Abstract
The multimeric size and the function of circulating von Willebrand factor are modulated via its proteolytic cleavage by the plasma metalloproteinase, ADAMTS13. It is unclear how ADAMTS13 activity is regulated within the vascular system. In the absence of a regulatory mechanism, ADAMTS13 activity might compromise platelet adhesion at sites of vascular injury. We hypothesized that at sites of vascular injury, ADAMTS13 activity could be regulated locally by coagulation proteinases. Initiation of coagulation in human plasma resulted in the disappearance of added full-length recombinant ADAMTS13. This loss was inhibited by hirudin. Using purified proteins, we showed that ADAMTS13 is proteolyzed at several cleavage sites by thrombin in a time- and concentration-dependent manner. Furthermore, this proteolysis ablated ADAMTS13 activity against purified von Willebrand factor. Preincubation of thrombin with soluble thrombomodulin, but not heparin, inhibited the proteolysis of ADAMTS13, suggesting the involvement of thrombin exosite I (and not exosite II) in ADAMTS13 recognition. Plasmin also cleaved ADAMTS13 into similar fragments, resulting in the loss of ADAMTS13 activity. This study demonstrates the susceptibility of ADAMTS13 to proteolytic inactivation and suggests possible roles for thrombin and plasmin at sites of vascular injury.
Introduction
Von Willebrand factor (VWF) is a large (2050 amino acid/∼250 kDa) multidomain glycoprotein whose functions are critical for normal hemostasis.1 VWF has 2 principal roles that influence the hemostatic process: (1) it acts as a carrier protein for coagulation factor VIII2 and (2) it mediates rapid adhesion of platelets to sites of vascular perturbation. The latter is one of the first hemostatic events following endothelial disruption and occurs through the specific binding of VWF to exposed subendothelial matrix proteins (principally collagen).3 Once immobilized, VWF affinity for the glycoprotein (GP) Ib-IX-V receptor complex on the surface of circulating platelets is significantly enhanced.4 This results in the tethering of platelets at sites of vascular damage and in the formation of a primary platelet plug. Tethered platelets are subsequently activated and expose phosphatidylserine-rich surfaces that are critical for efficient thrombin generation to occur.5
VWF is constitutively secreted into the blood by endothelial cells as multimers of varying size that differ predominantly in the number of component VWF units. A significant proportion is also stored within Weibel-Palade bodies, predominantly as “ultra-large” multimers (UL-VWF)6 that may exceed 2 × 104 kDa.1 This pool is released on demand in response to endothelial cell activation.7
The properties of circulating VWF are, in part, dependent on its molecular size. Larger VWF multimers not only bind circulating platelets more readily than smaller forms, but also undergo marked conformational changes in response to the rheologic forces exerted by the circulating blood.8 Under normal flowing conditions, VWF multimers circulate in a globular form. However, when VWF is exposed to increased shear forces, these molecules unravel into a “stringlike” conformation. This increases the number of exposed platelet/matrix binding sites and thus enhances the platelet tethering potential of the VWF molecule.
Thrombotic thrombocytopenic purpura (TTP) is a rare, life-threatening condition, linked to UL-VWF function.9 Patients with TTP develop platelet-rich thrombi in their microvasculature,10 which may lead to renal failure, neurologic dysfunction, and ultimately, death. Such clinical sequelae are, at least in part, related to the elevated plasma UL-VWF levels detected in affected individuals.11 These increased UL-VWF levels may promote platelet-rich thrombus formation by more readily tethering/aggregating circulating platelets.12 However, the precise stimulus that precipitates an episode of TTP remains unclear. The cause of increased UL-VWF levels in most patients with TTP has been attributed to deficiency in ADAMTS13.13 This enzyme cleaves VWF in the A2 domain at the Y1605-M1606 bond14 and so converts UL-VWF into smaller forms that demonstrate a reduced adhesive/prothrombotic potential. Consequently, TTP sufferers with congenital defects in ADAMTS13, or those with autoantibody inhibitors that compromise ADAMTS13 function, are unable to process UL-VWF normally.
ADAMTS13 is secreted into the blood as an active enzyme15 with a plasma concentration of approximately 1 μg/mL.16 It is a stable enzyme with a plasma half-life of approximately 2 to 3 days.17 Plasma ADAMTS13 is a 1349 residue protein with a molecular mass of 180 kDa to 200 kDa.18 The metalloproteinase domain contains a highly conserved active site that includes 3 histidine residues that coordinate a Zn2+ ion. The disintegrin-like domain and the first thrombospondin type-1 repeat (TSP1) domain precede the cysteine-rich domain and spacer domain, followed by 7 consecutive TSP1 and 2 C-terminal CUB domains.
It is uncertain how ADAMTS13 activity is both regulated and localized. ADAMTS13 cleaves VWF inefficiently in the absence of shear forces, or a denaturant (eg, urea), that “unravels” VWF and permits access of ADAMTS13 to the scissile bond.19 Although VWF is normally protected from proteolysis by its tertiary structure, it seems unlikely that ADAMTS13 activity in blood is not specifically regulated. In this study, we describe the proteolytic cleavage/inactivation of ADAMTS13 by thrombin and FXa. Moreover, we also demonstrate the proteolytic inactivation of ADAMTS13 by plasmin, another protease generated specifically at sites of vessel damage, following thrombin-dependent fibrin deposition.
Materials and methods
Expression and purification of recombinant ADAMTS13
Full-length ADAMTS13 cDNA (a gift from Dr F. Scheiflinger, Baxter, Austria) was cloned in-frame into the mammalian expression vector, pcDNA3.1myc/His, which fused a myc epitope and polyhistidine tag to the C-terminus of ADAMTS13. Stably transfected HEK293 cells were generated by standard techniques, using G418 for selection. Cleared serum-free conditioned medium was desalted and thereafter, ADAMTS13 was purified with a Ni2+-HiTrap Column (AmershamPharmacia, Buckinghamshire, United Kingdom), according to the manufacturer's instructions. Purified fractions were dialyzed into 20 mM Tris-HCl (pH 7.8) and analyzed by Western blotting using either anti-ADAMTS13 protease domain (a gift from Dr F. Scheiflinger18 ) or anti-myc epitope monoclonal antibodies (Invitrogen, Paisley, United Kingdom). Recombinant ADAMTS13 purity was assessed by nonreducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. ADAMTS13 concentration was determined with a BCA total protein assay kit (Perbio, Tattenhall, United Kingdom).
Proteolysis of ADAMTS13 in human plasma
To 800 μL citrated normal human plasma, or plasma defibrinated with Reptilase Reagent (Pentapharm, Basel, Switzerland), recombinant ADAMTS13 (80 nM) was added to increase the ADAMTS13 concentration. To 3 100-μL samples of ADAMTS13-spiked plasma, 8 pM lipidated human tissue factor (TF; Baxter, Vienna, Austria) and phosholipids (phosphatidylserine/phosphatidylcholine/phosphatidylethanolamine [PS/PC/PE], 20:60: 20) (Avanti, Alabaster, AL) were added. Thereafter, 0.4 mM fluorogenic thrombin substrate, Z-Gly-Gly-Arg-AMC-HCl (Bachem, Merseyside, United Kingdom), was added and samples were recalcified to initiate thrombin generation. Thrombin generation was monitored in real-time using a Fluoroscan plate reader (Thermo, Bassingstoke, United Kingdom) and quantified by comparison to a standard curve generated at the same time. From parallel samples lacking the thrombin substrate, subsamples (25 μL) were removed and analyzed by Western blotting using the anti-ADAMTS13 protease domain antibody. In some experiments, 5 mg/mL Gly-Pro-Arg-Pro–amide (Sigma, Poole, United Kindom) was added to whole plasma samples to prevent polymerization and cross-linking of fibrin. Hirudin (100 U/mL) was added to one sample to specifically inhibit thrombin.
Enzymatic proteolysis of ADAMTS13 by thrombin, FXa, and plasmin
ADAMTS13 (250 nM) was incubated with 0.9 nM to 90 nM human α-thrombin (Sigma), FXa (Calbiochem, Nottingham, United Kingdom) or plasmin (Calbiochem) in 20 mM Tris-HCl (pH 7.8), 5 mM CaCl2, 150 mM NaCl at 37°C. At specific time points (0-6 hours), ADAMTS13 fragmentation was analyzed by Coomassie staining and Western blotting (with anti-protease domain or anti-myc monoclonal antibody [mAb]). Experiments using heparin or thrombomodulin (TM) were similarly performed except that thrombin (50 nM) was preincubated for 5 minutes with equimolar or a 10-fold molar excess of unfractionated heparin (CP Pharmaceuticals, Wrexham, United Kingdom) or soluble rabbit TM (Cambridge Bioscience, Cambridge, United Kingdom), respectively. To assess the relative specificity of thrombin for ADAMTS13 over other ADAMTS family members, parallel experiments in which ADAMTS420 (a kind gift of Dr H. Nagase, Imperial College London, United Kingdom) was treated with 9 nM thrombin for 0 to 6 hours were analyzed by Western blotting using ADAMTS4 protease domain–specific polyclonal antibodies20 (also a gift of Dr H. Nagase).
Purification of VWF from human plasma
VWF was purified from normal human plasma as described by Bowen.21 Briefly, 600 mL fresh frozen plasma was cryoprecipitated. The pellet was dissolved in 10 mL 20 mM Tris-HCl (pH 8). Samples were cleared before purification by gel filtration using a Sepharose CL-2B HiPrep 26/60 column (AmershamPharmacia). VWF concentration in each fraction was determined using a specific VWF enzyme-linked immunosorbent assay (ELISA), as previously described.22 The purity of VWF was assessed by reducing SDS-PAGE and silver staining.
ADAMTS13 activity assays
To assess the effect of proteolysis on ADAMTS13 activity, 1 mM ADAMTS13 was incubated with and without 9 nM thrombin or plasmin for 16 hours at 37°C. ADAMTS13 activity assays were performed essentially as described14 and using a VWF collagen-binding assay (VWF-CBA; Technoclone, Surrey, United Kingdom). Briefly, 5 μg of each ADAMTS13 sample (with and without proteinase treatment) was preincubated with 10 mM BaCl2 for 10 minutes at 37°C. Thereafter, samples were incubated at 37°C with 8 nM purified human VWF in reaction conditions containing 1.5 M urea, 10 mM BaCl2, 5 mM NaCl, 0.5 mM CaCl2, and 15 mM Tris-HCl (pH 8.0). At 0, 1, 2, 4, and 6 hours, subsamples were removed for analysis of VWF proteolysis. Samples were mixed with 2× reducing SDS loading buffer and heated for 20 minutes at 95°C. Samples were separated on 5% polyacrylamide gels and electroblotted onto a polyvinylidene difluoride (PVDF) membrane. Proteolysis of VWF was examined by Western blotting using a polyclonal anti-VWF antibody (Dako, Cambridge, United Kingdom). Parallel samples were also analyzed in triplicate with a VWF-CBA, according to the manufacturer's instructions. ADAMTS13 activity was thus assessed indirectly by examining its effect on VWF collagen-binding function. VWF-CBA results were normalized by parallel sampling in the absence of recombinant ADAMTS13.
Results
Purification of recombinant ADAMTS13
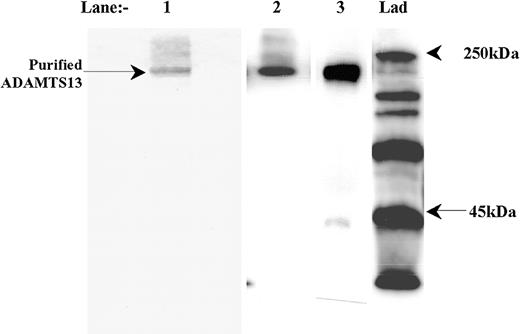
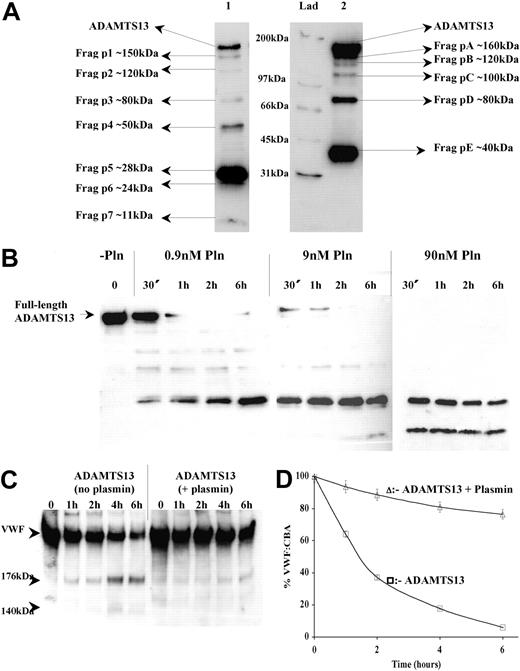
Recombinant myc/6xHis-tagged ADAMTS13 was expressed into conditioned medium from HEK293 cells that had been stably transfected with the vector, pcDNA3.1/ADAMTS13/myc/His. ADAMTS13 (∼200 kDa) was purified and concentrated by affinity chromatography; see result of SDS-PAGE (Figure 1, lane 1). Western blot analysis using an anti-ADAMTS13 protease domain antibody highlighted the presence of the same prominent band at approximately 190 kDa to 210 kDa (Figure 1, lane 2), in accordance with a previous report.18 However, analysis of this same sample using an anti-myc epitope antibody revealed the presence of an additional but lower intensity band at approximately 40 kDa (Figure 1, lane 3). This minor species was believed to be a C-terminal ADAMTS13 degradation fragment (containing the myc/His tag) that was copurified with the full-length enzyme.
Purification of recombinant human ADAMTS13. ADAMTS13 was expressed by HEK293 cells stably transfected with the expression vector, pcDNA3.1/ADAMTS13/myc/His. ADAMTS13 was purified from conditioned medium Ni2+-HiTrap columns. Purified material was electrophoresed on a 7.5% polyacrylamide gel under nonreducing conditions and stained with Coomassie (lane 1). Lanes 2 and 3: Western blot analysis under reducing conditions with the anti-ADAMTS13 protease domain antibody and anti-myc epitope antibody, respectively. Lad = marker ladder
Purification of recombinant human ADAMTS13. ADAMTS13 was expressed by HEK293 cells stably transfected with the expression vector, pcDNA3.1/ADAMTS13/myc/His. ADAMTS13 was purified from conditioned medium Ni2+-HiTrap columns. Purified material was electrophoresed on a 7.5% polyacrylamide gel under nonreducing conditions and stained with Coomassie (lane 1). Lanes 2 and 3: Western blot analysis under reducing conditions with the anti-ADAMTS13 protease domain antibody and anti-myc epitope antibody, respectively. Lad = marker ladder
ADAMTS13 is proteolyzed following activation of coagulation in human plasma
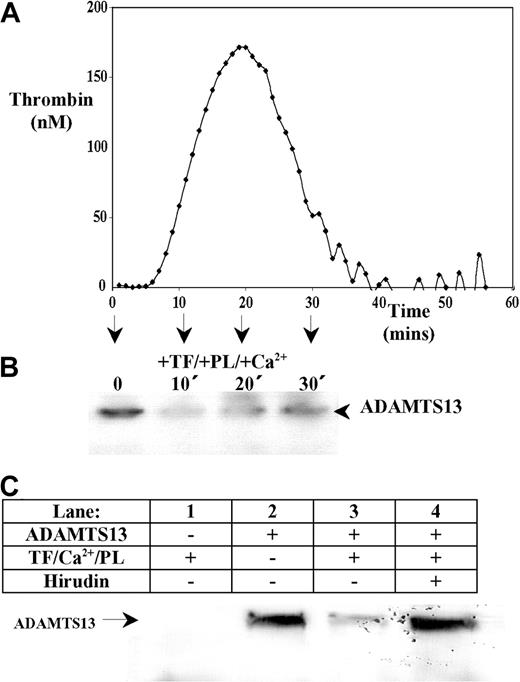
Western blotting to detect endogenous ADAMTS13 in human plasma was ineffective due to its low concentration. Addition of recombinant ADAMTS13 to defibrinated plasma allowed detection of a single approximately 200-kDa ADAMTS13 band. Adding relipidated TF, phospholipids, and Ca2+ initiated thrombin generation that was quantified in real-time using a fluorogenic substrate and an automated plate reader. This demonstrated that thrombin was generated in ADAMTS13-spiked plasma samples with a characteristic lag-time followed by a rapid propagation phase (Figure 2A). In parallel samples lacking the thrombin substrate, aliquots were removed at different time points (0-30 minutes), and the effect of coagulation on ADAMTS13 was examined by Western blotting using the anti-ADAMTS13 protease domain monoclonal antibody. In samples in which thrombin was generated (Figure 2B), a decrease in the intensity of the full-length ADAMTS13 band was observed. This demonstrated that activation of the coagulation cascade caused the rapid loss of full-length ADAMTS13. In the absence of a procoagulant stimulus (ie, in samples lacking TF, phospholipids, and Ca2+), no such depletion in full-length ADAMTS13 over the 30-minute time course (not shown). To further assess this loss of ADAMTS13, whole human plasma (containing fibrinogen) was supplemented with ADAMTS13 and also 5 mg/mL Gly-Pro-Arg-Pro–amide, to prevent fibrin polymerization and subsequent cross-linking. Analysis of these samples by Western blotting using the anti-protease domain mAb (Figure 2C) revealed that without supplemented ADAMTS13, no ADAMTS13 was detected in plasma (Figure 2C, lane 1). In the absence of procoagulant stimulus, recombinant ADAMTS13 was detected as a single band of approximately 200 kDa (Figure 2C, lane 2). Following a 30-minute incubation at 37°C with TF, Ca2+, and phospholipids, the intensity of the full-length ADAMTS13 band was markedly reduced (Figure 2C, lane 3), but this was completely inhibited by hirudin (Figure 2C, lane 4). This implied that the loss of ADAMTS13 from plasma was mediated in part via a mechanism involving thrombin.
ADAMTS13 is proteolyzed following thrombin generation in human plasma. Recombinant ADAMTS13 (final concentration 80 nM) was added to defibrinated normal pooled citrated human plasma. (A) Thrombin generation was initiated by the addition of 8 pM TF, Ca2+, and phospholipids (PLs; PS/PC/PE, 20:60:20). The amount of thrombin generated in defibrinated ADAMTS13-spiked plasma was monitored in triplicate in real-time by a Fluoroscan plate reader using a fluorogenic thrombin substrate. Readings were quantified with a standard curve generated in parallel. (B) Parallel plasma samples were also analyzed for ADAMTS13 fragmentation. At 0, 10, 20, and 30 minutes, ADAMTS13 degradation was assessed by Western blotting under reducing conditions using an anti-ADAMTS13 protease domain antibody. (C) Citrated whole human plasma (containing 5 mg/mL GPRP-amide) was incubated with either TF/PLs/Ca2+ (lane 1), 125 nM ADAMTS13 (lane 2), TF/PLs/Ca2+ and 125 nM ADAMTS13 (lane 3), or TF/PLs/Ca2+, 125 nM ADAMTS13, and 100 U/mL hirudin (lane 4) for 30 minutes at 37°C. Samples were analyzed by Western blotting using an anti-ADAMTS13 protease domain antibody.
ADAMTS13 is proteolyzed following thrombin generation in human plasma. Recombinant ADAMTS13 (final concentration 80 nM) was added to defibrinated normal pooled citrated human plasma. (A) Thrombin generation was initiated by the addition of 8 pM TF, Ca2+, and phospholipids (PLs; PS/PC/PE, 20:60:20). The amount of thrombin generated in defibrinated ADAMTS13-spiked plasma was monitored in triplicate in real-time by a Fluoroscan plate reader using a fluorogenic thrombin substrate. Readings were quantified with a standard curve generated in parallel. (B) Parallel plasma samples were also analyzed for ADAMTS13 fragmentation. At 0, 10, 20, and 30 minutes, ADAMTS13 degradation was assessed by Western blotting under reducing conditions using an anti-ADAMTS13 protease domain antibody. (C) Citrated whole human plasma (containing 5 mg/mL GPRP-amide) was incubated with either TF/PLs/Ca2+ (lane 1), 125 nM ADAMTS13 (lane 2), TF/PLs/Ca2+ and 125 nM ADAMTS13 (lane 3), or TF/PLs/Ca2+, 125 nM ADAMTS13, and 100 U/mL hirudin (lane 4) for 30 minutes at 37°C. Samples were analyzed by Western blotting using an anti-ADAMTS13 protease domain antibody.
Thrombin cleaves ADAMTS13 at several sites
Degradation of ADAMTS13 by thrombin and/or FXa was considered a possible mechanism for the removal of full-length ADAMTS13 from plasma. To circumvent the technical difficulties associated with blotting/detecting specific proteins in plasma samples, this was examined further using purified proteins. ADAMTS13 was incubated with 9 nM human thrombin for 16 hours in a reaction containing 150 mM NaCl and 5 mM Ca2+. Reducing SDS-PAGE and Coomassie staining revealed that without the addition of thrombin, full-length ADAMTS13 was clearly visible as a band of approximately 200 kDa (Figure 3A). In the same sample containing thrombin, the full-length ADAMTS13 band disappeared completely (Figure 3A). Furthermore, lower-molecular-weight bands that were not readily visualized in either the thrombin or ADAMTS13 preparations alone, became evident (Figure 3A, newly emerged arrows). These smaller bands represent degradation products arising from thrombin cleavage. This finding not only demonstrated that ADAMTS13 is susceptible to proteolysis by thrombin, but also that cleavage occurs at multiple sites.
Purified ADAMTS13 is cleaved by thrombin and FXa in a concentration- and time-dependent fashion. Purified ADAMTS13 (400 nM) was incubated with and without 9 nM thrombin (FIIa) in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ at 37°C for 16 hours. Samples were electrophoresed on a 4% to 15% polyacrylamide gel under both reducing (A) and nonreducing (B) conditions, followed by Coomassie staining. Thrombin was detected as a band of approximately 36 kDa (white arrowhead); full-length ADAMTS13 and its proteolytic fragments are identified by arrows. (C) Western blot analysis of ADAMTS13 with and without complete thrombin proteolysis (under reducing and nonreducing conditions) using the anti-ADAMTS13 protease domain antibody. (D) Western blot analysis of ADAMTS13 with and without complete thrombin proteolysis (under reducing and nonreducing conditions) using the anti-myc epitope antibody. (E) Varying concentrations of thrombin (0 nM-90 nM) were incubated with 250 nM ADAMTS13 at 37°C for 0 to 6 hours in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+. Samples were analyzed under reducing conditions by Western blotting using the anti-ADAMTS13 protease domain antibody. (F) FXa (50 nM) was incubated with 250 nM ADAMTS13 at 37°C for 0 to 6 hours in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+, and analyzed as in panel E.
Purified ADAMTS13 is cleaved by thrombin and FXa in a concentration- and time-dependent fashion. Purified ADAMTS13 (400 nM) was incubated with and without 9 nM thrombin (FIIa) in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ at 37°C for 16 hours. Samples were electrophoresed on a 4% to 15% polyacrylamide gel under both reducing (A) and nonreducing (B) conditions, followed by Coomassie staining. Thrombin was detected as a band of approximately 36 kDa (white arrowhead); full-length ADAMTS13 and its proteolytic fragments are identified by arrows. (C) Western blot analysis of ADAMTS13 with and without complete thrombin proteolysis (under reducing and nonreducing conditions) using the anti-ADAMTS13 protease domain antibody. (D) Western blot analysis of ADAMTS13 with and without complete thrombin proteolysis (under reducing and nonreducing conditions) using the anti-myc epitope antibody. (E) Varying concentrations of thrombin (0 nM-90 nM) were incubated with 250 nM ADAMTS13 at 37°C for 0 to 6 hours in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+. Samples were analyzed under reducing conditions by Western blotting using the anti-ADAMTS13 protease domain antibody. (F) FXa (50 nM) was incubated with 250 nM ADAMTS13 at 37°C for 0 to 6 hours in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+, and analyzed as in panel E.
Under nonreducing conditions (Figure 3B), after incubation with thrombin, a similar disappearance of the band corresponding to full-length ADAMTS13 was observed. Two major ADAMTS13 proteolytic fragments were resolved, one of approximately 150 kDa and another of approximately 40 kDa (Figure 3B), the latter band migrating with a similar molecular weight to that of thrombin. The presence of only 2 major cleavage products under nonreducing conditions suggested that most of the ADAMTS13 cleavage fragments, with the exception of the 40-kDa C-terminal species, remain covalently associated by disulfide bond linkages.
Western blot analysis of these same samples using the anti-ADAMTS13 protease domain antibody revealed that under reducing conditions, the protease domain was present within an approximately 28-kDa band (Figure 3C). Under nonreducing conditions and after treatment with thrombin, the protease domain was present in the approximately 150-kDa band (Figure 3C).
Analysis of these samples using the anti-myc antibody revealed that an approximately 40-kDa C-terminal fragment of ADAMTS13 was generated after thrombin treatment under both reducing and nonreducing conditions (Figure 3D). This implies that at the most C-terminal cleavage site, thrombin proteolyzes ADAMTS13 between unpaired cysteine residues and that the C-terminal fragment (containing all or most of the 2 CUB domains) is no longer covalently associated with the rest of the ADAMTS13 molecule.
Time- and concentration-dependent cleavage of ADAMTS13 by thrombin
Incubation of ADAMTS13 with varying concentrations of human thrombin (0.9 nM-90 nM or 0.1 U/mL-10 U/mL) for 0 to 6 hours, followed by Western blotting with the anti-ADAMTS13 protease domain antibody, revealed that ADAMTS13 was cleaved by thrombin in a time- and concentration-dependent fashion (Figure 3E). In the presence of 0.9 nM thrombin, no cleavage of the approximately 200-kDa full-length ADAMTS13 band was observed after a 1-hour incubation. At 2 hours, an additional faint band of approximately 150 kDa became evident and by 6 hours, this band and a band at approximately 28 kDa were also observed.
Using 9 nM thrombin, the rate of ADAMTS13 proteolysis was appreciably enhanced. After only 30 minutes, little full-length ADAMTS13 remained. Moreover, the 3 major fragmentation bands (∼150 kDa, ∼40 kDa, and ∼28 kDa, respectively) were prominent. Over time, the high Mr band disappeared, whereas the band of approximately 28 kDa became stronger. With 90 nM thrombin, there was complete loss of full-length ADAMTS13 within 30 minutes. Interestingly, the gain in the intensity of the 28 kDa band did not necessarily reflect the loss of intensity of the higher Mr bands. One possible cause of this might be the presence of an additional nonspecific or less favored cleavage site within the protease domain that with time, generated a fragment that was only marginally smaller than the 28-kDa band.
Experiments were also performed using FXa. These demonstrated that FXa caused ADAMTS13 fragmentation similarly to that caused by thrombin. However, the rate of ADAMTS13 proteolysis by FXa was less than that of thrombin (Figure 3F). Even at 50 nM FXa, proteolysis of ADAMTS13 was markedly slower than when 9 nM thrombin was used (Figure 3E).
Characterization of ADAMTS13 proteolysis by thrombin
To further analyze ADAMTS13 fragmentation, partially proteolyzed ADAMTS13 species were visualized by Western blotting. As before, we detected either N-terminus–containing fragments with the anti-ADAMTS13 protease domain antibody (Figure 4A) or C-terminus–containing fragments with the anti-myc antibody (Figure 4B). In these experiments, short incubations using limited quantities of thrombin were employed to facilitate visualization of proteolytic intermediates of ADAMTS13 (Figure 4A-B, lane 2). In addition, increased quantities of ADAMTS13 were treated with thrombin and blots developed for extended periods to better identify transient proteolytic intermediates that, over time, would be further cleaved into smaller species (Figure 4A, lane 3; Figure 4B, lane 4).
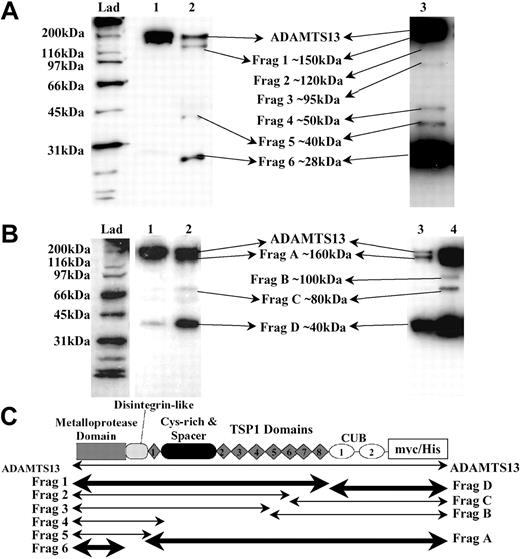
Identification of ADAMTS13 fragments generated by thrombin. Recombinant ADAMTS13 (250 nM) was incubated with 9 nM human thrombin at 37°C for 30 minutes in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+, causing partial proteolysis of ADAMTS13. Samples were analyzed by Western blotting. (A) ADAMTS13 was detected using the anti-ADAMTS13 protease domain antibody. ADAMTS13 (lane 1) was incubated with thrombin (lane 2). Increased quantities of ADAMTS13 (400 nM) were incubated with thrombin and extended development of blots (lane 3). (B) Probing of samples from panel A using the anti-myc antibody. ADAMTS13 (lane 1); ADAMTS13 treated with thrombin (lanes 2, 3, and 4). Lanes 3 and 4 represent the same sample; lane 3 contains 25% of the volume loaded in lane 4. (C) The predicted identities of the cleavage fragments in panel A (Frags 1-6) and panel B (Frags A-D) are represented. The locations of the predicted cleavage sites are approximate. Fragment boundaries and sizes were estimated from the molecular weights calculated from panel A and panel B. The fragments generated by cleavage at the favored cleavage sites are shown in bold.
Identification of ADAMTS13 fragments generated by thrombin. Recombinant ADAMTS13 (250 nM) was incubated with 9 nM human thrombin at 37°C for 30 minutes in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+, causing partial proteolysis of ADAMTS13. Samples were analyzed by Western blotting. (A) ADAMTS13 was detected using the anti-ADAMTS13 protease domain antibody. ADAMTS13 (lane 1) was incubated with thrombin (lane 2). Increased quantities of ADAMTS13 (400 nM) were incubated with thrombin and extended development of blots (lane 3). (B) Probing of samples from panel A using the anti-myc antibody. ADAMTS13 (lane 1); ADAMTS13 treated with thrombin (lanes 2, 3, and 4). Lanes 3 and 4 represent the same sample; lane 3 contains 25% of the volume loaded in lane 4. (C) The predicted identities of the cleavage fragments in panel A (Frags 1-6) and panel B (Frags A-D) are represented. The locations of the predicted cleavage sites are approximate. Fragment boundaries and sizes were estimated from the molecular weights calculated from panel A and panel B. The fragments generated by cleavage at the favored cleavage sites are shown in bold.
Limited treatment of ADAMTS13 with thrombin revealed at least 3 protease domain–containing fragments (Figure 4A, lane 2) of approximately 150 kDa, 40 kDa, and 28 kDa (designated Frag 1, Frag 5, and Frag 6, respectively). Frag 1 and Frag 6 were the major species. However, proteolysis using increased quantities of ADAMTS13 (Figure 4A, lane 3) indicated that at least an additional 3 intermediate species were generated by thrombin treatment (Frags 2, 3, and 4); the faintness of these bands, visible on the original blot, is suggestive of additional, yet less favored, thrombin cleavage sites. In Figure 4C, the approximate locations of Frags 1 to 6 are predicted based on their mobilities and identification by the detecting antibodies.
Similar analysis of C-terminal, myc epitope-containing fragments (Figure 4B) revealed a fragmentation pattern that corroborated and complemented the data obtained using the anti-ADAMTS13 protease domain antibody. The major species generated by thrombin were Frag A (∼160 kDa) and Frag D (∼40 kDa), again highlighting the potential preference for proteolysis at the most C-terminal and N-terminal sites. Interestingly, the approximately 40-kDa C-terminal fragment that was copurified with ADAMTS13 during its original preparation could not be resolved from Frag D. This might imply that both species have arisen via cleavage at the same proteolytically sensitive site.
Due to their predicted high molecular weight, we could not resolve the 170-kDa, 160-kDa, and 150-kDa fragments that would correspond to the C-terminal aspects of Frags 4, 5, and 6 (Figure 4C). These species probably comigrated as Frag A. Once again, proteolysis of increased quantities of ADAMTS13 indicated 2 additional intermediate species were generated (Frags B and C), very possibly representing the C-terminal portion of Frag 2 and Frag 3. Again, the location of these fragments is predicted in Figure 4C.
Proteolysis by thrombin ablates ADAMTS13 activity
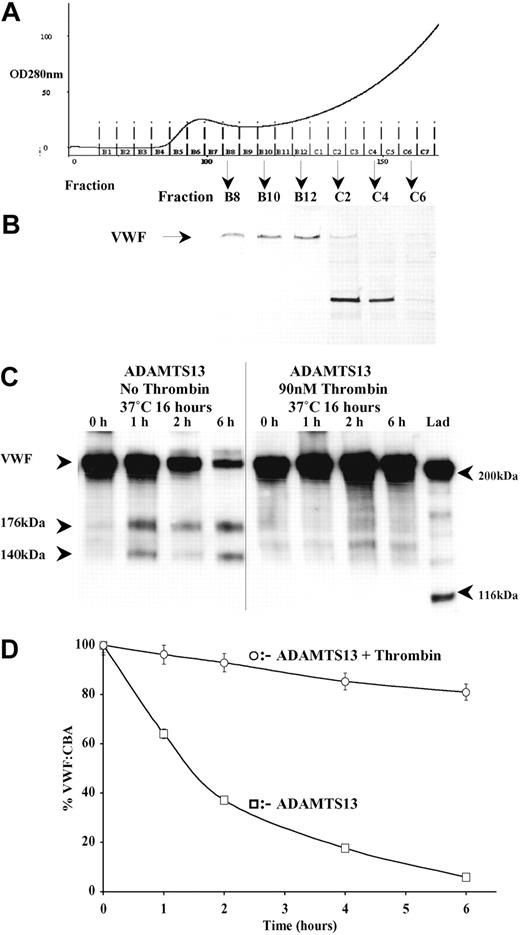
C-terminal truncation of ADAMTS13 past the spacer domain has previously been reported to abolish ADAMTS13 activity.23,24 It would seem that after proteolysis by thrombin, the protease domain to TSP1-8 remains covalently intact through disulphide bond linkages. Although an approximately 40-kDa C-terminal fragment of ADAMTS13, containing the CUB domains, is proteolytically removed by thrombin, the loss of these domains does not appear to significantly affect the VWF-cleaving activity of ADAMTS13 in static assays.23,24 Thus, to ascertain whether thrombin cleavage might simultaneously abrogate ADAMTS13 activity, we compared the ability of full-length and thrombin-proteolyzed ADAMTS13 to cleave purified human VWF. Human VWF was purified from plasma by cryoprecipitation followed by gel filtration (Figure 5A). The first protein fractions eluted (B8-C6) contained VWF. The purity of VWF was assessed by SDS-PAGE and silver staining (Figure 5B). Fractions B8 to B10 were free of major contaminating proteins and were thus used as an ADAMTS13 substrate. ADAMTS13 preincubated in the absence of thrombin was fully active and generated characteristic 176-kDa and 140-kDa VWF bands (Figure 5C). Moreover, the intensity of the 250-kDa VWF band diminished over the 6-hour time course. ADAMTS13 that had been pretreated with thrombin exhibited little or no VWF-cleaving activity. As the VWF Western blot detection method is qualitative, we also examined ADAMTS13 cleavage of VWF in a quantitative manner by monitoring loss of VWF function with a VWF-CBA (Figure 5D). These data corroborated the VWF fragmentation results. At 6 hours, the VWF in samples containing ADAMTS13 preincubated without thrombin exhibited approximately 5% original collagen-binding activity due to loss of VWF function arising from ADAMTS13-dependent proteolysis. In contrast, VWF incubated with thrombin-treated ADAMTS13 still exhibited approximately 90% of the original collagen-binding potential. Taken together, these data confirm that once proteolyzed by thrombin, ADAMTS13 is no longer able to efficiently cleave VWF and so inhibit its physiologic function.
Proteolysis of ADAMTS13 by thrombin abolishes ADAMTS13 enzymatic activity toward purified human VWF. Human VWF was purified from human plasma by gel filtration. After gel filtration (A), the purity of VWF in each fraction was assessed by reducing SDS-PAGE on a 7.5% polyacrylamide gel, followed by silver staining (B). Fractions B8-B12 contained VWF, visualized as a band of approximately 250 kDa (arrow), and were free of major contaminating proteins. Only the highest purity VWF from fractions B8-B10 were used as a substrate for ADAMTS13. ADAMTS13 that had been pretreated with and without 9 nM thrombin in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ for 16 hours at 37°C was assayed for activity. Subsamples were removed at 0, 1, 2, and 6 hours and assessed by Western blotting using an anti-VWF polyclonal antibody (C). ADAMTS13 generated characteristic VWF proteolytic fragments of 176 kDa and 140 kDa. Samples studied in panel C were also analyzed using a functional assay based on collagen binding (VWF-CBA) as outlined in “Materials and methods” (D). Results shown ± SEM, (n = 3).
Proteolysis of ADAMTS13 by thrombin abolishes ADAMTS13 enzymatic activity toward purified human VWF. Human VWF was purified from human plasma by gel filtration. After gel filtration (A), the purity of VWF in each fraction was assessed by reducing SDS-PAGE on a 7.5% polyacrylamide gel, followed by silver staining (B). Fractions B8-B12 contained VWF, visualized as a band of approximately 250 kDa (arrow), and were free of major contaminating proteins. Only the highest purity VWF from fractions B8-B10 were used as a substrate for ADAMTS13. ADAMTS13 that had been pretreated with and without 9 nM thrombin in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ for 16 hours at 37°C was assayed for activity. Subsamples were removed at 0, 1, 2, and 6 hours and assessed by Western blotting using an anti-VWF polyclonal antibody (C). ADAMTS13 generated characteristic VWF proteolytic fragments of 176 kDa and 140 kDa. Samples studied in panel C were also analyzed using a functional assay based on collagen binding (VWF-CBA) as outlined in “Materials and methods” (D). Results shown ± SEM, (n = 3).
Effect of thrombin cofactors on ADAMTS13 proteolysis
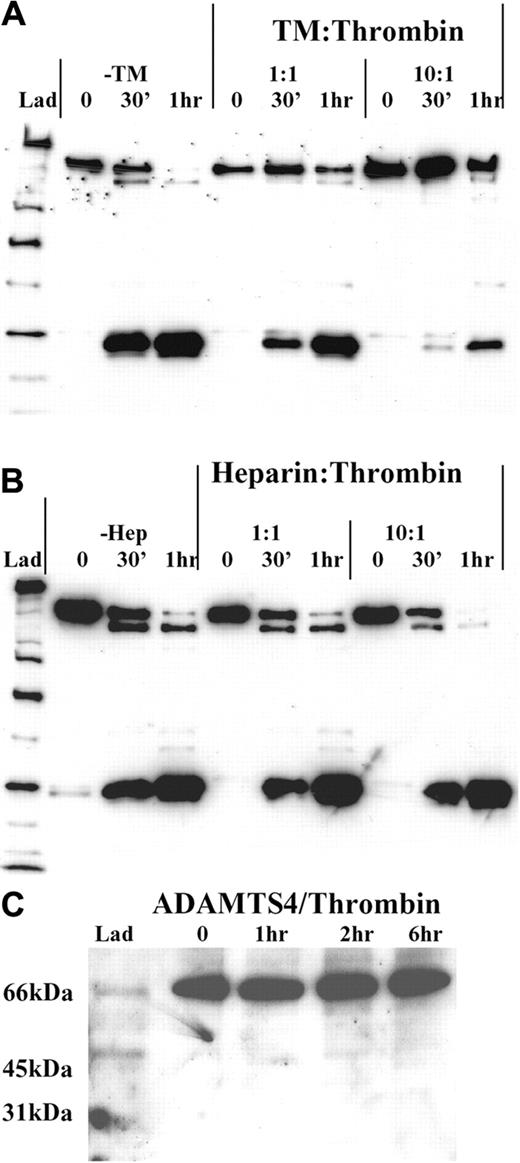
To further assess the nature of the interaction of thrombin with ADAMTS13 and also to examine the way in which thrombin-dependent ADAMTS13 inactivation might itself be regulated or localized to the site of vascular injury, we investigated the effect of thrombin cofactors (TM and heparin) on the rate of ADAMTS13 proteolysis. For these experiments, we preincubated 50 nM thrombin for 5 minutes with either equimolar (50 nM) or 10-fold molar excess (500 nM) of either soluble rabbit TM or heparin. Thereafter, we examined the extent of ADAMTS13 fragmentation over a 1-hour incubation period.
Thrombomodulin. Preincubation of thrombin with soluble rabbit TM markedly inhibited the ability of thrombin to proteolyze ADAMTS13 (Figure 6A). In the absence of TM, proteolysis went to near completion after one hour of incubation with thrombin. At this time, only trace amounts of full-length ADAMTS13 could be detected. However, with preincubation of thrombin with equimolar quantities of TM, the rate of full-length ADAMTS13 band depletion was reduced. The rate at which the smallest fragmentation band (Frag 6) was generated was also similarly impaired. As expected, the effect of a 10-fold molar excess of TM was more pronounced. Indeed, after a 30-minute incubation with thrombin, very little ADAMTS13 fragmentation was observed, by comparison to the no TM control. After one hour, we estimated that more than 50% of the remaining ADAMTS13 was still intact.
Cofactors of thrombin and ADAMTS13 proteolysis. Recombinant ADAMTS13 (250 nM) was treated with 50 nM thrombin that had been preincubated at 37°C with either equimolar or a 10-fold molar excess of either soluble rabbit TM (A) or heparin (B) in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+. ADAMTS13 fragmentation was monitored under reducing conditions using the anti-protease domain antibody. (A) Thrombin was preincubated with either 50 nM or 500 nM soluble rabbit TM for 5 minutes. Thereafter, the ability of each thrombin/TM mix to cleave ADAMTS13 was monitored at 0 minutes, 30 minutes, and 1 hour time points, and was compared with that of thrombin alone. (B) Thrombin was preincubated with either 50 nM or 500 nM heparin and analyzed as in panel A. (C) The specificity of thrombin for ADAMTS13 relative to other ADAMTS-family members was gauged by incubating 250 nM ADAMTS4 in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ with 9 nM thrombin for 0 to 6 hours. Samples were analyzed by Western blotting using a polyclonal anti-ADAMTS4 protease domain antibody.
Cofactors of thrombin and ADAMTS13 proteolysis. Recombinant ADAMTS13 (250 nM) was treated with 50 nM thrombin that had been preincubated at 37°C with either equimolar or a 10-fold molar excess of either soluble rabbit TM (A) or heparin (B) in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+. ADAMTS13 fragmentation was monitored under reducing conditions using the anti-protease domain antibody. (A) Thrombin was preincubated with either 50 nM or 500 nM soluble rabbit TM for 5 minutes. Thereafter, the ability of each thrombin/TM mix to cleave ADAMTS13 was monitored at 0 minutes, 30 minutes, and 1 hour time points, and was compared with that of thrombin alone. (B) Thrombin was preincubated with either 50 nM or 500 nM heparin and analyzed as in panel A. (C) The specificity of thrombin for ADAMTS13 relative to other ADAMTS-family members was gauged by incubating 250 nM ADAMTS4 in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ with 9 nM thrombin for 0 to 6 hours. Samples were analyzed by Western blotting using a polyclonal anti-ADAMTS4 protease domain antibody.
Heparin. Preincubation of thrombin with equimolar or a 10-fold molar excess of heparin, had no inhibitory effect on ADAMTS13 fragmentation, by comparison to samples lacking heparin (Figure 6B). Indeed, at a 10-fold excess, the rate of ADAMTS13 cleavage may have been slightly enhanced, when compared with the no heparin and equimolar heparin samples. The reason for this very moderate enhancement is uncertain but may be due to bridging of ADAMTS13 and thrombin by heparin.
Specificity of thrombin for ADAMTS13
To help establish whether thrombin proteolyzed ADAMTS13 in a specific fashion, we compared the susceptibility of another ADAMTS family member (ADAMTS4) to thrombin cleavage. In these experiments, no proteolysis of ADAMTS4 was detected with 9 nM thrombin within 6 hours (Figure 6C), suggesting that ADAMTS4 is resistant to thrombin-mediated proteolysis. This finding implies that ADAMTS13, which has a similar domain structure to ADAMTS4, is specifically recognized or cleaved by thrombin.
ADAMTS13 is rapidly proteolyzed and inactivated by plasmin
Thrombin and FXa are generated rapidly at the site of vascular injury. However, other proteases are also locally activated following initiation of coagulation that might also influence ADAMTS13 function. We examined the susceptibility of ADAMTS13 to cleavage and inactivation by plasmin. Following limited proteolysis of ADAMTS13 (9 nM plasmin for 30 minutes; Figure 7A), plasmin generated a similar, but not identical, fragmentation of ADAMTS13 to that observed for proteolysis by thrombin (Figure 4A-B), consistent with its limited specificity for cleavage after certain arginine residues.25 However, plasmin also cleaves after lysine, which potentially explains the presence of alternative ADAMTS13 fragments. Of particular interest was the additional cleavage site in the protease domain that generated Frag p7. This fragment was not generated by thrombin. Plasmin cleaved ADAMTS13 more rapidly than thrombin (compare Figure 7B and Figure 3, respectively). Indeed, at 0.9 nM plasmin, full-length ADAMTS13 was rapidly depleted/proteolyzed. The effect of plasmin proteolysis on ADAMTS13 activity was assessed using purified human VWF, as before. Pretreatment of ADAMTS13 with plasmin had the same effect on VWF-cleaving activity as did thrombin. No proteolysis of VWF by plasmin-treated ADAMTS13 was detected by Western blotting (Figure 7C). Furthermore, only a small reduction in VWF-CBA was observed in plasmin-treated samples, whereas untreated ADAMTS13 generated characteristic VWF cleavage products and rapidly abolished the VWF collagen-binding activity (Figure 7D).
Proteolytic inactivation of ADAMTS13 by plasmin. Recombinant ADAMTS13 (250 nM) was incubated with 9 nM human plasmin in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ at 37°C for 30 minutes, causing partial proteolysis of ADAMTS13 (A). Samples were analyzed by Western blotting using either anti-ADAMTS13 protease domain (lane 1) or anti-myc epitope antibodies (lane 2). The rate of plasmin-dependent ADAMTS13 proteolysis was assessed by incubating varying concentrations of plasmin (0 nM-90 nM) with 250 nM ADAMTS13 at 37°C for 30 minutes to 6 hours (B). Samples were analyzed under reducing conditions by Western blotting using the anti-ADAMTS13 protease domain antibody. To assess the effect of plasmin proteolysis on ADAMTS13 function, ADAMTS13 was pretreated with or without 9 nM plasmin for 16 hours at 37°C and thereafter assayed for activity by 2 methods, as outlined in “Materials and methods.” Subsamples were removed from each assay at 0, 1, 2, 4, and 6 hours and assessed by Western blotting using an anti-VWF polyclonal antibody (C). ADAMTS13 generated characteristic VWF proteolytic fragments of 176 kDa and 140 kDa that were visualized after 1 hour and increased in intensity over the 6-hour time course. There was also a concomitant decrease in the intensity of the 250-kDa VWF band. Analysis of VWF function as in Figure 6D using a VWF-CBA (D). Results shown ± SEM, (n = 3).
Proteolytic inactivation of ADAMTS13 by plasmin. Recombinant ADAMTS13 (250 nM) was incubated with 9 nM human plasmin in 20 mM Tris-HCl (pH 7.8) containing 150 mM NaCl and 5 mM Ca2+ at 37°C for 30 minutes, causing partial proteolysis of ADAMTS13 (A). Samples were analyzed by Western blotting using either anti-ADAMTS13 protease domain (lane 1) or anti-myc epitope antibodies (lane 2). The rate of plasmin-dependent ADAMTS13 proteolysis was assessed by incubating varying concentrations of plasmin (0 nM-90 nM) with 250 nM ADAMTS13 at 37°C for 30 minutes to 6 hours (B). Samples were analyzed under reducing conditions by Western blotting using the anti-ADAMTS13 protease domain antibody. To assess the effect of plasmin proteolysis on ADAMTS13 function, ADAMTS13 was pretreated with or without 9 nM plasmin for 16 hours at 37°C and thereafter assayed for activity by 2 methods, as outlined in “Materials and methods.” Subsamples were removed from each assay at 0, 1, 2, 4, and 6 hours and assessed by Western blotting using an anti-VWF polyclonal antibody (C). ADAMTS13 generated characteristic VWF proteolytic fragments of 176 kDa and 140 kDa that were visualized after 1 hour and increased in intensity over the 6-hour time course. There was also a concomitant decrease in the intensity of the 250-kDa VWF band. Analysis of VWF function as in Figure 6D using a VWF-CBA (D). Results shown ± SEM, (n = 3).
Discussion
In this study, we addressed how ADAMTS13 might be regulated. To date, no specific protease inhibitor has been identified that might localize and restrict the known role of ADAMTS13 activity in the prevention of unwanted VWF adherence and platelet aggregation to the intact endothelium. The absence of a specific inhibitor was considered surprising in view of the crucial role played by inhibitors in regulating proteases that participate in normal hemostasis. A single report has implicated interleukin-6 as a possible regulator of ADAMTS13; however, the mechanism for this and its significance in normal ADAMTS13 regulation remain uncertain.26 Accordingly, we have investigated proteolysis as a regulatory mechanism.
We have shown the rapid disappearance of full-length recombinant ADAMTS13 from defibrinated human plasma in which thrombin generation had been initiated (Figure 2). One explanation for this was the proteolytic depletion of ADAMTS13 by a component of the coagulation cascade. Further evidence for this was provided using whole plasma, in which the depletion of full-length ADAMTS13 was inhibited by hirudin. Thereafter, we confirmed that in vitro low concentrations of thrombin cleave purified ADAMTS13 at several sites (Figures 3 and 4), but does not cleave ADAMTS4 (Figure 6). Interestingly, FXa cleaved ADAMTS13 with a very similar fragmentation pattern to that of thrombin. However, FXa proteolyzed ADAMTS13 appreciably slower. As thrombin is more active against ADAMTS13 and given that the local concentrations of thrombin that are generated in vivo invariably exceed those of FXa, the proteolysis of ADAMTS13 by thrombin is more likely to be of physiologic importance.
It was apparent that the proteolysis of ADAMTS13 by thrombin occurs preferentially at 2 sites (Figure 3). The first appears to be located close to the end of the protease domain and the second, near to the start of the first CUB domain. ADAMTS13 fragments generated by cleavage at these sites were rapidly generated. With the exception of the most C-terminal site, thrombin cleaved at sites located between paired cysteine residues. Thus after proteolysis, the resulting ADAMTS13 cleavage fragments remain covalently associated, with the exception of the approximately 40-kDa C-terminal CUB domain–containing region (Figure 3).
The physiologic relevance of proteolytic removal of the CUB domains, located at the C-terminus of ADAMTS13, is uncertain. These domains are not required for VWF-cleaving activity measured under static conditions.23,24 Moreover, certain strains of mice have been identified that express an ADAMTS13 variant that lacks the CUB domains (and also TSP1-7 and TSP1-8),27 further suggesting that the functions of these domains are not critical in vivo. Although autoantibodies against the CUB domains have been found in patients with acquired TTP,28 it is unclear whether these compromise ADAMTS13 function. The recent study reporting that CUB domain–derived peptides inhibit VWF-cleaving activity under flowing, but not static, conditions might, however, imply a potentially important functional role to these domains.29 This latter finding suggests that removal of the CUB domains by thrombin might further contribute to ADAMTS13 inactivation in vivo. Further studies are now required to determine the precise cleavage site and physiologic effect of the proteolytic removal of the CUB domains.
ADAMTS13 is secreted into plasma, where it circulates as an active enzyme. At sites of endothelial disruption, VWF binds collagen and unfolds in response to shear forces. This in turn facilitates platelet tethering—critical for efficient platelet plug formation. If ADAMTS13 were to have no specific inhibitor, its VWF-cleaving activity would be predicted to oppose the essential tethering of platelets once the scissile bond in VWF is exposed by rheologic forces. The data presented here suggest an effective regulatory mechanism in which ADAMTS13 could be proteolytically inactivated by thrombin at sites of vascular perturbation, even at the low thrombin concentrations observed during the initial lag phase of coagulation. During initial platelet plug formation, platelets adhere to VWF and then exposed collagen. However, although this first platelet layer might resist ADAMTS13-mediated VWF cleavage/platelet release due to its tight association with collagen, subsequent layers of platelets will be more remote from collagen. In vivo imaging of thrombus development in mice has demonstrated that thrombin activity/fibrin deposition are generated very rapidly (within 20 seconds) after vascular injury.30 Interestingly, this study also reveals the “blebbing-off” of tethered platelets from the back of the developing thrombus prior to thrombin generation/fibrin deposition. This observation might be explained by or correspond to the action of uninhibited ADAMTS13 on VWF-bound platelets and highlight a need for local ADAMTS13 inactivation at the site of injury.
Thrombin has multiple physiologic substrates and roles.31,32 Following the initiation of coagulation, thrombin confers its prothrombotic activity by cleaving fibrinogen, feedback activation of up-stream clotting factors, and the activation of tethered platelets.33 Together, these functions promote rapid thrombus formation. We now propose a model describing a novel procoagulant function for thrombin. Uniquely, this involves the inactivation of an enzyme (ADAMTS13) to promote thrombus growth, as opposed to its well-characterized roles in activation of protease-activated receptor (PAR) molecules and clotting factors V, VIII, X, XI, and XIII.32 Recently, several studies have attempted to implicate moderate ADAMTS13 deficiency in the pathogenesis of certain thrombotic disorders.34-36 Our data suggest that reductions in ADAMTS13 activity might arise as a result of increased thrombin-dependent inactivation during the course of a thrombotic event. Therefore, additional studies are now required to ascertain whether partial deficiency of ADAMTS13 is a primary cause, or merely a consequence, of thrombosis.
Physiologically, the activity and specificity of thrombin is modulated, in part, by cofactors. Competition between cofactors for exosite binding have been proposed as a mechanism for directing the activities of thrombin.37 In binding TM, the specificity of thrombin toward fibrinogen, protein C, and PAR molecules is dramatically altered.38 This serves to restrict its procoagulant activities to the site of vascular injury. This effect is partly due to the occupancy of exosite I on thrombin with TM.39 We found that if thrombin was preincubated with TM, its ability to proteolyze ADAMTS13 was appreciably compromised (Figure 6). This might therefore implicate exosite I as a possible site of interaction with ADAMTS13. As preincubation of thrombin with heparin had no effect on the rate of ADAMTS13 cleavage, it seems unlikely that exosite II40 on thrombin plays a significant role in its interaction with ADAMTS13. Interestingly, there seemed to be a slight enhancement of ADAMTS13 proteolysis by thrombin in the presence of a 10-fold molar excess of heparin. This might be attributable to the bridging of thrombin and ADAMTS13 by heparin but further investigation is required to confirm this contention.
While examining the susceptibility of ADAMTS13 to proteolysis by coagulation proteinases, we found that plasmin cleaved ADAMTS13 at a faster rate than thrombin, and gave a similar, but not identical, fragmentation pattern. Physiologically, plasmin is generated following thrombin-dependent fibrin deposition and mediates fibrinolysis.41 Plasminogen activators bind to fibrin as it is deposited. Consequently, given the sensitivity of ADAMTS13 to proteolysis at low plasmin concentrations (Figure 7), such cleavage might also be of physiologic importance with respect to the modulation of ADAMTS13 activity at sites of hemostatic plug formation. Proteolysis of ADAMTS13 by plasmin might also suggest a role for VWF in tissue repair. Postinjury vessel regeneration is associated with plasmin activity. If this process should also involve a role for VWF, plasmin-dependent ADAMTS13 inactivation during tissue repair might serve to maintain VWF function. It is interesting to note that whereas plasmin proteolytically activates several different prometalloproteinases,42 it uniquely has an inactivating role against ADAMTS13.
Together, our data provide a plausible mechanism by which the thrombin-dependent inactivation of ADAMTS13 might be specifically localized to the site of hemostatic plug formation. Whereas thrombin (and plasmin) might rapidly inactivate ADAMTS13 at the site of vascular injury (and so favor platelet aggregation), on the surface of the adjacent uninjured endothelium, any thrombin would preferentially bind to TM. Consequently, thrombin-dependent ADAMTS13 proteolysis would be inhibited. ADAMTS13 activity in this location would serve to limit platelet aggregation from spreading beyond the site of injury.
Prepublished online as Blood First Edition Paper, September 23, 2004; DOI 10.1182/blood-2004-03-1101.
Supported by grants from the British Heart Foundation (RG/02/008, FS/03/042/15 450, and FS/03/001).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Antonella Adami for her assistance in producing ADAMTS13, and Ayesha Khan for her help with the thrombin generation experiments.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal