Abstract
The lymph nodes are generally the first extracutaneous manifestation in patients with cutaneous T-cell lymphoma (CTCL); however, their early involvement is difficult to assess. The aim of our study was to define the diagnostic and prognostic value of T-cell clonality analysis for a more precise assessment of lymph node involvement in CTCL. T-cell clonality was determined by 2 independent polymerase chain reaction (PCR) assays, namely a recently developed T-cell receptor-β (TCR-β) PCR technique as well as an established TCR-γ PCR. T-cell clonality was found in 22 of 22 lymph nodes with histologically detectable CTCL involvement as well as in 7 of 14 histologically noninvolved dermatopathic lymph nodes. The clonal T-cell populations in the lymph nodes were in all cases identical to those detected in the corresponding skin lesions, identifying them as the tumor cell population. T-cell clonality was not found in any of the 12 dermatopathic lymph nodes from 12 patients with inflammatory skin diseases. Clonal T-cell detection in 7 of 14 dermatopathic lymph nodes of patients with CTCL was associated with limited survival (74 months; confidence interval [CI], 66-82 months) as in patients with histologically confirmed lymph node involvement (41 months; CI, 35-47 months), whereas all patients without T-cell clonality in the lymph nodes (7 patients) were alive at the last follow-up. Thus, T-cell clonality analysis is an important adjunct in differentiating benign dermatopathic lymphadenitis from early CTCL involvement.
Introduction
Cutaneous T-cell lymphoma (CTCL) is a clonal lymphoproliferative malignancy primarily involving the skin. Mycosis fungoides (MF) and the leukemic variant called Sézary syndrome (SS) are the most frequent clinical types of CTCL. Patients with MF usually develop cutaneous patches and plaques and have an indolent course with a 5-year survival of approximately 87%.1-3 However, in a significant number of patients the disease may progress, and disseminated extracutaneous manifestations may develop, involving the lymph nodes, blood, and visceral organs such as the lung, spleen, and liver.4,5 The prognosis for patients with widespread manifestation of CTCL beyond the skin is poor with a 5-year survival rate of nearly 40%.6-8 Therefore, accurate evaluation in each individual case is crucial for an adequate, stage-adapted therapeutic approach.
Determining the status of peripheral lymph nodes, generally the first site of extracutaneous dissemination is particularly important for clinical staging of patients with CTCL.9-11 As a rule, patients with enlarged lymph nodes undergo lymph node biopsy; however, the neoplastic character of lymph node involvement is difficult to assess by histologic examination alone. Lymph nodes removed from patients with CTCL often show “dermatopathic lymphadenopathy,” a type of reactive lymphoid hyperplasia with expansion of the paracortical T-cell domain by melanin-containing macrophages and variable numbers of atypical lymphoid cells.12-15 The presence of palpable lymphadenopathy is thought to be associated with an unfavorable prognosis, regardless of whether the examined lymph node reveals only dermatopathic changes or clear lymphoma.16-18 In fact, it seems that many dermatopathic lymph nodes in CTCL may contain lymphoma cells, but their small number is obscured by superimposed reactive dermatopathic changes.
Recent developments in molecular techniques have provided the means for detecting monoclonal T-cell populations based on the detection of the rearranged T-cell antigen receptor (TCR) genes. Such techniques have been applied to support the diagnosis of either nodal or extranodal, eg, cutaneous T-cell lymphoma.19,20 Detection of a dominant T-cell clone in dermatopathic lymph nodes in MF by Southern blotting (SB) has been reported, suggesting that analysis of clonality may play an important role in the diagnosis of lymph node involvement in CTCL.21-26
The aim of this study was to determine the sensitivity, specificity, and predictive prognostic value of T-cell clonality in lymph nodes of patients with CTCL. A cohort of 36 patients was investigated using TCR-β and -γ polymerase chain reaction (PCR) in combination with Genescan analysis and DNA sequencing.27-30
Our findings suggest that TCR rearrangement analysis in lymph nodes of patients with CTCL is an important adjunct to the conventional staging criteria and may identify early lymph node involvement in CTCL. Furthermore, it appears that detection of clonal TCR rearrangements in lymph nodes is associated with an unfavorable prognosis, indicating that TCR rearrangement analyses may be of diagnostic as well as prognostic value in these cases.
Patients, materials, and methods
Study design
From January 1, 1991, to December 31, 2003, 36 patients with CTCL followed in our department of dermatology underwent lymph node biopsy and were included in this study. Informed consent was provided according to the Declaration of Helsinki. Those with mycosis fungoides (n = 29) had a diagnostic skin histology, and a cutaneous T-cell clone was detected by TCR-β or TCR-γ rearrangement studies.27,28 Patients with Sézary syndrome (n = 7) had erythroderma with a compatible skin histology and atypical circulating cells, and a peripheral blood T-cell clone. Diagnosis and staging of patients were completed after a comprehensive history and physical examination, complete blood cell counts including peripheral blood smears for Sézary cells, CD4/CD8 ratio according to Vonderheid et al,31 general chemistry panel including serum lactate dehydrogenase (LDH), chest radiograph, and ultrasound of abdomen and peripheral lymph nodes. Suspected visceral involvement was evaluated using imaging studies or by tissue and bone marrow biopsies when indicated. Only patients with clinically suspicious lymphadenopathy had a surgical lymph node biopsy. Blind lymph node biopsies were not performed.
After initial evaluation, all patients were staged according to tumor-node-metastasis (TNM) categories and an overall staging classification proposed by the MF Cooperative Group and the National Cancer Institute (Table 1).32
Staging system for MF and SS
Stage . | Tumor . | Lymph node . | Metastases . |
|---|---|---|---|
| IA | T1 | NO | M0 |
| IB | T2 | N0 | M0 |
| IIA | T1 or T2 | N1 | M0 |
| IIB | T3 | N0 or N1 | M0 |
| III | T4 | N0 or N1 | M0 |
| IVA | T1-T4 | N2 or N3 | M0 |
| IVB | T1-T4 | N0-N3 | M1 |
Stage . | Tumor . | Lymph node . | Metastases . |
|---|---|---|---|
| IA | T1 | NO | M0 |
| IB | T2 | N0 | M0 |
| IIA | T1 or T2 | N1 | M0 |
| IIB | T3 | N0 or N1 | M0 |
| III | T4 | N0 or N1 | M0 |
| IVA | T1-T4 | N2 or N3 | M0 |
| IVB | T1-T4 | N0-N3 | M1 |
T1 indicates patch/plaque = 10% of body surface; T2, patch/plaque = 10% of body surface; T3, skin tumor(s); T4, erythroderma; N0, normal nodes; N1, palpable nodes without histologic evidence of lymphoma; N2, no palpable nodes but histologic evidence of lymphoma; N3, palpable nodes with histologic evidence of lymphoma; M0, no visceral involvement; M1, histologically confirmed visceral involvement. This staging system was devised by Bunn and Lamberg in 1979.32
The purpose of this study was to define sensitivity, specificity, and any prognostic value of the analysis of T-cell clonality in lymph nodes suspected to be involved in CTCL. Clonality was analyzed without knowledge of the histopathologic diagnosis, and vice versa. Furthermore, 12 lymph node specimens of dermatopathic lymphadenopathy from patients with benign inflammatory dermatoses such as generalized atopic dermatitis, erythrodermic psoriasis, and generalized drug eruption were studied for comparison. None of these patients had developed malignant lymphoma during a follow-up period of 3 years.
Histologic studies
All lymph nodes were examined by an experienced hematopathologist (H.S.) using hematoxylin and eosin staining and immunohistologic stainings. The morphologic and immunohistologic criteria as outlined in the World Health Organization classification of lymphoid neoplasm were applied for diagnosis.33 Multiple sections were examined (Olympus AX70; camera: JVC/KXF-70; software: Diskus Version 4.20.35; Hilgers/Germany) to avoid sampling errors in the histopathologic staging. Sections (4 μm) of paraffin-embedded tissue specimens were immunostained using the immunoalkaline phosphatase–anti-alkaline phosphatase (APAAP) method.34 The primary antibodies were directed against T-cell receptor β-chain (clone βF1), CD3 (polyclonal CD3), CD4 (1F6), CD8 (C8-144), CD45RO (OPD4), and CD30 (Ber-H2). With the exception of βF1, which was purchased from T-Cell Sciences, (Cambridge, MA) and CD4 from Novocastra (Newcastle upon Tyne, United Kingdom), all other antibodies were purchased from DAKO (Glostrup, Denmark). To unmask antigen epitopes all primary antibodies were applied after high-pressure–cooking pretreatment of the sections in 10-mmol/L citrate buffer, pH 6.
PCR methodology
This study only examined specimens already acquired for routine diagnosis. PCR was done with DNA extracted from 25-μm paraffin sections after dewaxing and proteinase K digestion using a QIAGEN DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. To validate the molecular data, each skin and lymph node sample was investigated twice using the recently developed TCR-β PCR and an established TCR-γ PCR in combination with Genescan analysis and DNA sequencing.
Detailed techniques for the detection of rearranged TCR-β and -γ chain genes are published elsewhere.27-29 In brief, to detect rearranged TCR-β chain genes, a seminested PCR using family-specific Jβ-primers (JβFS1A and JβFS2A) for the first amplification and nested family-specific Jβ-primers (JβFS1 and JβFS2) for reamplification were applied in conjunction with a Vβ consensus primer.27 Seminested PCR was also applied for the amplification of rearranged TCR-γ chain genes using nested Vγ-primers in combination with 2 primers covering most Jγ-segments.28,29
The sensitivity of the TCR-γ and TCR-β PCR are described elsewhere.27,28 In these experiments, serial dilution experiments of DNA from T-cell lines (T-cell lines Hut, Molt 4) in tonsillar DNA (equivalent to lymph node DNA) have been performed and revealed a detection rate of 0.5% to 1% of clonally rearranged T-cells in a polyclonal background (0.5 ng to 1 ng T-cell line DNA in 100 ng tonsillar DNA) detectable after TCR-γ and TCR-β PCR and Genescan analysis. All PCR assays were performed for each case in duplicate, and only those cases with reproducible results were regarded as clonal.
Analysis of clonality
For CDR3 (complementarity-determining region 3) length diversity analysis, fluorescence-labeled PCR products were run on an ABI 310 genetic analyzer (Applied Biosystems, Weiterstadt, Germany). Vβ and Vγ primers of the reamplification were replaced by primers of the same sequence labeled at their 5′-end with 5-carboxyfluorescein (FAM). Aliquots of PCR products (1-2 μL) were separated on sequencing gel, and the results were analyzed using GENESCAN 672 software (Applied Biosystems).
Samples with 1 or 2 (biallelic) reproducible dominant peaks with or without polyclonal background (Figure 1A,C,D; Figure 2) were regarded as clonal and the different clonal populations were characterized by their different CDR3 size. Samples with multiple dominant peaks (oligoclonal) or samples with a polyclonal repertoire were regarded as nonclonal (Figure 1B). In all samples, the TCR rearrangement pattern of diagnostic skin biopsies was compared with that in corresponding lymph nodes. Two dominant T-cell populations in 1 patient were considered to be identical when they displayed the same CDR size. For confirmation, dominant PCR products were analyzed by DNA sequencing.
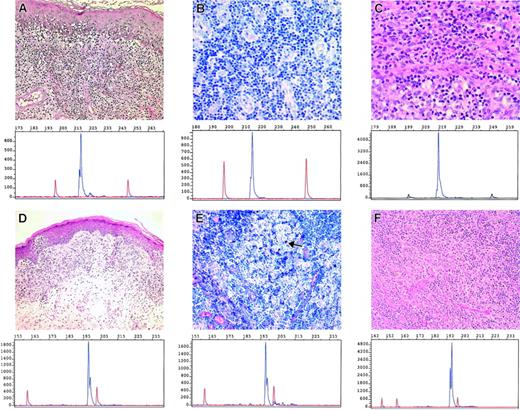
Correlation of TCR-γ PCR results and histologic evaluation in skin and corresponding lymph node. (A) This panel represents the skin lesion (MF) and (B) the dermatopathic lymph node of the same patient (no. 5) together with the corresponding PCR results after Genescan analysis. Whereas the skin lesion displays a clear clonal T-cell population (223 base pairs; bp), no clonally rearranged T cells were found in the dermatopathic lymph node (→ macrophages containing melanin). (C-D) These panels demonstrate the histology and PCR results (Genescan analysis) obtained from the skin (MF with typical Pautrier microabscess; C) and effaced lymph node (D) of patient no. 35. In contrast to patient no. 5, the same clonal T-cell population was detectable in both lesions (C and D, 200 bp). A,C: × 200. B,D: × 100.
Correlation of TCR-γ PCR results and histologic evaluation in skin and corresponding lymph node. (A) This panel represents the skin lesion (MF) and (B) the dermatopathic lymph node of the same patient (no. 5) together with the corresponding PCR results after Genescan analysis. Whereas the skin lesion displays a clear clonal T-cell population (223 base pairs; bp), no clonally rearranged T cells were found in the dermatopathic lymph node (→ macrophages containing melanin). (C-D) These panels demonstrate the histology and PCR results (Genescan analysis) obtained from the skin (MF with typical Pautrier microabscess; C) and effaced lymph node (D) of patient no. 35. In contrast to patient no. 5, the same clonal T-cell population was detectable in both lesions (C and D, 200 bp). A,C: × 200. B,D: × 100.
Correlation of TCR-γ PCR results in skin and dermatopathic and effaced lymph nodes. (A-C) Resembles skin and lymph node results of patient no. 7 and (D-F) of patient no. 12, respectively (A,D: skin; B,E: dermatopathic lymph node; C,F: effaced lymph node). Panels A and D show the typical MF histology with epidermotropic atypical lymphocytes present in the skin (with additional Pautrier abscess) in conjunction with the corresponding clonal T-cell populations as demonstrated by TCR-γ PCR and Genescan analysis. The same clonal T-cell populations were also found in the dermatopathic (B and E, → macrophages containing melanin) and effaced lymph node (C, F). Panels A to C (no. 7) revealed an identical peak size of 217 bp, and panels D to F (no. 12) revealed an identical peak size of 195 bp. A,D: × 200. B,C,E,F: × 100.
Correlation of TCR-γ PCR results in skin and dermatopathic and effaced lymph nodes. (A-C) Resembles skin and lymph node results of patient no. 7 and (D-F) of patient no. 12, respectively (A,D: skin; B,E: dermatopathic lymph node; C,F: effaced lymph node). Panels A and D show the typical MF histology with epidermotropic atypical lymphocytes present in the skin (with additional Pautrier abscess) in conjunction with the corresponding clonal T-cell populations as demonstrated by TCR-γ PCR and Genescan analysis. The same clonal T-cell populations were also found in the dermatopathic (B and E, → macrophages containing melanin) and effaced lymph node (C, F). Panels A to C (no. 7) revealed an identical peak size of 217 bp, and panels D to F (no. 12) revealed an identical peak size of 195 bp. A,D: × 200. B,C,E,F: × 100.
For DNA sequence analysis, dominant unlabeled PCR products were extracted from 6% polyacrylamide gel by elution. The Big Dye Terminator cycle sequencing reaction kit (Applied Biosystems) was used prior to electrophoresis to an automated sequencer (ABI 310; Applied Biosystems). The resulting sequences were aligned with the T-cell receptor sequences from the international ImmunoGeneTics database (http://imgt.cines.fr/cgi-bin/IMGTjcta.jv?livret=0), and the CDR3 region was identified.35
Assessment of clinical outcome
The 36 patients with CTCL included in the study were classified according to the course of the disease: complete remission, partial remission or stable disease, progressive disease, and exitus letalis due to lymphoma or causes unrelated to CTCL. Complete remission was defined as no evidence of residual lymphoma for at least 6 months; stable disease was defined as presence of lymphoma without progression, and change to a more advanced stage was seen as progressive lymphoma. Information on the causes of death was taken from the death certificate. The total follow-up period was calculated from the time of first diagnosis to the last presentation or death of the patient (data compiled in December 2003). The clinical outcome in each patient was recorded in months of survival after first diagnosis.
Statistical analysis
Statistical analyses were done in cooperation with our Institute of Medical Informatics, Biometrics, and Clinical Epidemiology. Survival time was calculated from the on-study date until death from CTCL-related disease or last follow-up. The Kaplan-Meier method was used to calculate the probability of survival as a function of time (Figure 3). The log-rank test was used to determine the significant differences between the survival curves. All data were analyzed with the Statistical Package for the Social Sciences (SPSS Science, Chicago, IL).36 In all analyses, the cut-off level of statistical significance was set at .05.
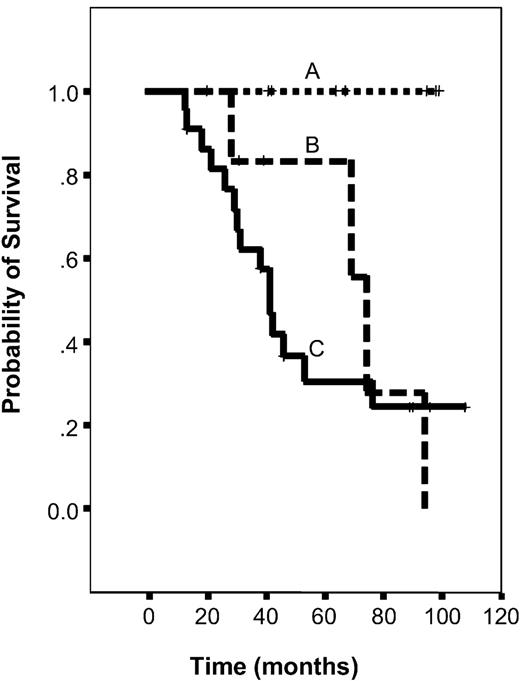
Graph showing Kaplan-Meier survival curves, given by stage of lymph node involvement in CTCL. Comparison of Kaplan-Meier survival estimates in patients with CTCL with different stages of lymph node involvement analyzed by histology and PCR (A-C). (A) Patients with no evidence of a lymph node manifestation of lymphoma by histology and PCR analysis. (B) Patients with no histologic lymph node involvement but with detection of a clonal TCR-rearrangement within the lymph node. (C) Patients with both histologic lymph node manifestation and detection of a clonal T-cell population.
Graph showing Kaplan-Meier survival curves, given by stage of lymph node involvement in CTCL. Comparison of Kaplan-Meier survival estimates in patients with CTCL with different stages of lymph node involvement analyzed by histology and PCR (A-C). (A) Patients with no evidence of a lymph node manifestation of lymphoma by histology and PCR analysis. (B) Patients with no histologic lymph node involvement but with detection of a clonal TCR-rearrangement within the lymph node. (C) Patients with both histologic lymph node manifestation and detection of a clonal T-cell population.
Results
Patients' characteristics
We included 36 patients with CTCL with suspected lymph node involvement in this study. Table 2 shows the clinical characteristics and disease stage identified by routine strategies without taking into account PCR results.
Relation between clinical characteristics and TCR-β and γ PCR results
Patient no. . | Age at diagnosis, y . | Sex . | Stage . | TCR-β skin . | TCR-β LN . | TCR-γ skin . | TCR-γ LN . | Follow-up, mo . | Course . | Status at last follow-up . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 33 | M | IIa | + | - | + | - | 42 | CR | Alive |
| 2 | 76 | F | IIa | + | - | - | - | 98 | CR | Alive |
| 3 | 59 | M | IIa | + | - | + | - | 67 | SD | Alive |
| 4 | 54 | F | IIa | + | - | + | - | 64 | CR | Alive |
| 5 | 57 | M | IIa | + | - | + | - | 41 | SD | Alive |
| 6 | 74 | M | IIb | + | - | + | - | 95 | CR | Alive |
| 7 | 81 | F | IIb | + | + | + | + | 31 | PD, IV | Alive |
| 8 | 52 | M | IIb | + | + | - | - | 28 | PD, IV | DOD |
| 9 | 72 | M | IIb | + | + | + | + | 20 | CR | D |
| 10 | 51 | F | IIb | + | + | + | + | 94 | PD, IV | DOD |
| 11 | 63 | M | III | + | + | + | + | 69 | PD, IV | DOD |
| 12 | 64 | M | III | + | + | + | + | 39 | PD, IV* | Alive |
| 13 | 77 | F | III | + | - | + | - | 99 | CR | Alive |
| 14 | 87 | F | III | + | + | + | + | 74 | PD, IV | DOD |
| 15 | 62 | M | IV | + | + | + | + | 26 | PD | DOD |
| 16 | 45 | F | IV | + | + | + | + | 96 | SD | Alive |
| 17 | 59 | M | IV | + | + | - | - | 46 | SD | D |
| 18 | 54 | M | IV | + | + | + | + | 42 | PD | DOD |
| 19 | 62 | M | IV | + | + | + | + | 30 | PD | DOD |
| 20 | 59 | F | IV | + | + | + | + | 13 | SD | Alive |
| 21 | 71 | M | IV | + | + | + | + | 108 | PD | D |
| 22 | 62 | F | IV | + | + | + | + | 76 | PD | DOD |
| 23 | 57 | M | IV | + | + | + | + | 41 | PD | DOD |
| 24 | 62 | F | IV | + | + | + | + | 90 | SD | Alive |
| 25 | 60 | F | IV | + | + | + | + | 46 | PD | DOD |
| 26 | 58 | M | IV | + | + | + | + | 21 | PD | DOD |
| 27 | 44 | F | IV | + | + | + | + | 89 | PD, IV* | Alive |
| 28 | 64 | F | IV | + | + | + | + | 41 | PD | DOD |
| 29 | 73 | M | IV | + | + | + | + | 38 | PD | DOD |
| 30 | 51 | F | IV* | + | + | + | + | 53 | PD | DOD |
| 31 | 75 | F | IV* | + | + | + | + | 12 | PD | DOD |
| 32 | 72 | M | IV* | + | + | + | + | 31 | PD | DOD |
| 33 | 43 | M | IV* | + | + | + | + | 38 | SD | Alive |
| 34 | 62 | M | IV* | + | + | + | + | 13 | PD | DOD |
| 35 | 59 | M | IV* | + | + | + | + | 18 | PD | DOD |
| 36 | 73 | M | IV* | + | + | + | + | 29 | PD | DOD |
Patient no. . | Age at diagnosis, y . | Sex . | Stage . | TCR-β skin . | TCR-β LN . | TCR-γ skin . | TCR-γ LN . | Follow-up, mo . | Course . | Status at last follow-up . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 33 | M | IIa | + | - | + | - | 42 | CR | Alive |
| 2 | 76 | F | IIa | + | - | - | - | 98 | CR | Alive |
| 3 | 59 | M | IIa | + | - | + | - | 67 | SD | Alive |
| 4 | 54 | F | IIa | + | - | + | - | 64 | CR | Alive |
| 5 | 57 | M | IIa | + | - | + | - | 41 | SD | Alive |
| 6 | 74 | M | IIb | + | - | + | - | 95 | CR | Alive |
| 7 | 81 | F | IIb | + | + | + | + | 31 | PD, IV | Alive |
| 8 | 52 | M | IIb | + | + | - | - | 28 | PD, IV | DOD |
| 9 | 72 | M | IIb | + | + | + | + | 20 | CR | D |
| 10 | 51 | F | IIb | + | + | + | + | 94 | PD, IV | DOD |
| 11 | 63 | M | III | + | + | + | + | 69 | PD, IV | DOD |
| 12 | 64 | M | III | + | + | + | + | 39 | PD, IV* | Alive |
| 13 | 77 | F | III | + | - | + | - | 99 | CR | Alive |
| 14 | 87 | F | III | + | + | + | + | 74 | PD, IV | DOD |
| 15 | 62 | M | IV | + | + | + | + | 26 | PD | DOD |
| 16 | 45 | F | IV | + | + | + | + | 96 | SD | Alive |
| 17 | 59 | M | IV | + | + | - | - | 46 | SD | D |
| 18 | 54 | M | IV | + | + | + | + | 42 | PD | DOD |
| 19 | 62 | M | IV | + | + | + | + | 30 | PD | DOD |
| 20 | 59 | F | IV | + | + | + | + | 13 | SD | Alive |
| 21 | 71 | M | IV | + | + | + | + | 108 | PD | D |
| 22 | 62 | F | IV | + | + | + | + | 76 | PD | DOD |
| 23 | 57 | M | IV | + | + | + | + | 41 | PD | DOD |
| 24 | 62 | F | IV | + | + | + | + | 90 | SD | Alive |
| 25 | 60 | F | IV | + | + | + | + | 46 | PD | DOD |
| 26 | 58 | M | IV | + | + | + | + | 21 | PD | DOD |
| 27 | 44 | F | IV | + | + | + | + | 89 | PD, IV* | Alive |
| 28 | 64 | F | IV | + | + | + | + | 41 | PD | DOD |
| 29 | 73 | M | IV | + | + | + | + | 38 | PD | DOD |
| 30 | 51 | F | IV* | + | + | + | + | 53 | PD | DOD |
| 31 | 75 | F | IV* | + | + | + | + | 12 | PD | DOD |
| 32 | 72 | M | IV* | + | + | + | + | 31 | PD | DOD |
| 33 | 43 | M | IV* | + | + | + | + | 38 | SD | Alive |
| 34 | 62 | M | IV* | + | + | + | + | 13 | PD | DOD |
| 35 | 59 | M | IV* | + | + | + | + | 18 | PD | DOD |
| 36 | 73 | M | IV* | + | + | + | + | 29 | PD | DOD |
+ indicates monoclonal; — not clonal; PD, progressive disease; SD, stable disease; CR, complete remission; D, died unrelated to disease; DOD, died of disease.
Patients with Sézary syndrome.
The study included 22 men and 14 women, ranging in age from 33 to 87 years (mean, 62 years). On the basis of TNM staging, 14 patients (39%) presented with patch or plaque-type lesions, cutaneous tumors, or erythroderma (stages IIa, IIb, III), while 22 patients (61%) also had histologically confirmed lymph node or visceral involvement (stage IV). The length of clinical follow-up ranged from 12 months to 9 years. The clinical outcome of patients at the last follow-up was as follows: 5 patients were in complete remission, 6 had partial remission or stable disease, 3 had progressive lymphoma, 19 had died of lymphoma, and 3 had died of causes unrelated to CTCL (Table 3).
Clinical outcome in relation to lymph node histology and TCR analysis
Course of disease . | Patients, no. . | Histology (N3), no. . | Clonal TCR-β-R, no. . | Clonal TCR-γ-R, no. . |
|---|---|---|---|---|
| CR | 5 | 0 | 0 | 0 |
| SD | 6 | 3 | 4 | 4 |
| PD | 3 | 1 | 3 | 3 |
| DOD | 19 | 15 | 19 | 18 |
| D | 3 | 3 | 2 | 2 |
| Total | 36 | 21 | 29 | 27 |
Course of disease . | Patients, no. . | Histology (N3), no. . | Clonal TCR-β-R, no. . | Clonal TCR-γ-R, no. . |
|---|---|---|---|---|
| CR | 5 | 0 | 0 | 0 |
| SD | 6 | 3 | 4 | 4 |
| PD | 3 | 1 | 3 | 3 |
| DOD | 19 | 15 | 19 | 18 |
| D | 3 | 3 | 2 | 2 |
| Total | 36 | 21 | 29 | 27 |
CR indicates complete remission; SD, stable disease; PD, progressive disease; DOD, died of disease; D, died, unrelated to disease; TCR-β-R, TCR-β rearrangement; TCR-γ-R, TCR-γ rearrangement; N3, lymph node involvement.
Identical clonal T-cell populations in lymph node and skin lesions
TCR rearrangement analysis of tumor skin samples by PCR enabled us to define the malignant T-cell clone by size of the PCR-product (Genescan analysis) and the DNA sequence. A cutaneous T-cell clone was found with TCR-β PCR in representative skin samples of 36 of 36 cases, and with TCR-γ PCR in 33 of 36 cases. In addition, the clonal nature of the dominant peaks was confirmed by CDR3 nucleotide sequencing as shown in Table 4.
Junctional sequences of the clonal TCR-β and TCR-γ rearrangements
Case . | 3′V sequence . | N-(D)-N sequence . | 5′J sequence . | V (D) J sequence . |
|---|---|---|---|---|
| 5 | TGTGCCACCTGGGACGG | TATAGTAGTGATTGGC | TCAAGACGTTT | Vγ2.1 JPγ2.1 |
| 7 | TCAAGACGTTT | CC | TTATTATAAGAAACTCTTT | Vγ4.2 Jγ2 |
| 11 | TTCTACATCTGCAGTGCTAG | GCCAGGACAATGCGACCGC | GAGACCCAGTACTTCGGG | Vβ29.1 Dβ1 Jβ2.5 |
| 12 S | TGTGCCACCTGGGATA | A | AAACTCTTT | Vγ8.1 Jγ2.1 |
| 12 LN | TGTGCCACCTGGGATA | A | AAACTCTTT | Vγ8.1 Jγ2.1 |
| 15 | TGCGCCAGCAGT | TTACTAAAACCAC | CTAACTATGGCTACACC | Vβ10.1 Dβ 2 Jβ1.2 |
| 16 | TTGATTTACTTC | CAGGGTAACAGT | CAACTAGACAAA | Vβ7.2 Dβ2 Jβ1.3 |
| 17 | TGTGCCAGTCA | TAAAGGGAGGGT | CTGATCTCTACG | Vβ30.4 Dβ2 Jβ2.1 |
| 18 | CAGTA | TTAGGCTAGCGGG | CCTACGACAGTACTTCGG | Vβ29.1 Dβ2 Jβ2.7 |
| 19 | TGTGCCAGTAGT | TCGGC | AGCAATCAGCCCCAGCATT | Vβ7.2 Dβ1 Jβ1.5 |
| 20 | TGTGCCACCTGGGATGGG | C | TATAAGAAACTCTTT | Vγ4.2 Jγ2.1 |
| 23 | TGTGCCAGCAGC | CCGCTACAGGGA | TCCTACGAGCAGTACTTCG | Vβ7.9 Dβ2 Jβ2.7 |
| 25* | TGTGATCTTGCTGAAGGAAGTA | GACTAAGCTCATAGTAACT | Vγ4.1 JPγ1.1 | |
| 25* | TGTGCCACCTGGGATAGG | ATTATTATAAGAAACTCTTT | Vγ8.1 Jγ2.1 | |
| 26 S | TGTGCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACCAA | Vβ5.4 Dβ1 Jβ2.7 |
| 26 LN | TGTGCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACCAA | Vβ5.4 Dβ1 Jβ2.7 |
| 27 | TGTGATCTTGCTGAAGGAAGTA | ATTATTATAAGAAACTCT | Vγ4.1 J γ2.1 | |
| 28 | CAGCTT | CGACAGGGC | CAGATACGCAGTATTTTGG | Vβ13.2 Dβ1 Jβ2.3 |
| 30 | TTCTACATCTGCAGTGCTAGAG | CTACGGCGGTAGCCTAT | GAGACCCAGTACTCGGGC | Vβ30.5 Dβ2 Jβ2.5 |
| 31 | AGTGCTGACTAGCGG | GGCTGGGGG | AGACCCAGTACTTCC | Vβ20.1 Dβ2 Jβ2-5 |
| 32 | AGCACC | AGTATCGCTAGCTCA | ACAATGAGCAGTTCTTCGG | Vβ30.4 Dβ2 Jβ2.1 |
| 33 S | TGTGCCACCTGGGATA | AGGAC | TATTATAAGAAACTCTTT | V γ8.1 J γ2.1 |
| 33 LN | TGTGCCACCTGGGA | AGGAC | TATTATAAGAAACTCTTT | Vγ8.1 J γ2.1 |
| 35 | TGTGCCACC | CC | TTATTATAAGAAACTCTTT | V γ4.1 J γ2.1 |
| MOLT† | TTATTATAAGAAACTCTTT | AGAGTCGACTAGCGATCCAAAA | AATGAGCAGTTCTTCGGG | Vβ20.1 Dβ2 J β2.1 |
| HUT† | TCTGCAGCGT | AGGCAGGAGGCGGTTGGC | GGCTACACCTTCGGTTC | Vβ30 Dβ2 J β1.2 |
Case . | 3′V sequence . | N-(D)-N sequence . | 5′J sequence . | V (D) J sequence . |
|---|---|---|---|---|
| 5 | TGTGCCACCTGGGACGG | TATAGTAGTGATTGGC | TCAAGACGTTT | Vγ2.1 JPγ2.1 |
| 7 | TCAAGACGTTT | CC | TTATTATAAGAAACTCTTT | Vγ4.2 Jγ2 |
| 11 | TTCTACATCTGCAGTGCTAG | GCCAGGACAATGCGACCGC | GAGACCCAGTACTTCGGG | Vβ29.1 Dβ1 Jβ2.5 |
| 12 S | TGTGCCACCTGGGATA | A | AAACTCTTT | Vγ8.1 Jγ2.1 |
| 12 LN | TGTGCCACCTGGGATA | A | AAACTCTTT | Vγ8.1 Jγ2.1 |
| 15 | TGCGCCAGCAGT | TTACTAAAACCAC | CTAACTATGGCTACACC | Vβ10.1 Dβ 2 Jβ1.2 |
| 16 | TTGATTTACTTC | CAGGGTAACAGT | CAACTAGACAAA | Vβ7.2 Dβ2 Jβ1.3 |
| 17 | TGTGCCAGTCA | TAAAGGGAGGGT | CTGATCTCTACG | Vβ30.4 Dβ2 Jβ2.1 |
| 18 | CAGTA | TTAGGCTAGCGGG | CCTACGACAGTACTTCGG | Vβ29.1 Dβ2 Jβ2.7 |
| 19 | TGTGCCAGTAGT | TCGGC | AGCAATCAGCCCCAGCATT | Vβ7.2 Dβ1 Jβ1.5 |
| 20 | TGTGCCACCTGGGATGGG | C | TATAAGAAACTCTTT | Vγ4.2 Jγ2.1 |
| 23 | TGTGCCAGCAGC | CCGCTACAGGGA | TCCTACGAGCAGTACTTCG | Vβ7.9 Dβ2 Jβ2.7 |
| 25* | TGTGATCTTGCTGAAGGAAGTA | GACTAAGCTCATAGTAACT | Vγ4.1 JPγ1.1 | |
| 25* | TGTGCCACCTGGGATAGG | ATTATTATAAGAAACTCTTT | Vγ8.1 Jγ2.1 | |
| 26 S | TGTGCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACCAA | Vβ5.4 Dβ1 Jβ2.7 |
| 26 LN | TGTGCCAGCAGC | TGGGACAGGGGTAG | CTCCTACGAGCAGTACCAA | Vβ5.4 Dβ1 Jβ2.7 |
| 27 | TGTGATCTTGCTGAAGGAAGTA | ATTATTATAAGAAACTCT | Vγ4.1 J γ2.1 | |
| 28 | CAGCTT | CGACAGGGC | CAGATACGCAGTATTTTGG | Vβ13.2 Dβ1 Jβ2.3 |
| 30 | TTCTACATCTGCAGTGCTAGAG | CTACGGCGGTAGCCTAT | GAGACCCAGTACTCGGGC | Vβ30.5 Dβ2 Jβ2.5 |
| 31 | AGTGCTGACTAGCGG | GGCTGGGGG | AGACCCAGTACTTCC | Vβ20.1 Dβ2 Jβ2-5 |
| 32 | AGCACC | AGTATCGCTAGCTCA | ACAATGAGCAGTTCTTCGG | Vβ30.4 Dβ2 Jβ2.1 |
| 33 S | TGTGCCACCTGGGATA | AGGAC | TATTATAAGAAACTCTTT | V γ8.1 J γ2.1 |
| 33 LN | TGTGCCACCTGGGA | AGGAC | TATTATAAGAAACTCTTT | Vγ8.1 J γ2.1 |
| 35 | TGTGCCACC | CC | TTATTATAAGAAACTCTTT | V γ4.1 J γ2.1 |
| MOLT† | TTATTATAAGAAACTCTTT | AGAGTCGACTAGCGATCCAAAA | AATGAGCAGTTCTTCGGG | Vβ20.1 Dβ2 J β2.1 |
| HUT† | TCTGCAGCGT | AGGCAGGAGGCGGTTGGC | GGCTACACCTTCGGTTC | Vβ30 Dβ2 J β1.2 |
S indicates DNA obtained from skin biopsy; LN, DNA obtained from lymph node biopsy.
Blank cells indicate no identifiable/truncated junctional region.
Biallelic rearrangement.
T-cell lines as positive controls.
After identifying the tumor clone in skin, we analyzed the corresponding lymph node specimen from each patient for the presence of clonal T-cell populations. Investigating at least 1 corresponding palpable lymph node during the disease course led to the detection of clonally rearranged T-cell populations in 29 (81%) of 36 lymph node biopsies by TCR-β and in 27 (75%) of 36 by TCR-γ PCR. The CDR3 size of the clonal T-cell population was determined by high-resolution Genescan analysis, which identifies PCR products that differ by only 1 base pair (bp) in length. Comparing the dominant peaks of both compartments, skin and corresponding lymph node demonstrated an identical CDR3 size of the clonal T-cell population in all cases, indicating the presence of the same clonal T-cell population (Figures 1C-D and 2). A clonal heterogeneity (different CDR size and sequence of the clonal T-cell population in different lesions) could not be detected in our patients.
Detection of clonal T-cell population in the lymph node correlates with histologic lymphoma manifestation
Lymphoma manifestation was demonstrated by conventional techniques in 22 (61%) of 36 palpable lymph nodes showing small or large clusters of atypical lymphocytes with convoluted nuclei and partial or total replacement of the lymph node architecture (Figures 1C and 2C,F).
TCR-β and TCR-γ PCR detected T-cell clonality in 22 of 22 and 21 of 22 nodes, respectively, indicating a high correlation between the T-cell clone and nodal lymphoma infiltration, as diagnosed by routine histology and immunohistochemical techniques (Cramer coefficient of correlations = 1 and 0.96, respectively). According to the histology the specificity and sensitivity of the TCR, PCR-analysis is estimated at 60% and 96.2%, respectively. Both the positive and negative predictive values of the analysis of lymph node T-cell clonality by PCR for diagnosis of lymphoma were high, estimated at 86%.
Detection of clonal T-cell population in dermatopathic lymph nodes
In 14 (39%) of 36 cases, dermatopathic changes were found by histology or immunohistology in the examined lymph nodes, particularly in the T-cell zones, however without clear evidence of lymphoma (Figures 1B and 2B,E). Analyzing clonality in these dermatopathic lymph nodes revealed a T-cell clone in 7 (50%) of 14 of the examined specimens by TCR-β and in 6 (43%) of 14 by TCR-γ PCR. Interestingly, a T-cell clone could not be detected in patients in the patch or plaque stage (0 of 5, stage IIa), whereas in 7 (78%) of 9 and 6 (67%) of 9 of the patients in the tumor (stage IIb) or erythrodermic stage (III), a clonal T-cell population was found by TCR-β and TCR-γ PCR. Further in 5 of 7 patients, who initially had only dermatopathic lymphadenopathy, lymph node specimens taken due to clinical progression clearly demonstrated lymphoma. Clonality analysis of these lymph nodes revealed a clonal T-cell population identical to that detected in the corresponding “dermatopathic stage” by TCR-β and TCR-γ PCR.
In 12 dermatopathic lymph nodes of control patients with inflammatory skin diseases, TCR-β and TCR-γ PCR showed a polyclonal or oligoclonal rearrangement pattern by Genescan analysis.
Lymph node clonality confirms and extends clinical classification
The results from the TCR-β and TCR-γ analyses were compared with the clinical stage of the patients at the time of lymph node biopsy. Patients classified as being in early stages of CTCL (IIa) did not show clonality in their lymph nodes (0 of 5). However, in patients with tumor or erythroderma stages (IIb and III), clonal T-cell populations were found in 78% and 67% by TCR-β and TCR-γ PCR: 4 of 5 cases in stage IIb and 3 of 4 cases in stage III. Since stage IV patients, including 7 with Sézary syndrome, had clonal TCR rearrangements in 22 of 22 and 21 of 22 of the cases, it seems that clonality confirmed the routine clinical staging here. Regarding the clinical outcome, patients without lymph node T-cell clonality had a better clinical course or died of causes unrelated to CTCL, while those with lymph node T-cell clonality had a poor clinical outcome. Six of 7 patients with a clonal T-cell population in their dermatopathic lymph nodes experienced a disease progression. Of these, 3 patients with stage IIb (nos. 7, 8, 10) and 2 patients with stage III (nos. 11, 14) developed histologically overt lymph node infiltration (stage IV), and 1 patient with stage III (no. 12) developed a Sézary syndrome (IVb) (Tables 2 and 3).
Lymph node clonality as an indicator of unfavorable prognosis
Thirty-six patients were divided into 3 groups: 1) patients with no evidence of lymph node involvement by routine histology (N1) and PCR analysis (not clonal), 2) patients without histologic lymph node involvement (N1) but with detection of a clonal T-cell population by PCR (clonal), and 3) patients with lymph node involvement by routine histology (N3) and detection of clonal TCR-rearrangement.
Patients in the first group (N1/not clonal) had a good clinical outcome (71% CR; 29% SD). All 7 patients were alive at the last follow-up. In contrast, only 2 of 7 patients in the second group (N1/clonal) were alive at that time (14% CR, 86% PD; median probability of survival of 74 months; confidence interval, 66-82 months). The difference between the 2 groups was significant (P = .0131, log-rank test).
Patients in the third group (N3/clonal) had a poor clinical outcome (SD 23%, PD 77%), only 5 of 22 patients were alive at the last follow-up. The median survival probability of these patients was 41 months (confidence interval, 35-47 months). This value differed from the median survival probability of the second group (N1/clonal, 74 months), but the difference was not significant (P = .6073, log-rank test) (Figure 3).
Discussion
The first extracutaneous manifestation in CTCL is usually the involvement of lymph nodes and appears clearly relevant for selecting the therapeutic schedule, the response of the patient, and their overall survival.37,38 Precise assessment of lymph node involvement in CTCL is difficult. Although the National Cancer Institute's histopathologic grading system, which classifies lymph nodes (LN0 to LN4) on the basis of an increasing number of atypical cells, appears to be clinically useful since it correlates with prognosis, it does not have a distinct cutoff for clearly differentiating between specific involvement and dermatopathic changes.39,40
This study has shown that lymph node involvement can be more objectively assessed by analyzing the rearrangement of T-cell antigen receptor genes in histologically defined lymph nodes. Patients with CTCL with palpable lymph nodes were investigated to assess both the feasibility of T-cell clone analysis in routine practice and its diagnostic value. Clonal T-cell populations could be detected in 81% and 75% of clinically suspected lymph nodes from patients with CTCL examined by TCR-β and TCR-γ PCR, respectively, and the tumor clone in the corresponding skin lesions was defined by CDR3 size determination and DNA sequencing at initial diagnosis. The dominant T-cell clones detected in skin and lymph node biopsies and assessed by Genescan analysis were identical in all cases, suggesting that dominant T-cell clones found in lymph nodes were derived from cutaneous tumor cells.
A lymph node–dominant T-cell population detected in 22 of 22 by TCR-β PCR and in 21 of 22 by TCR-γ PCR in lymph node biopsies containing histologically unambiguous CTCL demonstrated a strong correlation between routine histology or immunohistology and PCR analyses in these cases. In all cases the clonal T-cell population detected in lymph node was identical to the tumor cell clone in the skin. Overall, there was no significant difference between TCR-β and TCR-γ PCR based on the clonality detection rate. The slightly lower detection rate of TCR-γ PCR may be explained by T-cell clones having incomplete or deleterious rearrangements of the TCR-γ gene preventing primer binding to the target sequences.41 In contrast, most malignant and reactive T-cell populations express the α/β TCR protein, indicating a functional rearrangement of the TCR-β gene.42
In an attempt to detect early involvement of lymph nodes in patients with CTCL, we also investigated T-cell clonality in lymph nodes histologically assessed as dermatopathic. Clonal T-cell populations identical to those in skin were found in 50% of the specimens investigated by both TCR-β and -γ PCR, thus suggesting early CTCL involvement. Clonality could not be detected in the lymph nodes of patients with early CTCL stages IIa, but in 75% of the more advanced stages IIb and III, showing tumors or erythroderma. The fact that detectable T-cell clonality is not associated with dermatopathic lymphadenopathy itself was confirmed by examining 12 dermatopathic lymph nodes from patients with inflammatory skin disorders.
An important objective of our molecular staging was to determine the clinically relevant sensitivity threshold for detecting histopathologic occult CTCL. The clinical course observed in patients with or without clonal T-cells in their lymph nodes suggests that the presence of clonal T-cell populations may be a predictor for adverse outcome and reduced survival probability, indicating the clinical relevance of these clonality analyses. Noteworthy is that none of the patients without a T-cell clone developed progressive lymphoma during a median follow-up of 8 years. In contrast, 6 of 7 patients with clonal TCR rearrangement but histologically negative lymph nodes developed progressive lymphoma, and 4 of them died of lymphoma. These results clearly demonstrate that the detection of the identical T-cell clone in the skin and the lymph node represents not a physiologic recirculation of T-cells but reflects a real progression of the disease. Therefore, TCR rearrangement analysis by PCR in lymph nodes provides clinically relevant information beyond that obtainable by routine histopathology alone.
Our data support the few previous studies using Southern blotting to examine T-cell clonality in lymph nodes of patients with CTCL.21-24,43 Using the Southern blot technique, Lynch et al22 found T-cell clonality in 47% of dermatopathic lymph nodes and in 90% of histologically clearly effaced lymph nodes, indicating the correlation between T-cell clonality and palpable lymph nodes. Moreover, a poorer prognosis was associated with clonality detected in patients with dermatopathic lymph nodes. These data were consistent with those reported by Kern et al.24 However, in their population of patients with CTCL, 8 of 22 with a poor clinical outcome revealed no clonal TCR rearrangement by Southern blotting, using DNA extracted from lymph nodes. Our results based on TCR-β and TCR-γ PCR demonstrated a significantly higher detection rate than with Southern blotting, most probably due to the limited sensitivity of genomic Southern blotting (5%-10%). The need for large amounts of high-quality DNA, not available from paraffin-embedded tissue, limits this technique for routine application.44,45
Our results demonstrate that TCR-β and TCR-γ PCR analyses is an important adjunct for early diagnosis of lymph node involvement in CTCL prior to the disturbance of the normal lymph node architecture. The results of this study may also have an important effect on the therapy selected. In patients with dermatopathic lymph nodes and detection of a clonal T-cell population, not only treatment with local skin-directed therapies such as psoralen with UVA irradiation (PUVA) or topical nitrogen mustard should be considered but also the addition of systemic drugs like α-interferon or retinoids.46,47
In conclusion, this is the first study using PCR techniques to detect T-cell clonality in lymph nodes in a large cohort of patients with CTCL based on the clinical and histologic criteria. We could clearly demonstrate that TCR-PCR is highly suitable for early detection of clonal T-cell populations even in histologically uninvolved lymph nodes of patients with CTCL. Since these patients have a clinically worse course very similar to those with effaced lymph node architecture, they could benefit from a more extended treatment. Although our results have to be confirmed in larger prospective studies, they suggest that TCR-β/γ PCR analysis of lymph nodes is an important additional step in achieving accurate clinical staging.
Prepublished online as Blood First Edition Paper, September 30, 2004; DOI 10.1182/blood-2004-06-2220.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank H.-H. Müller from the Institute of Pathology for his excellent technical assistance.
An approval from our review board was not necessary because all samples derived from routine diagnostic procedures (paraffin-embedded material) were analyzed retrospectively and anonymously in this molecular study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal