Abstract
Tumors have several mechanisms to escape from the immune system. One of these involves expression of intracellular anticytotoxic proteins that modulate the execution of cell death. Previously, we have shown that the serine protease inhibitor (serpin) SPI-6, which inactivates the cytotoxic protease granzyme B (GrB), is capable of preventing cytotoxic T lymphocyte (CTL)–mediated apoptosis. Despite its potent antiapoptotic activity, SPI-6 does not prevent membranolysis induced by cytotoxic lymphocytes. We now provide evidence that several colon carcinoma cell lines do resist membranolysis and that this protection is dependent on SPI-6 but also requires expression of a closely related serpin called SPI-CI (serine protease inhibitor involved in cytotoxicity inhibition). Expression of SPI-CI is absent from normal colon but observed in placenta, testis, early during embryogenesis, and in cytotoxic lymphocytes. SPI-CI encodes a chymotrypsin-specific inhibitor and irreversibly interacts with purified granzyme M. Moreover, SPI-CI can protect cells from purified perforin/GrM-induced lysis. Our data therefore indicate that SPI-CI is a novel immune escape molecule that acts in concert with SPI-6 to prevent cytotoxic lymphocyte-mediated killing of tumor cells.
Introduction
Cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells induce cell death via 2 distinct mechanisms: the oligomerization of death receptors and secretion of cytolytic granules. These granules contain several important death effector molecules, such as the pore-forming protein perforin and a variety of serine proteases known as granzymes.1,2 Granzyme B (GrB) is the key granzyme in rapid induction of apoptosis.3,4 However, other proapoptotic enzymes, such as granzyme A (GrA)5-7 and granzyme C (GrC),8 can independently induce cell death with morphologic features reminiscent of apoptosis. Entry of these proteases into the target cells depends on the presence of perforin, but how this is achieved is debated. The original concept relied on the capacity of perforin to induce plasma membrane damage that provided access for entry of granzymes into target cells. It is clear, though, that GrB can enter target cells independent of perforin.9 Upon such receptor-mediated endocytosis, perforin is suggested to release granzymes from the endosomes by disruption of the endosomal membrane.9-11
Despite the multitude of granzymes, in vitro data using granzyme or perforin-deficient mice indicate that perforin is crucial in the induction of target cell lysis, whereas most granzymes are dispensable.12 This hypothesis was corroborated by Trapani et al, showing that tumor eradication is unaffected in GrA/GrB double knock-out mice,13,14 which are reportedly also diminished in the expression of granzymes C, D, F, and G.15 This suggests that perforin, possibly aided by an as yet undefined granzyme, is sufficient for effector cells to exert their lytic function in vivo. Importantly, recent data indicate that granzyme M (GrM) with perforin can induce target cell lysis.16 Because GrM is not deleted from GrA/GrB double knock-outs, it could be crucial for cytolysis in vivo. However, others have shown that GrA and GrB are essential in tumor rejection in mice, because deletion of these granzymes resulted in increased tumor growth.17 Similarly, clearance of some viral infections is ineffective in GrA/GrB double-knock-out mice,18 suggesting that both perforin and granzymes are required for efficient elimination of target cells.
Better insight into perforin-dependent killing can be attained by studying tumor cells that have acquired defense mechanisms against this cytotoxic pathway. Recently, we have shown that several human and murine tumors highly express the human serine protease inhibitor (serpin) PI-9 or its murine homolog SPI-6 19 and thereby prevent the induction of apoptosis via the perforin/GrB pathway.19,20 This suggests that PI-9/SPI-6 has an important role in immune escape of tumors. In agreement, PI-9 expression in non-Hodgkin lymphomas was linked with high-grade malignancy21 and also resulted in a poor prognosis in patients with anaplastic large cell lymphoma.22 The specificity of PI-9/SPI-6 for GrB, and of serpins in general, is defined by the C-terminal reactive center loop (RCL), which acts as a pseudosubstrate.23 On protease binding, the serpin is cleaved at the RCL between 2 amino acids designated P1 and P1′. This cleavage results in a conformational transition and formation of an irreversible serpin-protease complex that is protease-dead.24 This suicide inhibitory mechanism is very effective and, because PI-9 and SPI-6 were originally detected in CTLs and NK cells, they were suggested to protect effector cells from their own GrB.25-27 Combined, these observations indicate that expression of protease inhibitors by target cells can protect them from the lytic effects imposed by the immune system.
In view of the diversity of proapoptotic enzymes that are present in granules of cytotoxic lymphocytes, it is conceivable that malignant cells may have acquired expression of other serpins to counteract this variety of proteases. In this paper we describe a novel serpin that is expressed in colon carcinoma lines as well as in NK cells and CTLs. Importantly, this serpin can, in concert with SPI-6, prevent CTL-induced lysis in vitro. These data therefore reinforce the notion that perforin requires protease activity to kill target cells and provide compelling evidence for yet another potent immune-evasive mechanism.
Materials and methods
Mice and lines
C57BL/6Kh, GrB–/–, and GrAB–/– mice were bred at Central Animal Facility LUMC (Leiden, The Netherlands). Murine tumor lines AF11, CMT93, MBL2-Fas/FLIP (MFF), MC38, TC-1, XhoC3, C26, CC36, CD4+ T-cell clones 3A12 and 23.2, and CD8+ T-cell clones 1H11 and LP9 were cultured in Iscove modified Dulbecco medium (IMDM) containing 8% fetal calf serum (FCS), glutamine, penicillin/streptomycin (pen/strep), and β-mercaptoethanol (β-ME). AF11, MFF, and its derivatives are kept under puromycin selection. The adenovirus E1A-(line 5) and E1B-specific (100B6) CTL clones were cultured as previously described.19
Retroviral transduction of MFF was performed with an LZRS-based vector encoding enhanced green fluorescent protein (eGFP) alone, SPI-CI (serine protease inhibitor involved in cytotoxicity inhibition) (vesicular stomatitis virus (VSV)–tagged) plus ΔNGF-R (nerve growth factor receptor), or SPI-6 (VSV-tagged) plus eGFP. Transduced cells were sorted on basis of eGFP and/or ΔNGF-R expression.
SPI-CI cloning
PolyA-positive RNA was isolated from CMT93 using an mRNA isolation kit (Qiagen, Venlo, The Netherlands). Complementary DNA was synthesized from 5 μg polyA-positive RNA using SuperScript reverse transcriptase (Invitrogen, Breda, The Netherlands) and cloned into a SuperScript λ system. Murine SPI-CI was cloned by screening about 106 λ phages using a probe spanning the SPI-6 coding sequence. Positive plaques were purified, cloned into pZL1, and sequenced.
Reverse transcriptase–polymerase chain reaction (RT-PCR) and Northern blot
Multiple Tissue cDNA was purchased from BD Biosciences (Alphen aan den Rijn, The Netherlands), except for placenta and colon. Total RNA of placenta and colon was isolated using Trizol (Invitrogen). Synthesis of cDNA was performed using 2 μg total RNA and avian myeloblastosis virus (AMV) reverse transcriptase (Promega, Leiden, The Netherlands).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) polymerase chain reaction (PCR) (232 base pair [bp]) was performed with GAGCCAACGGGTCATCATCT and GAGGGGCCATCCACAGTCTT at an annealing temperature of 58°C. SPI-CI was amplified at an annealing temperature of 43°C with CCTGACTGCTCACAAGCC and AGTAAAACCCATAGCACT both in the 3′ untranslated region (3′UTR) (207 bp, 1622 to 1828). These were chosen for the low homology with SPI-6 and the fact that the corresponding stretch in SPI-6 is only 174 bp and can thus be discriminated.
For Northern blot analysis, 10 μg total RNA was loaded onto agarose and transferred to Hybond N+ membrane. SPI-6 and SPI-CI were detected using [32P]-labeled probes spanning the coding region of SPI-6 or specific 3′UTR sequences of SPI-6 (1392 to 1714) and SPI-CI (1511 to 1828). Probes were generated with PCR using 32P-labeled cytidine.
Western blot
Antibody against SPI-CI was generated using recombinant denatured His-tagged SPI-CI (1 mg per rabbit). Rabbits were boosted thrice, and serum was tested against an overlapping panel of peptides spanning the SPI-CI protein. The peptide VQMMOQTDTFMFAFVDEK was used to purify serum, resulting in a peptide-specific polyclonal serum. Lysates were prepared as described,19 separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF). Blots were blocked for 1 hour in phosphate-buffered saline (PBS)/0.2% Tween-20/5% milk and incubated overnight at 4°C with 2 μg/mL α–SPI-CI, 2 μg/mL α–SPI-6, or 1 μg/mL α–extracellular signal-regulated kinase-2 (α–ERK-2) antibody in block buffer. Blots were developed using horseradish peroxidase (HRP)–coupled goat anti–rabbit immunoglobulin G (IgG) and enhanced chemiluminescence (ECL).
LAK-cell and granule preparation
Mouse spleen cells were mechanically disrupted, and erythrocytes were lysed. Nylon wool nonadherent cells were cultured with 1000 U/mL interleukin-2 (IL-2). After 2 days, medium was removed, centrifuged, supplemented with another 1000 U/mL IL-2, and re-added to the adherent cells. After an additional 5 days, the resulting lymphokine-activated killer (LAK) cells were harvested. Separation of LAK cells into T, NKT, and NK cells was performed by fluorescence-activated cell sorting (FACS) using α-CD3 and α-NK1.1 (BD Biosciences).
LAK or CTL granules were isolated by resuspending cells in sucrose buffer (0.25 M sucrose 10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] [pH 7.4], 4 mM EGTA [ethylene glycol tetraacetic acid]) and forcing them to burst in an N2 bomb (350 psi). Lysates obtained were depleted of nuclei and bigger fragments and layered onto 48% Percoll/sucrose buffer. Gradients were run at 60 000g, and the lower fraction was collected. Percoll was then excluded by pelleting at 150 000g for 16 hours. Granules, which are layered on top of the Percoll using this method, were collected and stored at –80°C.
GrM activity, purification, and association
GrM was purified from RNK-16 cell granules using Superdex (Amersham Biosciences, Roosendaal, Netherlands) size exclusion chromatography as previously described.28 The purified fraction is free of Asp-ase and tryptase activity, but small impurities cannot be excluded.
For detection of endogenous Met-ase activity, granule isolates from CTL clones and LAK cells were incubated in HEPES buffer (0.1 M HEPES [pH 7.5], 0.5 M NaCl, 0.05% Triton X-100) for 1 hour at 37°C with Boc-Ala-Ala-Met-SBzl as substrate and Ellmann reagent (Sigma, Zwijndrecht, The Netherlands) as indicator. Substrate cleavage was followed using absorption at 405 nm.
For in vitro association between serpins and granzymes, SPI-CI and SPI-6 were in vitro transcribed/translated with 35S-methionine and incubated for 30 minutes with, respectively, purified GrM or GrB (ALEXIS, Breda, The Netherlands) in 20 mM Tris (tris(hydroxymethyl)aminomethane) (pH 7.5), 150 mM NaCl. Complexes were separated on reducing SDS-PAGE and visualized by autoradiography.
In vitro cytotoxicity
The 51Cr-release assay, measuring membrane integrity, was performed as described.19,29 In short, cells were labeled with 100 μCi (3.7 MBq) 51Cr-chromate for 1 hour and washed 3 times. Assays were performed with 2000 target cells (about 2000 cpm) per well. Target cells were loaded with 1 μg/mL adenovirus E1B or E1A peptide and incubated for 5 hours with a peptide-specific CTL clone or LAK cells at different effector-target (E/T) ratios.
For purified perforin/granzyme killing assays, cells were resuspended in RPMI/1% BSA. Subsequently, granzymes were added in HEPES/CaCl2 buffer (20 mM HEPES, 150 mM NaCl, 5 mM CaCl2 [pH 7.4]) and preincubated for 1 hour, after which human perforin, purified from YT granules as previously described,30 was added in HEPES buffer (20 mM HEPES, 150 mM NaCl [pH 7.4]) at sublytical doses for another 4 hours. Granule assays were performed essentially the same except that increasing amounts of granules were added to cells in HEPES/CaCl2 buffer. In both assays cell death was determined by propidium iodide (PI) exclusion.
RNAi constructs and transfections
To generate stable expression of siRNA, we used the pSuper vector system.31 Targeted sequence in the serpins was 5′-CTTGTGAAGTCCTCCAAAC-3′ (RS), which represents nucleotides 488 to 506 of SPI-CI (U96705) and 334 to 352 of SPI-6 (U96700).
For stable mRNA suppression, CMT93 was cotransfected using jetPEI (Qbiogene, Breda, The Netherlands) with a vector containing a puromycin-resistance marker and with RS plasmid in a 10-fold excess. After 36 hours, cells were selected with puromycin. Two weeks later, resistant cells were analyzed for SPI-CI and SPI-6 expression.
Results
SPI-6 does not protect against membranolysis
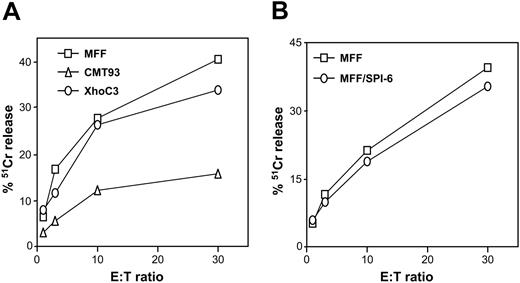
CMT93 is a colon carcinoma cell line that expresses high levels of the GrB-inhibiting serpin SPI-6.19 In agreement with the involvement of GrB in induction of apoptosis, we previously showed that high levels of SPI-6 in CMT93 correlate with resistance to CTL-induced apoptosis.19 We now show that CMT93 is also resistant to membrane disruption as measured in a 51Cr-release assay (Figure 1A), even though CMT93 is efficiently recognized by the CTL clone as determined by interferon-γ (IFN-γ) release.19 Accordingly, CMT93 grows efficiently in immunocompetent mice and therefore cannot be controlled by CTLs in vivo either.32 These data suggest a role for SPI-6 in protection of both apoptosis and cytolysis. Similarly, its human homolog PI-9 has been shown to protect against lysis by human NK-like cells.20 However, studies performed with GrB-deficient effector cells showed that this enzyme is dispensable for cytolysis.12 In line with this argument, we found that mouse T-cell lymphoma (MFF) overexpressing SPI-6 was not protected from CTL-induced cytolysis (Figure 1B), even though apoptosis was clearly inhibited.19 Apparently, SPI-6 expression is not sufficient to inhibit CTL-induced lysis. We therefore postulated that CMT93, which did escape from lysis, expresses an additional escape molecule, possibly another serpin.
CMT93 resists CTL-induced cytolysis. (A-B) Target cells were labeled with 51Cr-chromate for 1 hour, loaded with relevant E1B peptide, and incubated with the E1B-specific CTL clone at different effector-target ratios. After 5 hours, the released label was determined, which served as a measure for CTL-induced lysis.
CMT93 resists CTL-induced cytolysis. (A-B) Target cells were labeled with 51Cr-chromate for 1 hour, loaded with relevant E1B peptide, and incubated with the E1B-specific CTL clone at different effector-target ratios. After 5 hours, the released label was determined, which served as a measure for CTL-induced lysis.
Identification of a serpin expressed by CMT93
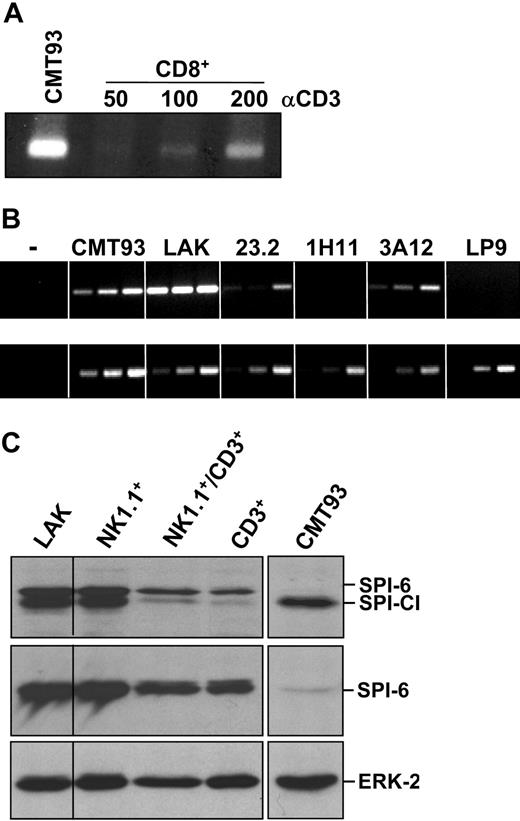
Northern blot analysis with a probe encompassing the coding sequence of SPI-6 provided the first hint for expression of a separate serpin, because this revealed an additional mRNA (X) in CMT93 compared with a CTL clone (Figure 2A). To characterize this mRNA, a CMT93 cDNA expression library was constructed and approximately 106 λ phages were screened with the SPI-6 probe. Isolates included SPI-6, but also a second cDNA, which we subcloned and sequenced. Alignment of this DNA sequence with SPI-6 revealed that it represents a serpin that is closely related to SPI-6, showing 80% sequence identity (Figure 2B). We have named this serpin SPI-CI for “serine protease inhibitor involved in cytotoxicity inhibition.” Interestingly, the putative RCL of SPI-CI was identical to R86, previously identified by Sun et al26 in a search for serpins expressed in the cytotoxic lymphocyte line R8. To confirm that SPI-CI represents the alternative mRNA in CMT93, we used the high degree of sequence dissimilarity in the 3′ untranslated region (3′UTR) of SPI-6 and SPI-CI. Northern blot analysis with 3′UTR probes of SPI-6 confirmed expression of SPI-6 in CTLs and CMT93 (Figure 2A). The 3′UTR of SPI-CI only hybridized with the faster-migrating mRNA in CMT93, confirming the identity of this mRNA as SPI-CI.
Identification of SPI-CI from CMT93. (A) Total RNA of a CTL clone (line 5) and CMT93 was hybridized on Northern blot with a probe spanning the coding sequence of SPI-6 (left panel). As compared with the CTL clone, an additional band is detected in CMT93 (X). To confirm the expression of SPI-CI in CMT93, Northern blots were incubated with either a probe against the 3′UTR of SPI-6 or a probe against the 3′UTR of SPI-CI (middle and right panels, respectively). (B) Alignment of SPI-CI and SPI-6 DNA sequences. The DNA sequence of SPI-CI shares 80% homology (shaded base pairs) with the DNA sequence of SPI-6. Arrows indicate the start and end of the coding sequence. (C) Alignment of the amino acid sequences of SPI-CI and SPI-6. SPI-CI shows 76% identity (shaded amino acids) with SPI-6. The RCL is boxed. (D) Reactive center loop of SPI-CI, SPI-6, and their predicted amino acid at the P1 position.
Identification of SPI-CI from CMT93. (A) Total RNA of a CTL clone (line 5) and CMT93 was hybridized on Northern blot with a probe spanning the coding sequence of SPI-6 (left panel). As compared with the CTL clone, an additional band is detected in CMT93 (X). To confirm the expression of SPI-CI in CMT93, Northern blots were incubated with either a probe against the 3′UTR of SPI-6 or a probe against the 3′UTR of SPI-CI (middle and right panels, respectively). (B) Alignment of SPI-CI and SPI-6 DNA sequences. The DNA sequence of SPI-CI shares 80% homology (shaded base pairs) with the DNA sequence of SPI-6. Arrows indicate the start and end of the coding sequence. (C) Alignment of the amino acid sequences of SPI-CI and SPI-6. SPI-CI shows 76% identity (shaded amino acids) with SPI-6. The RCL is boxed. (D) Reactive center loop of SPI-CI, SPI-6, and their predicted amino acid at the P1 position.
The open reading frame of SPI-CI encodes a protein of 377 amino acids with an estimated molecular weight of 42.8 kDa. At the protein level SPI-6 and SPI-CI were highly similar, being 76% identical with another 13% of conserved changes (Figure 2C). Of interest, protein sequences varied most in the RCL, which is essential for the inhibitory specificity of serpins. Using the Swiss-Pdb viewer33 we modeled the 3-dimensional (3-D) structure and RCL of SPI-CI. This analysis revealed that SPI-CI likely contains a leucine at its P1 position. In comparison, SPI-6 has a glutamic acid at its P1 position (Figure 2D). This suggests that SPI-CI and SPI-6 inhibit distinct serine proteases.
SPI-CI expression in transformed and normal cell types
To detect SPI-CI protein, we generated a polyclonal antibody. Western blot analysis revealed SPI-CI in CMT93 as well as in another C57BL/6-derived colon carcinoma cell line, MC38 (Figure 3A). SPI-CI protein was also expressed in C26 and CC36, 2 other chemically induced colon carcinoma lines derived from BALB/c mice. SPI-CI protein was not detected in several other noncolon tumor lines. This colon carcinoma–specific expression pattern prompted us to analyze the expression of SPI-CI by reverse transcriptase (RT)–PCR in normal murine colon and in several other tissue samples. SPI-CI mRNA expression was, however, not observed in normal colon, suggesting that SPI-CI expression in colon carcinomas is a result of tumorigenesis. SPI-CI was detected at high levels in placenta and early during embryogenesis (7-day-old embryo). Relatively low expression was found in spleen, lung, brain, and testis derived from adult mice (Figure 3B).
Expression of SPI-CI in tumor and tissue samples. (A) Protein expression of SPI-CI in murine tumor cell lines. Expression was detected by the polyclonal antibody against SPI-CI. (B) Complementary DNA of different murine tissue samples was analyzed by RT-PCR. (Top) SPI-CI PCR for 30, 35, and 40 cycles. (Bottom) GAPDH PCR for 22, 25, and 28 cycles.
Expression of SPI-CI in tumor and tissue samples. (A) Protein expression of SPI-CI in murine tumor cell lines. Expression was detected by the polyclonal antibody against SPI-CI. (B) Complementary DNA of different murine tissue samples was analyzed by RT-PCR. (Top) SPI-CI PCR for 30, 35, and 40 cycles. (Bottom) GAPDH PCR for 22, 25, and 28 cycles.
PI-9 and its murine ortholog SPI-6 are highly expressed in activated CTLs, and this is interpreted as an indication that these cells protect themselves from GrB-induced death.27 Because SPI-6 and SPI-CI expression clearly overlap, we set out to analyze SPI-CI mRNA levels in cytotoxic cell populations. Purified CD8+ T cells stimulated in vitro with platebound anti-CD3 and anti-CD28 antibody (Ab) showed clear, albeit relatively low, SPI-CI mRNA levels (Figure 4A). Similarly, T-cell clones displayed lower levels of SPI-CI mRNA in comparison with CMT93 cells. However, IL-2–activated NK-like lymphokine-activated killer (LAK) cells expressed SPI-CI mRNA at very high levels as compared with CMT93 (Figure 4B). Thus, SPI-CI is expressed in effector cells and with a clear preference for NK-like cells.
SPI-CI expression in effector cells. (A) Complementary DNA of in vitro–activated CD8+ T cells was analyzed for SPI-CI expression by RT-PCR for 40 cycles. Purified CD8+ T cells were cultured for 24 hours in the presence of the indicated (nanograms per milliliter) amounts of platebound α-CD3 Ab in combination with platebound α-CD28 Ab (5 μg/mL). (B) Complemenatary DNA of LAK cells and several T-cell clones was analyzed for SPI-CI expression by RT-PCR for 30, 35, and 40 cycles (top panel) and GAPDH PCR for 22, 25, and 28 cycles (bottom panel). (C) Expression of SPI-CI and SPI-6 protein in LAK cells. LAK cells were further separated by FACS using cell-specific antibodies into NK, NKT, and T cells and analyzed for SPI-CI (top panel), SPI-6 expression (middle panel), and ERK-2 (bottom panel) expression.
SPI-CI expression in effector cells. (A) Complementary DNA of in vitro–activated CD8+ T cells was analyzed for SPI-CI expression by RT-PCR for 40 cycles. Purified CD8+ T cells were cultured for 24 hours in the presence of the indicated (nanograms per milliliter) amounts of platebound α-CD3 Ab in combination with platebound α-CD28 Ab (5 μg/mL). (B) Complemenatary DNA of LAK cells and several T-cell clones was analyzed for SPI-CI expression by RT-PCR for 30, 35, and 40 cycles (top panel) and GAPDH PCR for 22, 25, and 28 cycles (bottom panel). (C) Expression of SPI-CI and SPI-6 protein in LAK cells. LAK cells were further separated by FACS using cell-specific antibodies into NK, NKT, and T cells and analyzed for SPI-CI (top panel), SPI-6 expression (middle panel), and ERK-2 (bottom panel) expression.
In analogy to mRNA expression, SPI-CI and SPI-6 protein were detected at high levels in LAK cells. LAK cells not only consist of NK cells but also contain NKT cells and CD8+ T cells.34 When LAK cells are separated into the different cell types by FACS, expression is primarily confined to NK1.1+ cells. However, SPI-CI was present, but at clearly lower levels, in the NKT and T-cell fraction (Figure 4C). Consistent with the SPI-6 data, SPI-CI may therefore serve a protective function in NK, NKT, and T cells. Importantly, mRNA expression of SPI-CI is much higher in LAK cells as compared with CMT93. Nevertheless, SPI-CI protein levels seem relatively comparable, suggesting that translational control or protein stability is different between these cells. In contrast, CTL clones express little SPI-CI mRNA, and protein cannot be detected using our antibody. CD8+ T cells can express SPI-CI, though, as can be seen in Figure 4C. SPI-CI, therefore, likely protects against a cytotoxic protease present in CTLs and LAK cells. Previously, Grossman et al35 showed that expression of several granzymes differed only slightly between LAK cells and CTLs. Using RT-PCR we found that most granzymes are expressed at higher levels (about 10-fold) in LAK cells as compared with our CTL clone (line 5), except for GrA, GrB, and GrM, which are similar, and GrK, which is higher in the clone (not shown). Because several of the elevated granzymes in LAK cells encode for chymotrypsin activities (GrD-GrG), their higher expression levels could be the underlying reason for elevated SPI-CI expression in LAK cells.
Serpin overexpression prevents target cell apoptosis but not cytolysis
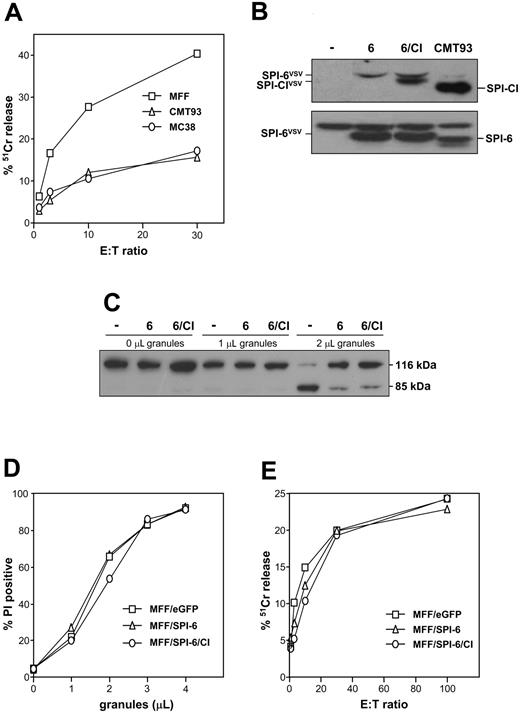
To test whether expression of SPI-CI in CMT93 and MC38 is causally related to cytolysis resistance, we first determined CTL sensitivity of MC38. Also, this SPI-CI– and SPI-6–expressing tumor is largely resistant to CTL-induced lysis (Figure 5A), supporting the hypothesis that these serpins together protect against cytolysis. To directly analyze this, we supertransduced MFF/SPI-6 cells with a vector encoding SPI-CI (Figure 5B). We first analyzed whether SPI-6 levels are sufficient to prevent GrB-mediated apoptosis by measuring processing of poly(adenosine diphosphateribose) polymerase (PARP), a downstream death substrate of GrB. Increasing amounts of purified LAK granules were used to exclude any variation that can be introduced by a difference in recognition of MFF lines by effector cells. In both MFF/SPI-6 and MFF/SPI-6/CI, PARP cleavage was inhibited to a similar extent (Figure 5C). Of note, SPI-CI expression by itself is not sufficient to prevent granule-induced apoptosis (not shown), confirming that the apoptosis inhibition observed in the double transfectant is due to SPI-6 and not SPI-CI. Next, we analyzed the membrane integrity after exposure to granules. Neither MFF/SPI-6 nor MFF/SPI-6/CI was resistant to lysis using this method (Figure 5D), indicating that expression of SPI-CI together with SPI-6 is not sufficient to resist cytolysis. Accordingly, these transfectants were also not protected in a classical 51Cr-release assay with a CTL clone as effector (Figure 5E).
Ectopic expression does not render MFF cells resistant. (A) Target cells were labeled with 51Cr-chromate for 1 hour, loaded with relevant E1B peptide, and incubated with the E1B-specific CTL clone at different effector-target ratios. After 5 hours, the released label was determined, which served as a measure of the percentage of CTL-induced lysis when compared with the control. (B) Ectopic expression of SPI-CI and SPI-6 in MFF cells. Transfected MFF cells were analyzed for SPI-CI (top panel) and SPI-6 (bottom panel) expression. Transfected SPI-6 and SPI-CI are VSV-tagged and therefore migrate slightly more slowly than their endogenous counterparts. (C) Target cells were incubated at 37°C with different concentrations of purified LAK granules. After 1 hour, cells were directly lysed in Laemmli buffer and analyzed for PARP cleavage. (D) Target cells were incubated at 37°C with different concentrations of purified LAK granules. After 1 hour, cells were harvested and membrane integrity was measured using propidium iodide. (E) Target cells were labeled with 51Cr-chromate for 1 hour, loaded with relevant E1A peptide, and incubated with the E1A-specific CTL clone at different effector-target ratios. After 5 hours, the released label was determined, which served as a measure for CTL-induced lysis.
Ectopic expression does not render MFF cells resistant. (A) Target cells were labeled with 51Cr-chromate for 1 hour, loaded with relevant E1B peptide, and incubated with the E1B-specific CTL clone at different effector-target ratios. After 5 hours, the released label was determined, which served as a measure of the percentage of CTL-induced lysis when compared with the control. (B) Ectopic expression of SPI-CI and SPI-6 in MFF cells. Transfected MFF cells were analyzed for SPI-CI (top panel) and SPI-6 (bottom panel) expression. Transfected SPI-6 and SPI-CI are VSV-tagged and therefore migrate slightly more slowly than their endogenous counterparts. (C) Target cells were incubated at 37°C with different concentrations of purified LAK granules. After 1 hour, cells were directly lysed in Laemmli buffer and analyzed for PARP cleavage. (D) Target cells were incubated at 37°C with different concentrations of purified LAK granules. After 1 hour, cells were harvested and membrane integrity was measured using propidium iodide. (E) Target cells were labeled with 51Cr-chromate for 1 hour, loaded with relevant E1A peptide, and incubated with the E1A-specific CTL clone at different effector-target ratios. After 5 hours, the released label was determined, which served as a measure for CTL-induced lysis.
Silencing of SPI-CI and SPI-6 renders CMT93 sensitive to cytolysis
Even though coexpression of both serpins is not sufficient to protect against cytolysis, this does not exclude a critical role for these serpins in protection. To analyze whether down-regulation of SPI-CI and SPI-6 indeed reverted the resistance of CMT93 for cytolysis, we made use of RNA interference. We used the pSuper vector to target SPI-CI and SPI-6 mRNA simultaneously, which is feasible due to the high sequence homology. The sequence used is specific for SPI-6 and SPI-CI, as no other matches were found using the BLAST database; nor did it affect a closely related serpin, NK9, which has 2 mismatches in the targeted sequence (not shown). The effectiveness of the RNAi SPI vector (RS) was ascertained using transient transfection of 293T cells with VSV-tagged SPI-6 in the absence or presence of RS. Expression of RS transcripts led to a clear reduction (more than 90%) in SPI-6 protein levels (not shown). CMT93 cells were therefore transfected with the RS or a control vector, and puromycin-resistant cells were screened for SPI-CI and SPI-6 protein. In agreement with the transient transfection experiments, RS was able to knock down SPI-6 protein. Moreover, it silenced SPI-CI, while it did not affect ERK-2 protein levels (Figure 6A) or NK9 mRNA levels (not shown). Consistent with this finding, Northern blot analysis with the 19-nucleotide siRNA target sequence as a probe revealed expression of 21- and 22-nucleotide siRNA in CMT93-RS (Figure 6B). This indicated that the transcript was generated and cleaved into a functional siRNA.
The silencing of SPI-CI and SPI-6 in CMT-93 affects cytolysis. (A) Expression of SPI-CI (top panel), SPI-6 (middle panel), and ERK-2 (bottom panel) after selection of CMT93 lines expressing siRNA constructs against SPI-CI and SPI-6 (RS) or a control vector (con). (B) Northern blot with total RNA from CMT93 and CMT93-RS, probed with the sense 19 nucleotide targeting both SPI-CI and SPI-6. (C) RS-transfected CMT93 (CMT93-RS), control-transfected CMT93 (CMT93-con), and CMT93 were labeled with 51Cr-chromate for 1 hour and incubated with an E1A-specific CTL clone in the presence of the relevant epitope. After 5 hours, the released label was determined, which served as a measure for lysis. (D) CMT93-RS and CMT93 were labeled with 51Cr-chromate for 1 hour and incubated with LAK cells. After 5 hours, the released label was determined, which served as a measure for lysis. Similar data were obtained when CMT-con was tested in comparison with CMT93-RS. (E) CMT93 and CMT93-RS were incubated at 37°C with different concentrations of purified LAK granules. After 1 hour, cells were harvested and membrane integrity was measured using propidium iodide. Similar data were obtained when CMT-con was tested in comparison with CMT93-RS.
The silencing of SPI-CI and SPI-6 in CMT-93 affects cytolysis. (A) Expression of SPI-CI (top panel), SPI-6 (middle panel), and ERK-2 (bottom panel) after selection of CMT93 lines expressing siRNA constructs against SPI-CI and SPI-6 (RS) or a control vector (con). (B) Northern blot with total RNA from CMT93 and CMT93-RS, probed with the sense 19 nucleotide targeting both SPI-CI and SPI-6. (C) RS-transfected CMT93 (CMT93-RS), control-transfected CMT93 (CMT93-con), and CMT93 were labeled with 51Cr-chromate for 1 hour and incubated with an E1A-specific CTL clone in the presence of the relevant epitope. After 5 hours, the released label was determined, which served as a measure for lysis. (D) CMT93-RS and CMT93 were labeled with 51Cr-chromate for 1 hour and incubated with LAK cells. After 5 hours, the released label was determined, which served as a measure for lysis. Similar data were obtained when CMT-con was tested in comparison with CMT93-RS. (E) CMT93 and CMT93-RS were incubated at 37°C with different concentrations of purified LAK granules. After 1 hour, cells were harvested and membrane integrity was measured using propidium iodide. Similar data were obtained when CMT-con was tested in comparison with CMT93-RS.
With CMT93-RS we determined the role of SPI-6 and SPI-CI in cytolysis resistance. CTLs efficiently lysed CMT93-RS, while CMT93 was largely resistant (Figure 6C). Similarly, LAK cells were able to induce 51Cr release from CMT93-RS but not from the parental CMT93 cells (Figure 6D). Moreover, purified LAK granules could kill CMT93-RS but not CMT93 (Figure 6E). These data indicate that CMT93 is not resistant due to a difference in recognition but fully resists the combined attack of perforin and granzymes. Therefore, we conclude that SPI-CI and SPI-6 protein are critical determinants in the resistance of CMT93 to NK- and CTL-induced cytolysis.
SPI-CI inhibits GrM and prevents perforin/GrM-induced lysis
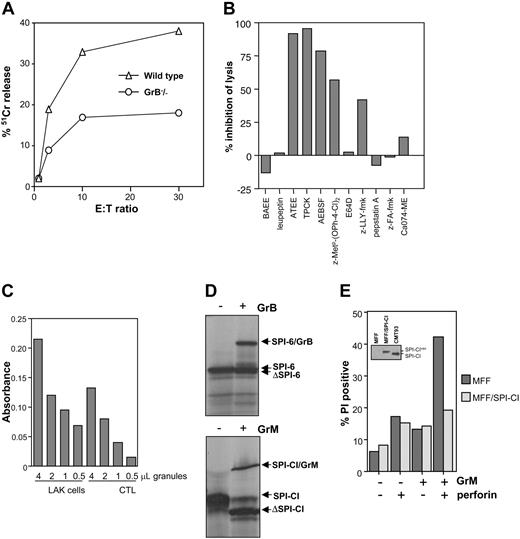
To analyze which granzyme is targeted by SPI-CI, we first analyzed whether lysis of target cells still occurred using GrB cluster knock-out LAK cells. These effector cells are (largely) deficient for GrB, GrC, GrD, GrF, and GrG 15 but are reportedly still capable of lysing cells.12 In agreement, CMT93-RS is lysed by these LAK cells, albeit less efficiently than wild-type LAK cells (Figure 7A). The remaining lytic activity observed is not exerted by GrA, because GrA/GrB double knock-out induced lysis of CMT93-RS with comparable efficiency (not shown). This indicates that the deleted granzymes do play a role in lysis of CMT93-RS but that other activities are also involved. To determine which activity is responsible for this lytic effect and is possibly targeted by SPI-CI, we decided to use a range of chemical inhibitors to prevent lysis by GrB cluster knock-out LAK cells. This revealed that chymotrypsin activity was responsible for the lysis observed, because all chymotrypsin inhibitors protected CMT93-RS from death (Figure 7B). In this light, it is notable that a recent publication by Kelly et al16 showed that GrM, one of the chymotrypsins not deleted in GrA/GrB knock-out, can induce target cell lysis in concert with perforin. In agreement, the GrM-specific inhibitor z-MetP-(OPh-4-Cl)236 also largely protected CMT93-RS from LAK cell–induced killing (Figure 7B). Previously, GrM was suggested to be an NK-specific granzyme with little expression in CTLs. However, RT-PCR analysis of LAK cells and CTL clones used in this study indicated that these effectors contain comparable levels of GrM mRNA (not shown). To ascertain that this indeed relates to GrM protease activity, we isolated granules from both populations. Using the typical substrate Boc-Ala-Ala-Met-SBzl,28 we found that both cell types indeed contain Met-ase activity, although it is less pronounced (about 2-fold lower) in CTLs (Figure 7C). Importantly, GrA/GrB knock-out LAK cells displayed similar Met-ase activity (not shown), indicating that this assay is not influenced by the presence of GrB, which has previously been reported to contain some Met-ase activity as well.37 It is therefore likely that both effector populations can employ this activity to kill target cells, which is in line with previous observations for human effector cells.38 Inhibition of this activity by a serpin would involve serpin cleavage and formation of a covalent serpin-protease complex. We therefore used a previously described method to purify GrM from granules derived from the rat NK line RNK-16.28 Superdex size fractionation of proteins derived from these granules results in separation of GrM from the rest of the known granzymes.28 These fractions contain clear Met-ase activity and are devoid of tryptase and Asp-ase activity (not shown). We used these fractions to analyze whether the purified Met-ase can irreversibly associate with 35S-labeled in vitro–translated SPI-CI. SPI-CI is clearly cleaved by the Met-ase present (Figure 7D), indicating that it is a substrate for this protease. Moreover, this contact is a classical serpin-protease interaction, because a covalent complex between protease and SPI-CI can be detected, similar to what is observed for GrB/SPI-6 (Figure 7D). These data therefore suggest that SPI-CI targets GrM and can potentially protect cells from the effects induced by GrM. To formally prove this, we used MFF cells transduced with retrovirus encoding SPI-CI (Figure 7E, inset). These cells were incubated with sublytical doses of perforin (100 U/mL) together with purified GrM as described.16 Control MFF cells were effectively lysed using this approach (Figure 7E), as were control-transduced MFF cells (not shown). MFF/SPI-CI cells, however, completely resisted the attack by purified GrM (Figure 7E). We therefore conclude that GrM is cytotoxic to cells and that this cytotoxicity is counterbalanced by SPI-CI in the target cell.
GrM is targeted by SPI-CI. (A) CMT93-RS is lysed by GrB cluster knock-out LAK cells. CMT93-RS was incubated for 5 hours with different E/T ratios of wild-type and GrB knock-out LAKs, and membranolysis was determined using chromium release. (B) Several chymotrypsin inhibitors inhibit lysis of CMT93-RS by GrB cluster knock-out LAK cells. Lysis assays were performed as described in panel A, and inhibitors used are as follows: for tryptase: BAEE (10 mM; Sigma) and leupeptin (0.1 mM; Sigma); for chymotrypsin: ATEE (10 mM; Sigma), TPCK (10 μM; Sigma), AEBSF (0.4 mM; Sigma), and GrM inhibitor z-MetP-(OPh-4-Cl)2 (0.1 mM; kind gift from Jim Powers); for calpain: E64D (40 μM; Sigma) and z-LLY-fmk (20 μM; Enzyme Systems Products, Livermore, CA); for Asp proteases: pepstatin A (25 μg/mL; Calbiochem, San Diego, CA); and for cathepsin B: z-FA-fmk (10 μM; Enzyme Systems Products) and Ca074-ME (25 μM, Calbiochem). None of the inhibitors displayed toxicity on the LAK cells as determined by trypan blue exclusion. (C) Met-ase activity in granule isolates of CTLs and LAK cells was determined using the preferred substrate Boc-Ala-Ala-Met-SBzl. Activity is depicted as absorbance at 405 nm. (D) In vitro association between purified GrB and in vitro–translated SPI-6 and purified GrM and in vitro–translated SPI-CI. Complexes are visualized on reducing SDS-PAGE. (E) Lysis of MFF and MFF/SPI-CI using purified perforin in combination with purified GrM. GrM was added to cells either with or without perforin, and cell death was determined using PI exclusion. One representative experiment of 3 is shown.
GrM is targeted by SPI-CI. (A) CMT93-RS is lysed by GrB cluster knock-out LAK cells. CMT93-RS was incubated for 5 hours with different E/T ratios of wild-type and GrB knock-out LAKs, and membranolysis was determined using chromium release. (B) Several chymotrypsin inhibitors inhibit lysis of CMT93-RS by GrB cluster knock-out LAK cells. Lysis assays were performed as described in panel A, and inhibitors used are as follows: for tryptase: BAEE (10 mM; Sigma) and leupeptin (0.1 mM; Sigma); for chymotrypsin: ATEE (10 mM; Sigma), TPCK (10 μM; Sigma), AEBSF (0.4 mM; Sigma), and GrM inhibitor z-MetP-(OPh-4-Cl)2 (0.1 mM; kind gift from Jim Powers); for calpain: E64D (40 μM; Sigma) and z-LLY-fmk (20 μM; Enzyme Systems Products, Livermore, CA); for Asp proteases: pepstatin A (25 μg/mL; Calbiochem, San Diego, CA); and for cathepsin B: z-FA-fmk (10 μM; Enzyme Systems Products) and Ca074-ME (25 μM, Calbiochem). None of the inhibitors displayed toxicity on the LAK cells as determined by trypan blue exclusion. (C) Met-ase activity in granule isolates of CTLs and LAK cells was determined using the preferred substrate Boc-Ala-Ala-Met-SBzl. Activity is depicted as absorbance at 405 nm. (D) In vitro association between purified GrB and in vitro–translated SPI-6 and purified GrM and in vitro–translated SPI-CI. Complexes are visualized on reducing SDS-PAGE. (E) Lysis of MFF and MFF/SPI-CI using purified perforin in combination with purified GrM. GrM was added to cells either with or without perforin, and cell death was determined using PI exclusion. One representative experiment of 3 is shown.
Discussion
We provide compelling evidence that tumors can fully resist cytolytic activities of lymphocytes even under stringent in vitro conditions. Our data indicate that this inhibition is dependent on SPI-6 and SPI-CI. Cytolysis-resistant cell lines showed simultaneous expression of SPI-6 and SPI-CI, while tumor cells solely expressing SPI-6 do not resist membranolysis. More important, combined silencing of SPI-6 and SPI-CI renders the otherwise resistant cells sensitive.
The physiologic expression profile of SPI-CI is reminiscent to that of SPI-6.26 Previously, Sun et al26 identified the RCL of SPI-CI (R86) in CTLs, and later it was described to be expressed in placenta.39 We now extend these observations by showing that SPI-CI is highly expressed in NK-like cells, to a lesser extent in CTLs, and in immune-privileged sites, like placenta, testis, and brain. Moreover, SPI-CI is expressed in colon carcinoma lines, which is not due to endogenous expression in colon but appears to be a result of tumorigenesis.
Perforin is central to all models of granule-mediated cytotoxicity. In the granzyme-entry model, it is thought that perforin releases internalized granzymes from endosomes and allows them to process substrates inside the target cell. In the more classical model, an important role for perforin lies in the fact that it (temporarily) disrupts the plasma membrane, allowing ion fluxes and possibly protein fluxes that will eventually lead to target cell death. Proof and disproof for either model exists, but so far there is no reason to assume that they could not coexist. The central role for perforin was even more firmly established when the GrA/GrB double knock-out was analyzed (GrA-, GrB-, GrC-, GrD-, GrF-, GrG-deficient). Effector cells derived from this mouse were still capable of killing,12 initially suggesting that perforin by itself can induce changes in target cells that would lead to death. However, these effectors are not completely devoid of granzymes, and recent evidence indicates that GrM can induce lysis of target cells as well.16 Our data now clearly indicate that this occurs in a relevant effector-target setting and that it can be prevented by the expression of a serine protease inhibitor (SPI-CI) or by chemical chymotrypsin inhibitors (Figure 7). This formally proves that protease activity is required for perforin-dependent lysis. In this light, it is important to note that older data from the groups of Hudig and Bleackley already provided evidence that inhibition of chymotrypsin activity can diminish perforin-dependent cytolysis.40,41 The current data now provide a clearer insight into this activity and suggest that it is exerted by GrM. GrM is expressed in both CTLs and LAK cells as indicated by mRNA expression and Met-ase activity present in granules from both effectors (Figure 7C). Previous reports suggested that human GrM expression in vivo predominates in NK-like cells, even though human CTL clones can express the enzyme. It therefore remains to be determined whether expression is also observed in in vivo–activated murine CTLs. At this point we cannot exclude, though, that other chymotrypsins are also involved in cytolysis. In fact, expression of SPI-CI and SPI-6 in MFF cells is not sufficient to prevent cytolysis (Figure 5D-E). This can be due to the levels of SPI-CI attained in these cells, which is significantly lower as compared with CMT93. However, it appears more likely that additional inhibitors may be present in CMT93 and MC38, because SPI-CI expression in MFF is sufficient to protect these cells from purified GrM-induced death (Figure 7E).
Intriguingly, PI-9, the human ortholog of SPI-6, is sufficient to protect MCF-7 tumor cells from human LAK cell–induced cytolysis.20 This mouse/human difference may reflect the broader range of granzymes present in murine as compared with human granules35 and is also manifested in the fact that the murine genome contains more ovalbumin serpins.39 As a result target cells would require less protease inhibitors to prevent human granule-induced lysis. Alternatively, this may result from PI-9's capacity to inhibit other proteases than GrB.42
Although the data point to the involvement of serine proteases in cytolysis, we formally cannot exclude that SPI-CI serves as an inhibitor that allows for the expression of a protective protease in target cells. Such a protease would prevent perforin from eliciting its cytolytic effect. Balaji et al43 have shown that CTLs protect themselves using this mechanism, because expression of cathepsin B on the surface of CTLs degrades perforin and disallows suicide. Although cathepsin B is not present on the surface of CMT93, these cells might have acquired comparable proteolytic mechanisms for which they require SPI-CI. We believe that this is unlikely, though, not only because of the observed inhibition of GrM-induced lysis but also because processing of perforin is not observed in our experimental setting. That is, incubation of CMT93 with granules containing perforin did not result in detectable degradation of perforin (not shown), while these granules did efficiently kill CMT93-RS (Figure 6E).
In conclusion, our data show that SPI-CI is a candidate immune escape molecule that together with SPI-6 allows colon carcinoma lines to resist CTL- and NK-induced lysis. Importantly, orthopoxviruses have previously been shown to encode serpin SPI-1 and SPI-2,44 which cooperatively protect infected cells from lysis. Deletion of SPI-1 and SPI-2 together was required to revert this inhibition, while cells infected with virus encoding single serpin deletions were still largely protected from lysis, suggesting that both serpins are involved in inhibition. Nevertheless, SPI-2 overexpression does not protect cells from lysis.45 This suggests that these serpins are necessary but not sufficient. This is highly reminiscent of what we observe with SPI-6 and SPI-CI deletion in CMT93 and overexpression in MFF. Moreover, because SPI-2 encodes an inhibitor of caspases and possibly GrB and SPI-1 encodes a chymotrypsin inhibitor, this setting is comparable to what is observed with SPI-6 and SPI-CI. The 2 murine serpins described here may thus represent the orthologs of these orthopox serpins, and it will be interesting to analyze whether SPI-1 can indeed target GrM as well. Further analysis into the in vivo role of the different pathways is awaited, and it will be especially interesting to see whether the GrA/GrB cluster/GrM knock-out will still be capable of dealing with viral infections and tumorigenesis. In analogy, in vivo tumor studies with serpin-overexpressing tumors will further increase our fundamental understanding of the pathways that CTLs and NK cells use to induce target cell death in vivo.
Prepublished online as Blood First Edition Paper, September 28, 2004; DOI 10.1182/blood-2004-03-0791.
Supported by grants from the Dutch Cancer Society (RUL 2001-2531 [M.B., G.M.d.R.] and RUL1998-1860 [I.G.M.K.]), and the Netherlands Organization for Scientific Research (M.T.G.A.R., J.P.M.).
M.B. and I.G.M.K. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Tom Sayers for providing and advising us on the RNK-16 granules and his encouragement on the “dutch game,” Tim Ley for kindly providing the PCR primer sets necessary to analyze the granzyme expression in our cells, Jim Powers and Amy Campbell for kindly sharing the GrM-specific inhibitor z-MetP-(OPh-4-Cl)2, and Markus Simon for providing GrB and GrA/GrB double knock-out mice. We also thank the people from the Tumor Immunology Laboratory for culturing and providing T-cell clones.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal