV strain P3HR1 and constitutively express EBNA2 fused to the estrogen receptor (ER), which translocates to the nucleus upon estrogen addition and drives proliferation.34 Incubation of EREB2-5 cells in the presence of estrogen rapidly led to IL-18Rα protein expression on the cell surface (Figure 2A). To assess whether induction of IL-18Rα by EBNA2 required de novo protein synthesis, we recorded the accumulation of IL-18Rα mRNA in estrogen-treated EREB2-5 cells in the presence and in the absence of the protein synthesis inhibitor, cycloheximide. Figure 2B shows that IL-18Rα mRNA was detectable already 6 hours after estrogen addition. While cycloheximide alone also led to accumulation of a small amount of IL-18Rα transcripts, it almost completely abrogated their induction by EBNA2. Given that other viral proteins are not required (Figure 1), these data indicate that activation of IL-18Rα expression by EBNA2 depends on de novo synthesis of a cellular protein(s). LMP1 and EBNA2 have overlapping functions as transactivators of cellular genes. We asked whether LMP1, together with the P3HR1-encoded latent proteins, could replace EBNA2 in induction of IL-18Rα expression. We made use of the cell line LMPtet-EREB (ie, EREB2-5 cells stably transfected with a tetracycline-controlled expression plasmid for LMP1). We induced LMP1 expression and/or EBNA2 expression in these cells and monitored the appearance of IL-18Rα on the cell surface. Figure 2C shows that LMP1 expression in the absence of EBNA2 is not sufficient to induce IL-18Rα expression in EREB2-5 cells and that it does not interfere with induction of IL-18Rα by EBNA2. These data confirm again that the IL-18Rα gene is not a target of LMP1.Acandidate cellular gene to mediate induction of IL-18Rα expression by EBNA2 is the transcription regulator c-myc, because the c-myc gene is activated by EBNA249 through an RBPJκ-independent mechanism. In addition, constitutive expression of c-myc can substitute for EBNA2 in maintenance of proliferation in EREB2-5 cells.33 We therefore asked whether it was also sufficient to drive IL-18Rα expression. The cell line P493-6 was established by stable transfection of EREB2-5 cells with a tetracycline-controlled expression plasmid for c-myc. We induced c-myc expression and/or EBNA2 expression in these cells and analyzed IL-18Rα RNA and protein expression by Northern blotting and flow cytometry. Figure 2D shows that c-myc cannot activate IL-18Rα in P493-6 cells in the absence of EBNA2. Rather, IL-18Rα induction by EBNA2 appears reduced on the protein and RNA levels when c-myc is overexpressed. We conclude that, even in the P3HR1 background, c-myc cannot replace EBNA2 in IL-18Rα induction and that the latter does not result from activation of the proliferative program that is triggered by c-myc. EBNA2 and the cellular receptor, Notch, have overlapping functions beyond their capacity to interact with RBPJκ. Thus, the activated form of Notch, Notch-IC, can induce the EBNA2 target genes CD21 and CD23 in EREB2-5 cells,50 and it can partially substitute for the growth-promoting functions of EBNA2 in the presence of LMP1.36 To investigate whether Notch could activate IL-18Rα expression, we used the cell line 1414, derived from EREB2-5 by stable transfection of a tetracycline-controlled expression plasmid for Notch-IC. Figure 2E shows that induction of Notch-IC is unable to trigger IL-18Rα expression in the absence of EBNA2.
EBNA2 but not LMP1, c-myc, or Notch induces IL-18Rα expression in nontransformed B cells. (A) Flow cytometric analysis of IL-18Rα expression (Figure 1B) in EREB2-5 cells treated with estrogen for the indicated times. (B) EREB2-5 cells were treated with estrogen (abbreviated E) and/or cycloheximide (CHX). Total RNA was prepared at the indicated time points, blotted, and probed with IL-18Rα cDNA. (C) Flow cytometric analysis of IL-18Rα expression in LMPtet-EREB cells treated with estrogen and/or tetracycline for 24 hours, as indicated. (D) Flow cytometric analysis of IL-18Rα expression in P493-6 cells treated with estrogen (EBNA2 on) and/or tetracycline (c-myc off) for 24 hours, as indicated. (E) Flow cytometric analysis of IL-18Rα expression in 1414 cells treated with estrogen (EBNA2 on) and/or tetracycline (Notch-IC off) for 24 or 96 hours, as indicated.
EBNA2 but not LMP1, c-myc, or Notch induces IL-18Rα expression in nontransformed B cells. (A) Flow cytometric analysis of IL-18Rα expression (Figure 1B) in EREB2-5 cells treated with estrogen for the indicated times. (B) EREB2-5 cells were treated with estrogen (abbreviated E) and/or cycloheximide (CHX). Total RNA was prepared at the indicated time points, blotted, and probed with IL-18Rα cDNA. (C) Flow cytometric analysis of IL-18Rα expression in LMPtet-EREB cells treated with estrogen and/or tetracycline for 24 hours, as indicated. (D) Flow cytometric analysis of IL-18Rα expression in P493-6 cells treated with estrogen (EBNA2 on) and/or tetracycline (c-myc off) for 24 hours, as indicated. (E) Flow cytometric analysis of IL-18Rα expression in 1414 cells treated with estrogen (EBNA2 on) and/or tetracycline (Notch-IC off) for 24 or 96 hours, as indicated.
Taken together, these results show that activation of the IL-18Rα gene is an activity highly specific to EBNA2 that is not shared by the known viral or cellular proteins with similar functions.
EBNA2 activates IL-18Rβ in a manner very similar to IL-18Rα
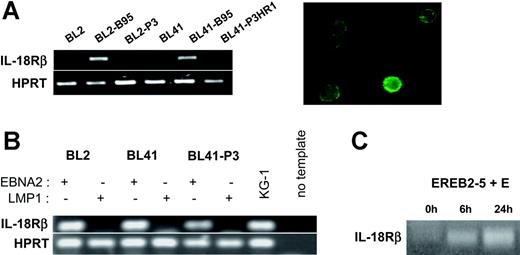
The high-affinity IL-18 receptor is composed of IL-18Rα, which binds IL-18, and IL-18Rβ, which mediates signaling.24 Hence, the biologic consequences of induction of IL-18Rα expression by EBNA2 depend on the presence of IL-18Rβ in the EBV-infected cells. We therefore analyzed the expression of IL-18Rβ in the same cellular settings used for the analysis of IL-18Rα. RT-PCR from total RNA of the EBV-negative BL cell lines DG75, BL2, BL30, BL31, BL41, and BL70 demonstrated the absence of transcripts specific for IL-18Rβ. The B95-8 convertants, however, clearly expressed IL-18Rβ mRNA and surface protein, while P3HR1 convertants did not (Figure 3A and data not shown). This expression profile was strikingly similar to that observed for IL-18Rα. Upon transfection into the EBV-negative BL cell lines BL2 and BL41, EBNA2 induced IL-18Rβ mRNA in the absence of other viral proteins and independently of its capacity to bind RBPJκ. LMP1, even in the P3HR1 background, could not substitute for EBNA2 (Figure 3B and data not shown). Induction of EBNA2 expression in EREB2-5 cells led to accumulation of IL-18Rβ RNA with similar kinetics as for IL-18Rα; this effect was inhibited by cycloheximide (Figure 3C and data not shown). Using the cell lines P493-6 and 1414, we found that the IL-18Rβ gene is neither inducible by c-myc nor by Notch in the presence of P3HR1 (data not shown). These data demonstrate that the genes coding for the 2 chains of the IL-18 receptor are both specifically targeted by EBNA2.
EBNA2 induces IL-18Rβ expression. (A) (Left) RT-PCR for IL-18Rβ and the housekeeping gene, HPRT, in BL2, BL2-B95, BL2-P3, BL41, BL41-B95, and BL41-P3HR1 cells. (Right) Green fluorescence micrograph of BL41-B95 cells, reacted with a goat polyclonal antibody to IL-18Rβ, followed by biotinylated antibodies to goat IgG and FITC-coupled streptavidin. (B) RT-PCR for IL-18Rβ and HPRT in BL2, BL41, and BL41-P3 cells transfected with expression plasmids for EBNA2 or LMP1, as indicated. (C) RT-PCR for IL-18Rβ in EREB2-5 cells treated with estrogen (E) for the indicated times.
EBNA2 induces IL-18Rβ expression. (A) (Left) RT-PCR for IL-18Rβ and the housekeeping gene, HPRT, in BL2, BL2-B95, BL2-P3, BL41, BL41-B95, and BL41-P3HR1 cells. (Right) Green fluorescence micrograph of BL41-B95 cells, reacted with a goat polyclonal antibody to IL-18Rβ, followed by biotinylated antibodies to goat IgG and FITC-coupled streptavidin. (B) RT-PCR for IL-18Rβ and HPRT in BL2, BL41, and BL41-P3 cells transfected with expression plasmids for EBNA2 or LMP1, as indicated. (C) RT-PCR for IL-18Rβ in EREB2-5 cells treated with estrogen (E) for the indicated times.
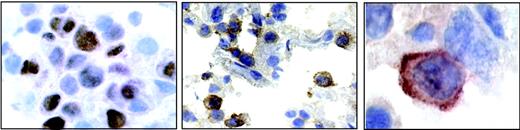
A section from patient 4. (Left) Nuclear staining for EBNA2. (Middle) Positive staining for IL-18Rα in a subset of immunoblastic cells. (Right) Highermagnification micrograph showing membrane staining for IL-18Rα.
A section from patient 4. (Left) Nuclear staining for EBNA2. (Middle) Positive staining for IL-18Rα in a subset of immunoblastic cells. (Right) Highermagnification micrograph showing membrane staining for IL-18Rα.
IL-18 receptor expression is linked to EBNA2 expression in vivo
In vivo, EBNA2 is expressed mainly in freshly infected B cells,51 in posttransplantation EBV-associated lymphoproliferative disease, and in EBV-positive lymphomas in AIDS patients, with particularly high incidence in primary central nervous system lymphomas.52 To assess whether EBNA2 expression was linked to IL-18Rα expression in vivo, we examined 13 cases of diffuse large B-cell lymphoma (6 cases of primary central nervous system lymphoma from AIDS patients and 7 cases from other anatomic sites) for the presence of EBV and for the expression of EBNA2 and IL-18Rα. The results are listed in Table 1. Eight of 13 cases were EBV positive as judged by in situ hybridization with a probe specific for EBER1 RNA, as previously published.44 The typical nuclear staining for EBNA2 was seen in 5 cases (4 cerebral and 1 peripheral lymphoma), with a range of EBNA2-positive cells among neoplastic cells from 35% to 70%. A representative example of EBNA2 staining (patient 4) is shown in Figure 4. IL-18Rα expression was observed in 5 of 13 lymphomas tested, where 10% to 65% of the tumor cells stained positive for IL-18Rα (Figure 4). Four of 5 immunoblastic lymphomas expressing EBNA2 stained positive for IL-18Rα. Seven of 8 EBNA2-negative cases stained negative for IL-18Rα. The significant correlation between EBNA2 and IL-18Rα staining (P < .01; χ2 test) supports the notion that EBNA2 is linked to IL-18Rα expression in vivo.
Immunohistochemical staining for EBNA2 and IL-18Rα in a series of immunoblastic lymphomas
Patient no. . | Histologic subtype . | Anatomic site . | EBER* . | EBNA2† . | IL-18Rα† . |
|---|---|---|---|---|---|
| 1 | DLBL (I-BL) | Brain | + | — | — |
| 2 | DLBL (IB) | Brain | + | + (70%) | + (50%) |
| 3 | DLBL (IB) | Brain | + | + (70%) | + (20%) |
| 4 | DLBL (IB) | Brain | + | + (65%) | + (60%) |
| 5 | DLBL (IB) | Brain | + | + (70%) | + (10%) |
| 6 | DLBL (IB) | Brain | ND | — | + (65%) |
| 7 | DLBL (IB) | Lymph node | + | — | — |
| 8 | DLBL (IB) | Testis | — | — | — |
| 9 | DLBL (IB) | Skin | — | — | — |
| 10 | DLBL (CB) | Lymph node | — | — | — |
| 11 | DLBL (IB) | Lymph node | — | — | — |
| 12 | DLBL (IB) | Lymph node | + | + (35%) | — |
| 13 | DLBL (IB) | Lymph node | + | — | — |
Patient no. . | Histologic subtype . | Anatomic site . | EBER* . | EBNA2† . | IL-18Rα† . |
|---|---|---|---|---|---|
| 1 | DLBL (I-BL) | Brain | + | — | — |
| 2 | DLBL (IB) | Brain | + | + (70%) | + (50%) |
| 3 | DLBL (IB) | Brain | + | + (70%) | + (20%) |
| 4 | DLBL (IB) | Brain | + | + (65%) | + (60%) |
| 5 | DLBL (IB) | Brain | + | + (70%) | + (10%) |
| 6 | DLBL (IB) | Brain | ND | — | + (65%) |
| 7 | DLBL (IB) | Lymph node | + | — | — |
| 8 | DLBL (IB) | Testis | — | — | — |
| 9 | DLBL (IB) | Skin | — | — | — |
| 10 | DLBL (CB) | Lymph node | — | — | — |
| 11 | DLBL (IB) | Lymph node | — | — | — |
| 12 | DLBL (IB) | Lymph node | + | + (35%) | — |
| 13 | DLBL (IB) | Lymph node | + | — | — |
Results of the immunohistochemical analysis of EBER1, EBNA2, and IL-18Rα.
DLBL indicates diffuse large B-cell lymphoma; I-BL, immunodeficiency-associated Burkitt lymphoma (World Health Organization classification of tumors); IB, immunoblastic variant; ND, not determined; CB, centroblastic variant.
EBER status was determined in a standard in situ hybridization assay.
The percentages of EBNA2-positive or IL-18Rα-positive neoplastic cells are indicated in parentheses.
IL-18 modifies gene expression in EBV-infected BL cells
Little is known about the signaling capacity of IL-18R in B cells. In primary human B cells, IL-18 signals to NF-κB and synergizes with IL-12 in IFN-γ production only after preactivation by IL-12.26 We treated the EBV-positive BL cells BL2-B95, which express IL-18R (Figure 1) with various amounts of IL-18. We did not observe enhanced activity of the NF-κB reporter genes Igκ3-ConA-Luc and p(IL6κB)3-50hu.luc, or of the human IFN-γ promoter, or enhanced IFN-γ production, even after pretreatment with IL-12 (data not shown).
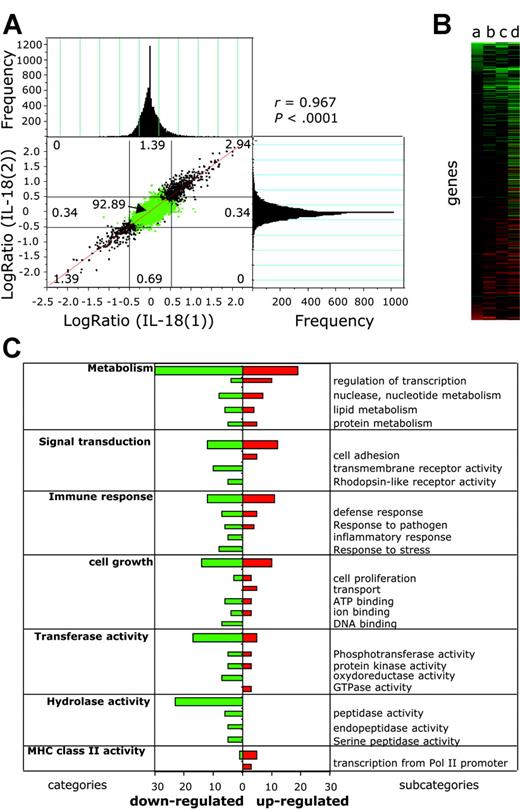
To assess the capacity of EBV-infected BL cells to react to IL-18 alone, we compared the gene expression profile of IL-18-treated BL2-B95 cells with that of BSA-treated cells using cDNA microarrays (Figure 5). To measure the reproducibility of the results and to exclude false positive and false negative results, we replicated the experiment (ie, we performed 4 microarray hybridizations using probes from separate duplicates of RNA prepared from BSA-treated [experiments BSA(1), BSA(2)] and IL-18-treated [experiments IL-18(1), IL-18(2)] cells compared with the reference [R]). Statistical significance of the correlation between experiments IL-18(1) and IL-18(2) was determined (Figure 5A) by plotting differential gene expression of one experiment against the value from the other experiment.45 An excellent correlation between experiments was observed (r = 0.967), which was statistically significant (P < .0001), showing the homogeneity and the consistency of the cyanine-3 (Cy3) and and cyanine-5 (Cy5) signals over different experiments. The frequency of genes for a given log ratio is shown in the histograms (Figure 5A). The sensitivity and reproducibility of the DNA microarrays is highlighted by the fact that common regulated genes were detected in separate DNA microarray experiments. In the whole data set we could not find a single major difference (genes regulated in opposite directions) between the experiments, showing a very low variance between microarrays. Good intramicroarray reproducibility on different cDNAs from the same gene was also found (eg, coregulation of major histocompatibility complex [MHC] class II DQ, clone M24364, clone U83582, and clone M60028).
IL-18 modifies gene expression in EBV-infected BL cells. Differential gene expression in IL-18-treated versus BSA-treated BL2-B95-8 cells was analyzed using DNA microarrays. (A) Correlation analysis was performed by plotting differential gene expression (log ratio) from experiment IL-18(1) against that of experiment IL-18(2) (see “Results”). Negative values represent down-regulated genes, and positive values represent up-regulated genes. Histograms represent the frequency of genes with differential expression (log ratio). Correlation coefficient between experiments (r = 0.967) and statistical significance (P < .0001) was determined. (B) Hierarchical clustering analysis of genes differentially regulated between IL-18- and BSA-treated cells. Values a to d (see “Results”) are plotted for genes having a significant (P < .05) difference of expression. (C) Genes having a significant (P < .05) difference of expression between BSA- and IL-18-treated cells were grouped in functional categories according to the Gene Ontology Database (GO; http://www.godatabase.org). Bars represent the number of IL-18-regulated genes in the indicated functional clusters and subcategories.
IL-18 modifies gene expression in EBV-infected BL cells. Differential gene expression in IL-18-treated versus BSA-treated BL2-B95-8 cells was analyzed using DNA microarrays. (A) Correlation analysis was performed by plotting differential gene expression (log ratio) from experiment IL-18(1) against that of experiment IL-18(2) (see “Results”). Negative values represent down-regulated genes, and positive values represent up-regulated genes. Histograms represent the frequency of genes with differential expression (log ratio). Correlation coefficient between experiments (r = 0.967) and statistical significance (P < .0001) was determined. (B) Hierarchical clustering analysis of genes differentially regulated between IL-18- and BSA-treated cells. Values a to d (see “Results”) are plotted for genes having a significant (P < .05) difference of expression. (C) Genes having a significant (P < .05) difference of expression between BSA- and IL-18-treated cells were grouped in functional categories according to the Gene Ontology Database (GO; http://www.godatabase.org). Bars represent the number of IL-18-regulated genes in the indicated functional clusters and subcategories.
The gene expression differences between IL-18- and BSA-treated cells (log ratio, log2(IL-18/BSA)) were calculated from the differences log2(IL-18(1)/R(1) - log2(BSA(1)/R), log2(IL-18(2)/R(1) - log2(BSA(1)/R), log2(IL-18(1)/R - log2(BSA(2)/R), and log2(IL-18(2)/R(1) - log2(BSA(2)/R) (values a to d), the variance being 2σ.2 The mean and SD were calculated, and genes having a significant (P < .05) difference of expression between IL-18- and BSA-treated cells were plotted using hierarchical clustering (Figure 5B). Of the 12 814 unique human expressed genes of the DNA microarray, 0.57% were down-regulated (73 genes) and 0.71% up-regulated (91 genes) using a P < .05 across all experiments, whereas 32 genes and 26 genes were respectively up-regulated and down-regulated using a P < .01. Up-regulated genes include CD83, CD86, RANTES (regulated on activation normal T cells expressed and secreted), and the MHC alleles DQα1, DQβ1, and DNα. Down-regulated genes include HLA-DR.
Using the Gene Ontology database (GO, http://www.godatabase.org), IL-18-regulated genes were clustered into functional categories (Figure 5C). Expression of genes from 7 families having a frequency of regulated genes above 6 was altered by IL-18. These include metabolism, cell growth, signal transduction, transferase and hydrolase activities, immune response, and MHC class II receptor activity-regulated genes.
These data imply that EBV infection leads to expression of a functional IL-18 receptor that signals in response to IL-18.
Discussion
The transcription factor EBNA2 activates, on one hand, cellular genes that are involved in intrinsic growth and survival control, like cyclinD2,53 c-myc,49 and c-fgr.54 On the other hand, it induces expression of cell surface and soluble proteins that modulate the cell's interaction with the environment, like the receptors CD21,5 CD23,6 and BLR2/CCR79 and the cytokines TNF-α and LT-α.10
EBNA2 cannot bind DNA directly but contacts responsive promoters through interactions with cellular factors that include the sequence-specific repressor RBPJκ (CBF1),42,55 Spi-1/PU.1,56,57 and activating transcription factor (ATF).58 It also can modulate transcription through interactions with components of the basal transcription machinery59-61 and probably through chromatin modification, because it binds the SWI/SNF chromatin-remodeling complex62,63 and can form complexes with the histone deacetylase HDAC2 independently of its ability to bind RBPJκ.64
We have shown here that EBNA2 induces expression of the 2 chains of the IL-18 receptor in B cells. This activity is independent of its capacity to bind to RBPJκ. No other EBV proteins but de novo synthesis of a cellular protein(s) are required for this effect.
The EBV latent membrane protein LMP1 and the cellular proteins c-myc and Notch have overlapping functions with EBNA2, particularly as transcriptional activators. LMP1 can partially substitute for EBNA2 in cell survival35 and in induction of the surface markers CD21, CD23, intercellular adhesion molecule 1 (ICAM-1), and lymphocyte function antigen-1 (LFA-1).65,66 Notch can partially36 and c-myc fully33 replace EBNA2 in growth promotion in the presence of the P3HR1 genome. We report here that neither LMP1, c-myc, nor Notch can induce expression of IL-18R even if the full set of EBV latent proteins (except EBNA2) is coexpressed. The IL-18R genes hence belong to the hitherto relatively small group of genes identified as specific targets of EBNA2, which includes TNF-α, LT-α,10 and BLR2/CCR7.9
The c-myc gene is transcriptionally activated by EBNA2 in an RBPJκ-independent fashion10 and hence was a good candidate to mediate IL-18 activation by EBNA2. However, its overexpression in P493-6 cells was not sufficient to drive IL-18R expression in the absence of EBNA2 (Figure 2D). Rather, c-myc overexpression reduced IL-18R induction by EBNA2 in P493-6 cells, in keeping with the previous observation that c-myc down-regulates several B-cell activation markers in this cell line.67
Taken together, our data demonstrate that the IL-18R genes are activated through a mechanism that is highly specific to EBNA2 among the EBV latent genes and among the known cellular genes with similar functions as EBNA2.
Presence of the 2 chains of the IL-18 receptor has been shown to be sufficient for high-affinity binding of IL-18.24 However, signaling capacity of IL-18 in B cells has been demonstrated only after pretreatment with IL-12.26,68 In our hands, IL-18 neither enhanced the activity of NF-κB reporter genes nor of the IFN-γ promoter and did not lead to enhanced IFN-γ secretion in BL2-B95 cells even after pretreatment with IL-12, in keeping with previous reports showing that EBV-transformed B-cell lines lack a functional IL-12 receptor.69,70 However, using microarray expression profiling, we could show here that IL-18 significantly modifies gene expression in BL2-B95-8 cells (Figure 5). In a total of 12 800 genes analyzed in 4 DNA microarray experiments, we found that IL-18 treatment significantly and reproducibly alters the expression of at least 58 genes (P < .01). This result implies a signaling capacity of the EBNA2-induced IL-18 receptor, because IL-18 does not bind to other surface molecules.71 Analysis of the regulatory elements of the IL-18-regulated genes will contribute to elucidate the signal transduction pathways of IL-18R in B cells.
While we could not find IL-18 to be secreted by the BL cells themselves in vitro (data not shown), IL-18 may be available to EBV-infected cells in vivo as part of T-cell-dependent and -independent immune reactions to EBV. Thus, elevated levels of IL-18 are found in lymphoid tissues during infectious mononucleosis,28 and substantial amounts of murine IL-18 accumulate in experimental tumors of LMP1-expressing BL cells in athymic mice.29 In patients, EBNA2 is mainly expressed in newly EBV-infected B cells51,72 and in EBV-positive lymphomas of immunocompromised patients, particularly of the central nervous system.52 We examined here a series of cerebral immunoblastic lymphomas from AIDS patients and report a correlation between EBNA2 expression and IL-18Rα receptor expression (Figure 4). This supports the notion that EBNA2 can induce IL-18Rα expression in vivo. Our observations prompt us to investigate whether IL-18R expression has a role in the establishment and maintenance of EBV-infected B cells, in their EBNA2-dependent immunogenicity, and in their immune surveillance.
Prepublished online as Blood First Edition Paper, October 21, 2004; DOI 10.1182/blood-2004-08-3196.
Supported by Association pour la Recherche sur le Cancer (G.K., B.H.), and by the Alliance des Recherches sur le Cancer (ARECA) network.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank G. Lenoir, J. Feuillard, F. Baran-Marszak, M. Rowe, B. Kempkes, and U. Zimber-Strobl for cell lines; J. E. Sims, A. Israel, H. Gruffat, A. Sergeant, G. Haegeman, K. Barbulescu, and W. Hammerschmidt for plasmids; and J. J. Hauw for providing tissue specimens from patients with DLBL.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal