Abstract
Abnormalities of hemoglobin synthesis are usually inherited but may also arise as a secondary manifestation of another disease, most commonly hematologic neoplasia. Acquired hemoglobin disorders can be seen in any population and are not restricted to areas of the world with high incidences of inherited hemoglobinopathies. In fact, the acquired hemoglobinopathies may be more readily recognized where inherited hemoglobin abnormalities are rare and less likely to cause diagnostic confusion. Acquired α-thalassemia is the best characterized of the acquired red blood cell disorders in patients with hematologic malignancy, and it is almost always associated with a myelodysplastic syndrome (MDS). At least 2 molecular mechanisms for acquired α-thalassemia are now recognized: acquired deletion of the α-globin gene cluster limited to the neoplastic clone and, more commonly, inactivating somatic mutations of the trans-acting chromatin-associated factor ATRX, which cause dramatic down-regulation of α-globin gene expression. Here we review the clinical, hematologic, and molecular genetic features of α-thalassemia arising in a clonal myeloid disorder, and we discuss howATRX might affect gene expression in normal and abnormal hematopoiesis through epigenetic mechanisms.
Introduction
For more than 4 decades, the synthesis of hemoglobin during erythropoiesis has served as an excellent model for understanding mammalian gene regulation. A diverse repertoire of naturally occurring mutations of the globin genes has enabled researchers to use this system to establish many of the general principles underlying human molecular genetics and, in particular, to advance our understanding of molecular hematology.1 Although nearly all globin mutations characterized to date have been cis-acting defects, it has recently emerged that globin synthesis may also be altered by trans-acting mutations involving erythroid-specific and general transcriptional regulators.2-4
It has long been recognized that patients with hematologic malignancy and abnormal erythropoiesis may also acquire changes in hemoglobin structure or synthesis, but the molecular basis of such abnormalities remained obscure. However, recent studies of patients with myelodysplasia who acquire thalassemia have identified somatic mutations in a known trans-acting regulator of globin gene expression, ATRX (α-thalassemia mental retardation X-linked, named after the phenotype associated with germline mutations in the gene).5,6 Although these new observations will undoubtedly increase our understanding of how globin gene expression is normally controlled, they may also point to common genetic or epigenetic mutations that contribute to the development or progression of myelodysplasia.
Normal structure and expression of globin gene clusters
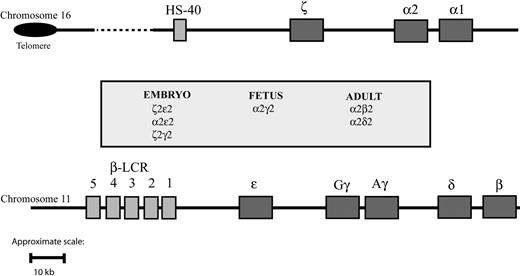
The synthesis of hemoglobin is controlled by 2 developmentally regulated multigene clusters: the α-like globin cluster on chromosome 16 (5′-ζ-α2-α1-3′) and the β-like globin cluster on chromosome 11 (5′-ϵ-Gγ-Aγ-δ-β-3′) (Figure 1). Coordinated expression of the genes in each cluster at all stages of development is dependent on critical regulatory elements located upstream of the genes. In the β-cluster, there are 5 such elements, collectively referred to as the locus control region (LCR), 7 whereas in the α cluster, there is a single known element referred to as HS-40.8,9 Evidence gathered over many years using indirect assays of chromatin structure and function (eg, studies of nuclear position, timing of replication, chromatin modifications, patterns of DNA methylation) indicates that these 2 clusters have translocated to, and evolved in, different chromosomal environments.1,10,11 Despite this, in healthy persons, the synthesis of α- and β-globin chains is finely balanced during terminal erythroid differentiation, giving rise to red blood cells of consistent size (reflected in the mean corpuscular volume [MCV]) and hemoglobin content (mean corpuscular hemoglobin [MCH]). These 2 multigene clusters are among the most intensively studied of all mammalian genes, and current understanding of their genetic and epigenetic regulation has been comprehensively reviewed elsewhere.12
Schematic representation of the human globin gene clusters. Theα-like globin cluster (top) is situated near the telomeric region of the short arm of chromosome 16 and includes the ζ, α2, and α1 globin genes, which are under the control of an upstream remote regulatory region, HS-40 (so-called because it is a DNase I hypersensitive site located approximately 40 kb upstream of the 5′ end of the ζ globin gene). The β-like globin cluster (bottom) is interstitial and located on the short arm of chromosome 11; expression of the genes in this cluster is under the control of a group of remote regulatory elements/DNase I hypersensitive sites collectively known as the locus control region (LCR). Pseudogenes and the θ globin gene, whose functional significance is unknown, are not represented. To make visualization of them easier, the globinlike genes are represented over a larger space of genome than they actually span.
Schematic representation of the human globin gene clusters. Theα-like globin cluster (top) is situated near the telomeric region of the short arm of chromosome 16 and includes the ζ, α2, and α1 globin genes, which are under the control of an upstream remote regulatory region, HS-40 (so-called because it is a DNase I hypersensitive site located approximately 40 kb upstream of the 5′ end of the ζ globin gene). The β-like globin cluster (bottom) is interstitial and located on the short arm of chromosome 11; expression of the genes in this cluster is under the control of a group of remote regulatory elements/DNase I hypersensitive sites collectively known as the locus control region (LCR). Pseudogenes and the θ globin gene, whose functional significance is unknown, are not represented. To make visualization of them easier, the globinlike genes are represented over a larger space of genome than they actually span.
Molecular basis of inherited forms of α-thalassemia
We now know of more than 1000 naturally occurring inherited mutations that affect the structure or the synthesis of the α- and β-like globin chains.13,14 Many different mutant alleles have been selected to reach high frequencies in tropical and subtropical regions of the world because heterozygotes are, to some extent, protected from the effects of falciparum malaria.1,15 α-thalassemia, characterized by hypochromic microcytic anemia, occurs when there is a reduction in the synthesis of the α-globin chains of adult hemoglobin (HbA, α2β2) relative to β-globin synthesis; excess β-chains then form homotetramers (HbH, β4). HbH can be detected by supravital staining (demonstrating the presence of characteristic erythrocyte inclusions), chromatography, or gel electrophoresis.9,12 The level of HbH broadly reflects the degree of reduction in α-globin synthesis.
More than 50 different cis-acting mutations of the α cluster have been described.13 Deletions removing one (haplotype–α/) or both (haplotype––/) linked α-globin genes are the most common mutations.13 Less frequently, point mutations are found in the structural genes or in their local cis-acting sequences (eg, initiation codons, splice sites, translational termination codons, polyA addition sites).9,12,13 In 9 families, inherited or sporadic deletions of various combinations of conserved upstream regulatory elements, all including HS-40, have been shown to cause the typical hematologic features of α-thalassemia.16 In addition to these cis-acting defects, inherited mutations in an X-linked gene encoding a trans-acting chromatin-associated protein, called ATRX, also result in mild α-thalassemia associated with a severe form of syndromal mental retardation (ATR-X syndrome; Mendelian Inheritance in Man catalog 301040).17-19 ATR-X syndrome is a rare condition that, to date, has been identified in approximately 100 families from many regions of the world. The mechanism by which ATRX mutations down-regulate α-globin expression is unknown (see “What role does ATRX play in regulating gene expression?”).
α-thalassemia in the context of hematologic malignancy
Although abnormal patterns of hemoglobin synthesis are nearly always inherited, occasionally persons with previously normal hematologic function develop aberrant hemoglobin synthesis as an acquired abnormality, usually within the context of hematologic malignancy.20 Such perturbations may be of little clinical or hematologic consequence, such as when the only change is a minor alteration in the level of fetal hemoglobin (HbF, α2γ2) or the minor adult hemoglobin A2 (α2δ2).21-23 Alternatively, hemoglobin synthesis may become abnormal, as in the rare neoplasm juvenile chronic myelocytic leukemia (also known as juvenile myelomonocytic leukemia), in which hemoglobin synthesis often reverts from a normal adult pattern to a fetal pattern.24,25 In addition to these examples, α- and β-thalassemia have been reported as acquired defects, and the associated abnormal red blood cell production, a consequence of inefficient erythropoiesis or hemolysis, may exacerbate the anemia of the associated hematologic malignancy.26-28
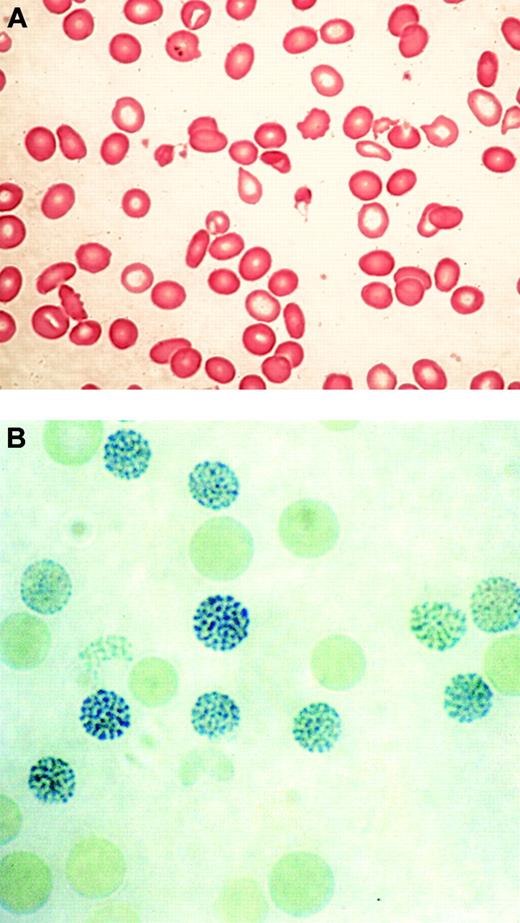
In 1960, 2 groups described a series of previously healthy patients with clonal hematopoietic disorders who acquired an unusual form of thalassemia during the course of their illnesses.29,30 This syndrome was characterized by marked hypochromic and microcytic anemia (Figure 2A) and the presence of HbH, demonstrable by gel electrophoresis and supravital staining of peripheral red blood cells (Figure 2B).29,30 Patients with similar conditions were soon found to have reduced α-globin–β-globin chain synthesis ratios, demonstrating unequivocally that α-thalassemia could occur as an acquired abnormality in patients with hematologic malignancy.28,31
Typical blood findings in ATMDS. (A) Peripheral blood smear (May-Grünwald-Giemsa) from a patient with ATMDS who had not undergone transfusion. Many red blood cells are hypochromic or microcytic, but others appear relatively normal and well preserved, presumably the descendants of nonthalassemic clones. (B) Classic hemoglobin H (HbH) inclusions in a patient with ATMDS seen after supravital staining (brilliant cresyl blue). Images were visualized under a Zeiss upright optical microscope with Trinoc ocular unit (Carl Zeiss GmbH, Oberkochen, Germany) equipped with Zeiss 10 × ocular lenses and an Olympus C3000Zoom CCD 3.3 megapixel digital camera on a C-style mount (Olympus UK, Middlesex, United Kingdom). Digital images were transferred to an 800 MHz iBook computer with 12-inch video screen running OS 10.3.2 and iPhoto (Apple Computers, Cupertino, CA) and were processed with Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA). Processing involved adjustments in image contrast, hue, and conversion from RGB to CMYK colorspace only; no other alterations were made. Figure 2A was viewed without a coverslip or immersion oil at 400 × total magnification, obtained using the ocular described above along with a Zeiss Plan 40 ×/0.75 objective lens. Figure 2B was viewed without a coverslip at 1000 × total magnification under a Zeiss Ph3 100 ×/1.25 oil immersion objective lens.
Typical blood findings in ATMDS. (A) Peripheral blood smear (May-Grünwald-Giemsa) from a patient with ATMDS who had not undergone transfusion. Many red blood cells are hypochromic or microcytic, but others appear relatively normal and well preserved, presumably the descendants of nonthalassemic clones. (B) Classic hemoglobin H (HbH) inclusions in a patient with ATMDS seen after supravital staining (brilliant cresyl blue). Images were visualized under a Zeiss upright optical microscope with Trinoc ocular unit (Carl Zeiss GmbH, Oberkochen, Germany) equipped with Zeiss 10 × ocular lenses and an Olympus C3000Zoom CCD 3.3 megapixel digital camera on a C-style mount (Olympus UK, Middlesex, United Kingdom). Digital images were transferred to an 800 MHz iBook computer with 12-inch video screen running OS 10.3.2 and iPhoto (Apple Computers, Cupertino, CA) and were processed with Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA). Processing involved adjustments in image contrast, hue, and conversion from RGB to CMYK colorspace only; no other alterations were made. Figure 2A was viewed without a coverslip or immersion oil at 400 × total magnification, obtained using the ocular described above along with a Zeiss Plan 40 ×/0.75 objective lens. Figure 2B was viewed without a coverslip at 1000 × total magnification under a Zeiss Ph3 100 ×/1.25 oil immersion objective lens.
Defining the clinical and hematologic features of the acquired form of α-thalassemia
In light of these reports, a registry of patients with acquired α-thalassemia (http://www.imm.ox.ac.uk/groups/mrc_molhaem/home_pages/Higgs/index.html) was established in the early 1980s, and it currently includes 68 verified cases; findings for approximately half have been published.20,28-61 Minimal criteria for inclusion in the ATMDS registry include 3 components. First, HbH must be demonstrated by electrophoresis, chromatography, or supravital staining (the latter technique is the most sensitive). Second, some form of hematologic neoplasia must be present. Finally, given that inherited α-thalassemia is so common, it is important to exclude any congenital forms of thalassemia predating the hematologic neoplasm. Because not all inherited forms of thalassemia are fully characterized, there should be no family history of thalassemia, unless the specific inherited mutation in that pedigree has been defined and carefully excluded in the patient. Using these criteria, it has been possible to construct a profile of this rare group of patients.
Acquired α-thalassemia is not limited to the geographic regions in which the inherited forms of α-thalassemia are common (eg, the Mediterranean basin, Southeast Asia, Africa, and Melanesia).1 In fact, most patients have been of northern European descent (49 of 62 for whom ethnicity is known); 5 Mediterranean and 8 Asian (5 Japanese, 1 Chinese, 1 Filipino, and 1 Indian) patients have also been reported. It is possible that this distribution reflects ascertainment bias. If microcytic, hypochromic red blood cell indices or other markers of thalassemia were observed in a patient with hematologic malignancy from an area in which inherited forms of thalassemia are common, it is likely that these findings would be ascribed to a previously unrecognized form of inherited thalassemia, and such an assumption would usually be correct. However, rare cases of acquired α-thalassemia originating from these areas of the world might have been missed. By contrast, thalassemic red blood cell indices in a patient with hematologic malignancy from outside the malaria belt (such as from northern Europe) are unexpected and should prompt further evaluation.
A second, striking, finding is that most patients reported to date with the acquired form of α-thalassemia (57 of the 66 for whom sex is known) have been male. In addition, most have been elderly, with a median age at diagnosis of 68 years. Only 10 patients have been younger than 50 years at presentation; the youngest was a 10-year-old girl with congenital Fanconi anemia who acquired myelodysplastic syndrome (MDS) (she exhibited HbH only after developing MDS; HbH was no longer detectable after her condition progressed to acute myeloid leukemia [AML]).60 Although a median age in the seventh decade of life is typical for malignant myeloid disorders in general, the strong male preponderance (greater than 6:1 male-female ratio) in the subset of patients with acquired α-thalassemia is remarkable. Several previous reports have described a slight male preponderance for hematologic malignancy in general and malignant myeloid disorders in particular, but the male-female sex ratio is certainly less than 2:1.62,63 No consistent occupational or environmental exposure has been described for patients with the acquired form of α-thalassemia, but, given the limited history available for most patients, specific predisposing factors cannot be excluded.
As more cases have been evaluated, it is now clear that acquired α-thalassemia most commonly develops in patients with MDS. Some patients were originally described as having underlying marrow disorders such as diGuglielmo syndrome, atypical erythroid hyperplasia, or erythroleukemia29,30,34,36-38 before the introduction of the widely used 1976 and 1982 definitions of MDS of the French-American-British (FAB) Cooperative Group.64 Careful review of these older reports suggests that most, if not all, of these patients would have been classified as having MDS if they had been evaluated more recently. In fact, one of the authors of the original report describing the first 3 cases of acquired HbH disease in erythroleukemia in 1960 confirmed in 1994 that these patients would have been described as having MDS in the present era (multiple oral communications between D.R.H. and David A. Galton [Hammersmith Hospital; Leukaemia Research Fund, University of London Royal Postgraduate Medical School, London, United Kingdom], 1991-1994).65 In some published reports, such as an instance of atypical chronic myeloid leukemia with HbH disease described in 1963 (only 3 years after the discovery of the Philadelphia chromosome and several years before the advent of routine karyotyping), MDS is a strong possibility, but there is not enough information to make a conclusive retrospective diagnosis.32 In several other cases that have come to our attention, the quality of archival material has been insufficient to allow a diagnosis more specific than chronic myeloid disorder. Despite these limitations, among the 68 registry cases, we have classified 54 as having MDS or probable MDS. All FAB subtypes are represented, as is secondary MDS associated with a history of radiotherapy or alkylating agent exposure. Therefore, it seems reasonable that HbH disease appearing for the first time in a patient with chronic myeloid disorder should be referred to as α-thalassemia myelodysplastic syndrome (ATMDS; Mendelian Inheritance in Man catalog 300448).28
Although the term ATMDS is accurate in most cases, 65 acquired α-thalassemia is not unique to MDS and has been reported in patients with other malignant hematologic disorders. For example, a patient with acquired HbH disease associated with acute lymphoblastic leukemia (ALL) was reported in 1982 in whom the hematologic diagnosis appears secure, at least on the basis of the immunohistochemical studies that were available at the time; there was no suggestion of MDS.49 Acquired α-thalassemia has also been reported in association with myelofibrosis, essential thrombocythemia, and de novo AML.
Among the 54 patients with definite or probable MDS, there is documentation of eventual leukemic transformation in 17 patients, a figure consistent with the 25% rate of development of leukemia reported for MDS in general.66 Persistence of HbH after leukemic transformation was demonstrated only in a single instance. Two additional registry patients (one report published; see Tanaka et al42 ) had AML at presentation without a clear preceding MDS phase; in these patients, as in the lone patient with acute lymphocytic leukemia (ALL), circulating HbH-containing red blood cells were easily demonstrable despite heavy marrow infiltration by malignant blasts.
The causes of death in patients with ATMDS are typical for a nonselected group of elderly MDS patients. Complications of leukemic transformation, neutropenia-associated infection, and cardiovascular disease are the most common terminal events.
Red blood cell findings and HbH inclusions in patients with ATMDS
Most patients with MDS have unexplained macrocytic or normocytic anemia; sometimes the latter is the result of pseudonormalization by red blood cell transfusion. In contrast, typical findings in ATMDS show hypochromic, microcytic red blood cells and striking anisopoikilocytosis (Figure 2A).12 Two distinct populations of red blood cells have been observed—poorly hemoglobinized ghost cells scattered among more normal-appearing cells—that may be the consequence of transfusion or the progeny of nonthalassemic erythropoietic clones.
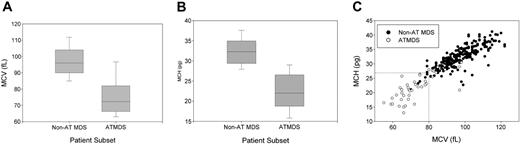
In contrast to MDS in general, in which the proportion of patients with microcytic anemia in all published series is less than 10%, most patients with ATMDS have hypochromic or microcytic red blood cell indices, reflecting what is seen on the peripheral blood smear. The average MCV is 75.2 fL for patients with ATMDS at the time of presentation with HbH, whereas the average MCV is 97.3 fL for MDS patients in general (Figure 3A). Similarly, the MCH is 22.4 pg in patients with ATMDS and 32.3 pg for MDS without thalassemia (Figure 3B). The MCH may be a slightly more specific indicator of acquired thalassemia than the MCV (Figure 3C).
Distribution of red blood cell indices in patients with α-thalassemia MDS (ATMDS) compared with a more general population with MDS. (A) MCV in ATMDS (n = 55) and MDS (n = 282) patients. Values are from the time of initial presentation with HbH or (if presentation data were unavailable) as close as possible to that time. Boxes enclose the 25th to 75th percentiles, and the middle bar denotes the median; vertical bars outline the 10th (bottom) to 90th (top) percentiles. (B) MCH in ATMDS and MDS. Boxes denote the same statistical distribution as in panel A. (C) Scattergram of MCV and MCH distribution in patients with ATMDS (○) versus MDS (•). (dashed lines) Eleven (23%) of 47 patients with ATMDS for whom MCH was available had MCH of 27 pg or greater, whereas 15 (27%) of 55 had MCV greater than 80 fL. In contrast, almost all MDS patients with MCH below 27 pg and MCV below 80 fL had evidence of acquired thalassemia. The non-ATMDS data source is personal communication from Drs Alison May, Graham Tennant, and Ala Al-Sabah (University Hospital of Wales in Cardiff; e-mail communication, January 19, 2004) regarding 282 nonthalassemic MDS patients; similar data were communicated by Dr David Oscier (Royal Bournemouth Hospital, Bournemouth, United Kingdom; oral communication to D.R.H. and Charles Craddock, Spring 1994) with respect to 101 MDS patients in Bournemouth and Southampton.
Distribution of red blood cell indices in patients with α-thalassemia MDS (ATMDS) compared with a more general population with MDS. (A) MCV in ATMDS (n = 55) and MDS (n = 282) patients. Values are from the time of initial presentation with HbH or (if presentation data were unavailable) as close as possible to that time. Boxes enclose the 25th to 75th percentiles, and the middle bar denotes the median; vertical bars outline the 10th (bottom) to 90th (top) percentiles. (B) MCH in ATMDS and MDS. Boxes denote the same statistical distribution as in panel A. (C) Scattergram of MCV and MCH distribution in patients with ATMDS (○) versus MDS (•). (dashed lines) Eleven (23%) of 47 patients with ATMDS for whom MCH was available had MCH of 27 pg or greater, whereas 15 (27%) of 55 had MCV greater than 80 fL. In contrast, almost all MDS patients with MCH below 27 pg and MCV below 80 fL had evidence of acquired thalassemia. The non-ATMDS data source is personal communication from Drs Alison May, Graham Tennant, and Ala Al-Sabah (University Hospital of Wales in Cardiff; e-mail communication, January 19, 2004) regarding 282 nonthalassemic MDS patients; similar data were communicated by Dr David Oscier (Royal Bournemouth Hospital, Bournemouth, United Kingdom; oral communication to D.R.H. and Charles Craddock, Spring 1994) with respect to 101 MDS patients in Bournemouth and Southampton.
Based on these hematologic data from the registry, we can now say that ATMDS is most frequently found in patients with MDS who have the typical abnormal red blood cell morphology of MCV less than 80 fL and MCH less than 27 pg (Figure 3C). However, these criteria are not diagnostic, because approximately 25% of patients with ATMDS have MCVs of 80 fL or greater or MCHs of 27 pg or greater. Furthermore, microcytic anemia may be observed in MDS patients with no evidence of α-thalassemia, so the diagnosis of ATMDS is ultimately based on demonstrating HbH inclusions in a proportion of the red blood cells. It would be useful to know how commonly α-thalassemia occurs in patients with MDS, but there have been no large systematic studies of the prevalence of HbH inclusions in unselected MDS patients. Therefore, the precise sensitivity of red blood cell indices as diagnostic markers is unknown.
In the past, many patients with normal red blood cell indices were identified as having ATMDS because brilliant cresyl blue (BCB) supravital staining of peripheral blood, performed to evaluate the reticulocyte count, unexpectedly demonstrated characteristic HbH inclusions. Most of the registry cases of ATMDS with normocytic or macrocytic red blood cell indices were discovered serendipitously in this way, but many clinical laboratories now measure the reticulocyte count by flow cytometric methods, 67 which do not routinely detect HbH inclusions. Therefore, we believe it is now essential to perform an independent assessment for HbH inclusions when a diagnosis of ATMDS is suspected based on the red cell criteria set out here. Because many clinical facilities no longer offer supravital staining, our laboratory provides this service free of charge for patients thought to have ATMDS if sent 1 mL fresh blood by courier.
The proportion of HbH-containing cells in the peripheral blood of patients with ATMDS varies among patients, but the level in a given patient typically remains stable unless leukemia develops. However, one patient who initially had 50% HbH-containing cells on repeated analyses was found to have less than 1% HbH-containing cells 5 months later without any change in the marrow appearance, presumably as a result of clonal evolution in the absence of leukemic transformation.6 When HbH is assayed by electrophoresis or high-performance liquid chromatography, the quantity detected is usually less than the proportion of red blood cells containing HbH inclusions. Rarely, trace amounts of hemoglobin Bart's (γ4) have been detected.12
In a subset of patients with ATMDS documented in the registry, a very small proportion (less than 1%) of HbH-containing cells remained stable over long periods of time. This, together with the disappearance of HbH in some patients, suggests that acquired α-thalassemia does not give affected clones any growth advantage. Loss of HbH after leukemic transformation suggests either that the thalassemic clone is overgrown by another neoplastic clone or that, when thalassemic clones develop additional mutations and become frankly leukemic, they almost always lose the ability to differentiate into mature erythroid progeny and to manifest a thalassemic phenotype.
Does the coexistence of α-thalassemia exacerbate the anemia and clinical picture of MDS? It is difficult to study this systematically because of inconsistencies in the reporting of transfusion status, but anecdotal evidence indicates that the development of thalassemia is associated with a worsening of the MDS-associated anemia.41 Not only does the presence of thalassemia cause additional dyserythropoiesis or hemolysis, a proportion of the hemoglobin produced consists of high-affinity HbH, further reducing the capacity for oxygen delivery.41,68
Characterizing the abnormal pattern of gene expression in ATMDS
Investigators in several studies have tried to determine at what level α-globin production is down-regulated in ATMDS (eg, transcription, mRNA processing, or translation). The αβ globin mRNA ratio has been studied only in a few patients with ATMDS and was reduced to less than 0.5 in all patients analyzed.28,31 The α-globin–β-globin chain synthesis ratio is strikingly abnormal in ATMDS; 25 patients have been analyzed, and the ratio was reduced to less than 0.2 in 13 patients and was less than 0.75 in 23 cases (normal range, 0.9-1.2). In contrast, the αβ globin chain synthesis ratio in MDS without thalassemia has been reported to be elevated—in some cases as high as 2.4.69
An α/β-synthetic ratio of less than 0.2 is less than one would see in patients with only a single functional α-globin gene (genotype––/–α).12 Although it is possible to imagine 2 different acquired cis-acting mutations underlying such a genotype in ATMDS, a trans-acting defect causing down-regulation of all 4 α-globin genes has long seemed a more likely explanation.12 Additional support for the presence of a trans-acting defect came from experiments in which bone marrow from patients with ATMDS was fused with murine erythroleukemia cells, resulting in an interspecific hybrid cell line retaining a single copy of the human (ATMDS) chromosome 16.53 Transfer of human chromosome 16 from the patients with ATMDS into a mouse erythroid background in this fashion normalized α-globin synthesis, though the admixture of abnormal clonal and normal hematopoietic cells characteristic of MDS meant that creation of the hybrid from a residual normal progenitor cell could not be ruled out.12
Identifying the molecular defect underlying ATMDS
Despite this preliminary evidence for a trans-acting mutation, the α-globin cluster and its surrounding region on chromosome 16 was still a reasonable first target for mutation screening. Such a defect might be observed if part of chromosome 16 were lost as a consequence of the genomic instability common in MDS. However, no consistent cytogenetic abnormalities have been associated with ATMDS, and when chromosome 16 has been examined in detail, it has appeared entirely normal (with a single exception described in the next paragraph). Of 28 successful cytogenetic studies summarized in the registry (there were at least 4 failures because of insufficient metaphases for analysis), results of 13 (46%) were abnormal, with karyotypes typical for MDS in general (frequent deletions involving chromosomes 5, 7, and 20).
In almost all recent cases of ATMDS, the α-globin gene cluster itself has been carefully analyzed by Southern blot hybridization and direct sequencing. In only a single instance has a lesion in this region been found: an MDS patient with a complex karyotype was recently found to have lost the telomeric region of 1 of 2 copies of chromosome 16 (acquired genotype: ––/αα) in a clonal fashion.61 The patient had relatively mild anemia (his hemoglobin level was 10.4 g/dL in the absence of transfusion), and only 0.11% HbH-containing red blood cells were detected, but the red blood cells were microcytic and hypochromic (MCV, 64 fL; MCH, 21.7 pg). Despite the relatively well-maintained hemoglobin level, the red blood cell morphology was more abnormal than one would expect to see with a constitutional loss of only 2 of the 4 α-globin genes. With this exception, in all other ATMDS cases, the globin genes themselves have been intact (genotype αα/αα), and no lesions of the major regulatory element (HS-40) have been found.12
A major clue to identifying the trans-acting molecular defect underlying ATMDS came from a cDNA microarray study that used peripheral blood neutrophil RNA from a patient with newly diagnosed ATMDS. This material was compared with pooled granulocyte RNA from several healthy persons.5 The experiment demonstrated that the ATRX gene at Xq13.3 was one of the genes with the lowest expression in the patient with ATMDS compared with controls, a result that was confirmed by reverse transcription–polymerase chain reaction (RT-PCR). An ATRX point mutation (G>A) was then found in the canonical splice donor site of intron 1 in this patient with ATMDS, which likely resulted in nonsense-mediated decay of the ATRX transcript. It was perhaps fortunate that the first patient analyzed in this way had a null mutation in ATRX because many of the point mutations later described would not be expected to affect the level of ATRX mRNA. The patient's ATRX mutation was restricted to myeloid cells, because buccal cells and a lymphoblastoid cell line derived from the patient had normal genotypes. This finding is consistent with previous studies showing that MDS is always associated with abnormal myeloid progenitors but that lymphoid lineages are only rarely part of the neoplastic clone.70
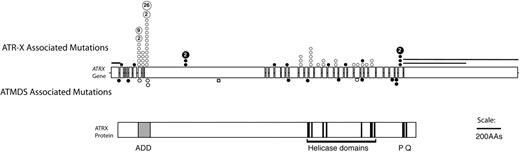
Because ATRX is an X-linked gene and because congenital ATR-X syndrome is frequently associated with mild α-thalassemia, ATRX had previously been considered a good candidate to be an ATMDS-associated gene. However, Southern blot and DNA sequence analyses of the ATRX gene in several patients with ATMDS had been unremarkable, 12 most likely because unfractionated archival DNA is not ideal material for detecting acquired mosaic mutations. In light of the encouraging finding in the single case described above, archival material including unfractionated blood and marrow from ATMDS registry patients was screened by denaturing high-performance liquid chromatography, which is sensitive to low-level mosaicism, and also by sequencing of the 10-kb ATRX mRNA. This revealed 11 additional patients with point mutations or splicing abnormalities in ATRX.5,6 The mutations clustered in and near the ADD (ATRX, DNMT3, DNMT3L) zinc-finger domain that is characteristic of the ATRX protein and de novo methyltransferases, and several mutations were also scattered throughout the putative ATRX helicase domain (Figure 4).71 These ATRX domains are highly conserved and are also common sites of mutation in the inherited ATR-X syndrome, suggesting that they are functionally important.6,72,73
Schematic representation of the spectrum of ATRX mutations that have been described in boys with ATR-X syndrome with those found in ATMDS. The ATRX gene (top, with introns not to scale) is aligned with ATRX protein (bottom) to allow comparison of the mutation site with functionally important protein domains such as the ADD (ATRX, DNMT3, DNMT3L) zinc-finger domain (ADD includes a C2-C2 type of zinc finger and the closely located PHD motif), helicase domains, P-box, and Q-box. ATR-X–associated mutations are represented above the ideogram of the ATRX gene, and mutations associated with ATMDS are denoted below. (•) Mutations predicted to cause protein truncation (frameshifts and nonsense mutations) and null mutations. (○) Amino acid changes, including in-frame insertions and deletions. Recurrent mutations in ATR-X syndrome are indicated by a number inside the circle, representing the number of families in whom the mutation has been identified. Circles denote point mutations and extended lines denote the extent of large deletions in 2 pedigrees with ATR-X syndrome. Modified with permission from Steensma et al.6
Schematic representation of the spectrum of ATRX mutations that have been described in boys with ATR-X syndrome with those found in ATMDS. The ATRX gene (top, with introns not to scale) is aligned with ATRX protein (bottom) to allow comparison of the mutation site with functionally important protein domains such as the ADD (ATRX, DNMT3, DNMT3L) zinc-finger domain (ADD includes a C2-C2 type of zinc finger and the closely located PHD motif), helicase domains, P-box, and Q-box. ATR-X–associated mutations are represented above the ideogram of the ATRX gene, and mutations associated with ATMDS are denoted below. (•) Mutations predicted to cause protein truncation (frameshifts and nonsense mutations) and null mutations. (○) Amino acid changes, including in-frame insertions and deletions. Recurrent mutations in ATR-X syndrome are indicated by a number inside the circle, representing the number of families in whom the mutation has been identified. Circles denote point mutations and extended lines denote the extent of large deletions in 2 pedigrees with ATR-X syndrome. Modified with permission from Steensma et al.6
At present, the molecular defect underlying ATMDS remains unexplained in only 5 of the 20 registry patients from whom genomic DNA is available. One of these 5 was the only woman analyzed, while the other 4 had less than 10% HbH-containing cells and HbH was undetectable by electrophoresis and chromatography. It is possible that these 5 patients actually have ATRX mutations but that current mutation detection techniques are insensitive to very small ATRX-mutant clonal populations in a large normal DNA background. Another possibility is that mutations in genes besides ATRX may be responsible for ATMDS in a subset of patients.
What role does ATRX play in regulating gene expression?
Our findings have demonstrated that inherited and acquired mutations in ATRX cause α-thalassemia, suggesting that ATRX normally plays a central role in the regulation of α-globin gene expression. The widespread developmental abnormalities in ATR-X syndrome also suggest that ATRX plays a more general role in gene regulation.19 However, at present, the mechanism by which ATRX controls gene expression is unclear. Nevertheless, many different lines of evidence suggest that ATRX is a chromatin-associated protein that may act as a transcriptional cofactor and influence the epigenetic control of gene expression.
The field of epigenetics describes how gene regulatory information not directly encoded in the underlying DNA can be stably transmitted from one cell to its somatic or germline progeny.74 In the past decade, there has been considerable progress in our understanding of epigenetic mechanisms as the protein-DNA complex that comprises chromatin has been characterized. It appears that epigenetic information may be established and maintained through DNA methylation, the covalent modifications of histones (the so-called histone code), nonhistone proteins that contribute to the maintenance of genes in an active or an inactive state, and factors that remodel chromatin by altering the positions of nucleosomes on DNA. It has also been noted that, as with ATRX, mutations in other genes whose products are predicted to play a part in these epigenetic pathways often cause complex multisystem disorders and may contribute to the development of neoplasia.75
Further clues to the normal biologic function of ATRX
At present, the precise normal role of ATRX in the cell is uncertain, and we are trying to pull together a series of disparate observations. In particular, documenting the pattern of ATRX expression, comparing it with similar proteins, establishing its localization in the cell, and identifying some of its protein partners may direct us to an understanding of its normal function(s).
ATRX is a member of the SWI2/SNF2 family of molecular motors that remodel chromatin using the hydrolysis of adenosine triphosphate (ATP) as a source of energy.17 Chromatin remodeling may alter the access of DNA to trans-acting factors and, therefore, may influence a wide variety of cellular activities involving DNA and RNA (eg, transcription, DNA replication, repair, recombination, methylation, cell division). The recent isolation of endogenous ATRX has confirmed that it is an ATPase and that, like other members of this family, it has translocase activity, which is to say the protein can move along double-stranded DNA in the presence of ATP in vitro.76
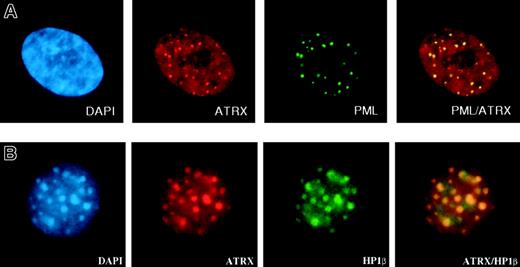
Within cells, the ATRX protein is localized in the nucleus and is concentrated in 3 distinct nuclear subcompartments—those associated with heterochromatin, those at rDNA arrays, and those in promyelocytic leukemia (PML) nuclear bodies (Figure 5A), normal subnuclear structures characteristically disrupted in PML (AML subtype M3). ATRX may also be present at more diffuse euchromatic loci, including the α-globin complex, but, to date, technical hurdles have hampered the search for ATRX at these sites. The proteins that have been shown unequivocally to interact with ATRX are a reflection of its sublocalization. First, an important interaction appears to be with HP1, a protein that colocalizes with ATRX in heterochromatin (Figure 5B).77 HP1 was originally described in Drosophila melanogaster, where it was identified as an epigenetic modifier of gene expression.78 ATRX binds HP1 by way of the latter's chromoshadow domain, and HP1 in turn binds to the N-terminal tail of histone H3 when this tail is methylated at the lysine 9 residue (H3-K9), a histone modification characteristic of heterochromatin.79 Methylation of this lysine 9 residue depends on the activities of specific histone methyltransferases (SUV39H1 and SUV39H2; for a review, see Maison and Almouzni80 ). Disrupting H3-K9 methylation delocalizes HP1 and ATRX from heterochromatin (David J. Garrick [MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, Oxford, United Kingdom], oral communication, July 2004), strongly suggesting that ATRX is targeted to heterochromatin through HP1.
Immunofluorescence results. (A) ATRX localizes to subnuclear PML bodies (in human fibroblasts). Reprinted with permission from Xue et al.76 (B) ATRX colocalizes with HP1β at heterochromatin (in murine L929 cells). Courtesy Drs D. Garrick and T.L. McDowell; used with permission. Images were captured with a Sensys CCD camera (Photometrics, Tucson, AZ) mounted on an Olympus BX51 microscope (Olympus) equipped with an Olympus UPlanApo 100 ×/1.35 oil immersion objective lens. Coverslips were mounted in Vectashield (Vector Laboratories, Peterborough, UK) containing 0.5 μg/mL DAPI. Images were acquired with MacProbe version 4.3 software (Applied Imaging, Newcastle, UK) and processed with Adobe Photoshop 7.0 (Adobe Systems).
Immunofluorescence results. (A) ATRX localizes to subnuclear PML bodies (in human fibroblasts). Reprinted with permission from Xue et al.76 (B) ATRX colocalizes with HP1β at heterochromatin (in murine L929 cells). Courtesy Drs D. Garrick and T.L. McDowell; used with permission. Images were captured with a Sensys CCD camera (Photometrics, Tucson, AZ) mounted on an Olympus BX51 microscope (Olympus) equipped with an Olympus UPlanApo 100 ×/1.35 oil immersion objective lens. Coverslips were mounted in Vectashield (Vector Laboratories, Peterborough, UK) containing 0.5 μg/mL DAPI. Images were acquired with MacProbe version 4.3 software (Applied Imaging, Newcastle, UK) and processed with Adobe Photoshop 7.0 (Adobe Systems).
Second, a well-established interaction occurs between ATRX and Daxx, one of many proteins localized to PML bodies that influence gene expression, the cell cycle, or apoptosis.76,81 Recent work has shown that localization of ATRX within PML bodies depends specifically on its interaction with Daxx.76,81 Additionally, Daxx has been shown to be recruited to heterochromatin in a cell-cycle–dependent fashion through its interaction with ATRX.82 The precise role of PML bodies in the nucleus is unresolved, and it is unclear whether they play a direct role in transcription or are merely a “sink” for various nuclear proteins.83-85 Although other potential interactions with ATRX have been reported (SSRP1, 76,82 EZH2, 86 and others), none of these has been fully confirmed.
The most important next step is to define a specific function for ATRX at one or more of these loci. ATRX clearly plays a role in regulating gene expression, but how does it do so? At present, the clearest example of a biologic function concerns the role of ATRX in DNA methylation. Genetic studies in diverse organisms have defined the epigenetic markers of silent heterochromatin and have shown that gene silencing depends on a dynamic and complex interaction between chromatin remodeling, histone methylation at H3-K9, nonhistone proteins such as HP1, DNA methylation, and RNA interference.80 It was of particular interest, therefore, when it was shown that CpG methylation at heterochromatic loci is perturbed in patients with constitutional mutations in ATRX.87 This is the first evidence that ATRX serves a purpose at these loci, and it suggests that, directly or indirectly, ATRX is involved in the establishment or maintenance of DNA methylation. Somewhat disappointingly, no change in DNA methylation or chromatin modification at the α-globin loci has been observed in primary erythroid cells carrying an ATRX mutation (Takahito Wada and R.J.G., unpublished data, 2001-2002) or ATMDS (D.P.S., R.J.G., and D.R.H., unpublished data, 2003-2004).
Despite these advances in our understanding of the localization and function of ATRX protein, it is frustrating that none of these insights has revealed how ATRX influences α-globin gene expression. The fact that ATRX mutations affect α-but apparently not β-globin expression may be informative. These 2 clusters are embedded in different chromosomal environments, 11 which supports the view that ATRX influences gene expression through one or more of the epigenetic features that distinguish these different regions. Clearly, ATRX plays a role at heterochromatin; its presence may be important to prevent silencing of the α-globin genes during terminal differentiation of erythroid cells when most genes are being switched off. Mutations in ATRX might cause premature and inappropriate silencing of α-globin expression during terminal erythropoiesis, when transcriptional activity in the cluster is normally the greatest. This may occur by the spreading of heterochromatin and associated gene silencing, perhaps from the nearby chromosome 16p telomere (approximately 150 kb from the α gene cluster) or, alternatively, by facilitating the recruitment of factors contributing to a repressive heterochromatic environment to the α-globin complex. In either case, one prediction of this model is that the expression of genes adjacent to the α-globin cluster might also be perturbed by ATRX mutations, and this possibility is being actively pursued. Alternatively, ATRX may act directly at euchromatic loci, such as α-globin, to facilitate transcription through chromatin-remodeling activity, thereby allowing the access of key transcription factors to regulatory elements. A current research priority is to determine whether ATRX is detectable at the α-globin locus or its upstream regulatory sites; preliminary chromatin immunoprecipitation experiments have not yet identified ATRX at these loci (Eduardo Anguita, Marco De Gobbi, R.J.G., and D.R.H., unpublished data, 2002-2004).
In hematopoiesis, ATRX appears to exert its effects during terminal differentiation of erythroid cells. Differentiation along the erythroid lineage seems to be specified correctly in hematopoietic progenitors, and, apart from abnormal globin synthesis, which occurs late in differentiation, the ATRX-mutant red blood cells appear relatively normal. ATRX is widely expressed throughout all stages of development in some tissues (eg, brain and lymphoid cells), but ATRX is more highly expressed in terminally differentiating rather than proliferating cells (Helen Turley and R.J.G., unpublished data, 1998).88 In patients with ATR-X syndrome who have abnormally developed genitalia (eg, male pseudohermaphroditism), testis specification occurs, but subsequent maturation of the cellular components appears to have failed. It is possible that many of the genes whose regulation is perturbed by ATRX mutations are those expressed in terminal differentiation.
Unresolved issues
The discovery of ATRX gene mutations in ATMDS has highlighted several key unanswered questions. Solutions to these puzzles may provide important insights into how ATRX normally regulates gene expression, and they may also uncover some common molecular defects underlying MDS that have more general importance.
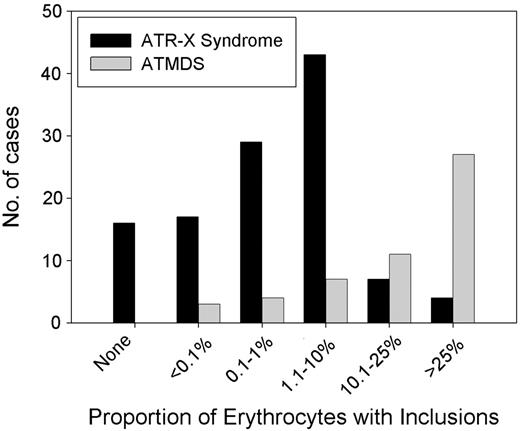
It is curious that the hematologic phenotype in patients with ATMDS is, in general, more severe than that seen in boys who have congenital ATR-X syndrome (Figure 6).6 For example, a boy with a germline c.576G>C; p.L192F ATRX mutation had 0.1% HbH-containing erythrocytes, whereas a patient with ATMDS who had acquired the identical mutation (restricted to myeloid cells) had 50% HbH-containing cells. As described here, patients with ATMDS usually have very low α-globin–β-globin synthesis ratios (less than 0.2 in 52% of patients) and large amounts of HbH (median, 30%) compared with boys with inherited ATR-X syndrome, who generally have only mildly reduced α-synthesis–β-synthesis ratios (range, 0.3-0.98 in 10 patients tested; median, 0.64) with low and sometimes undetectable amounts of HbH (53% of boys have 1% or less HbH, and 90% have less than 10%).6 Given that the spectrum of ATRX mutations is similar in these congenital and acquired forms of HbH disease (Figure 4), the suggestion is that the different hematologic effects of ATRX mutations in erythroid cells depends on when the mutation occurs during development or, more likely, on the cellular context in which the mutation occurs.
Distribution of the proportion of HbH in the peripheral blood in ATMDS compared with ATR-X syndrome. Results of supravital staining have been reported in 61 patients with ATMDS, and the number of inclusion-containing cells has been quantified in 54 patients. Of these, 17 (31%) patients have had 50% or more circulating erythrocytes containing HbH inclusions, 22 (44%) have had 10% to 50% of such cells, and 15 (28%) patients have had 10% or less (median, 30%). Only 8 (17%) of 48 quantified cases have had 25% or more HbH when measured by electrophoretic or chromatographic methods (data not shown), and in 10 patients the amount of HbH was below the level of detection despite the presence of small numbers of inclusions on supravital staining (mean, 15%). Several patients with ATR-X syndrome have no HbH, whereas all patients with ATMDS have some. This is because, by definition, patients with ATMDS must have some evidence of HbH, but ATR-X syndrome may be diagnosed from other characteristic nonthalassemic features of the multisystemic disorder in the presence of an ATRX gene mutation.
Distribution of the proportion of HbH in the peripheral blood in ATMDS compared with ATR-X syndrome. Results of supravital staining have been reported in 61 patients with ATMDS, and the number of inclusion-containing cells has been quantified in 54 patients. Of these, 17 (31%) patients have had 50% or more circulating erythrocytes containing HbH inclusions, 22 (44%) have had 10% to 50% of such cells, and 15 (28%) patients have had 10% or less (median, 30%). Only 8 (17%) of 48 quantified cases have had 25% or more HbH when measured by electrophoretic or chromatographic methods (data not shown), and in 10 patients the amount of HbH was below the level of detection despite the presence of small numbers of inclusions on supravital staining (mean, 15%). Several patients with ATR-X syndrome have no HbH, whereas all patients with ATMDS have some. This is because, by definition, patients with ATMDS must have some evidence of HbH, but ATR-X syndrome may be diagnosed from other characteristic nonthalassemic features of the multisystemic disorder in the presence of an ATRX gene mutation.
Could there be a relatively common pattern of genetic or epigenetic changes in the erythroblasts of patients with MDS that drastically enhances the deleterious effects of ATRX mutations on α-globin expression? Myeloid cells of patients with MDS often have many different genetic mutations, the full repertoire of which is only just beginning to be defined, and recent reports demonstrate widespread MDS-associated epigenetic abnormalities reflected in the patterns of DNA methylation and histone acetylation.89-91 This question might be analyzed further by studying patients with the common, inherited forms of α-thalassemia who, by chance, go on to develop MDS. To our knowledge, there are no systematic reports of such patients in the literature. Anecdotally, however, MDS patients with inherited forms of α-thalassemia do not appear to have an especially severe hematologic phenotype or very high levels of HbH-containing cells. This suggests that the abnormal genetic and epigenetic programs in hematopoietic progenitors in MDS do not directly influence α-globin expression. It seems more likely that, in the context of MDS, mutations directly or indirectly exacerbate the deleterious effects of an ATRX mutation. Given the presumed rarity of ATRX mutations, these interacting mutations might occur frequently in all types of MDS.
Additional mutations might explain the rare phenotype in some patients with ATMDS who have a very mild hematologic phenotype (eg, less than 1% HbH and minimal anemia). To date, we have not found ATRX mutations in any of these patients. Although this could be explained by the limited sensitivity of current detection techniques, it is also possible these patients have acquired mutations in one or more of the proposed modifying genes that, by itself, have only minimal effect on α-globin expression. A severe ATMDS phenotype may occur only when loss of ATRX function is combined with one or more molecular defects in such modifiers. Our knowledge of the sublocalization of ATRX and its interacting protein partners (Figure 7; see also “Further clues to the normal biologic function of ATRX”) provides several candidates for potential modifying genetic and epigenetic factors that may be commonly altered in MDS, and we are currently systematically screening for mutations in these partners.
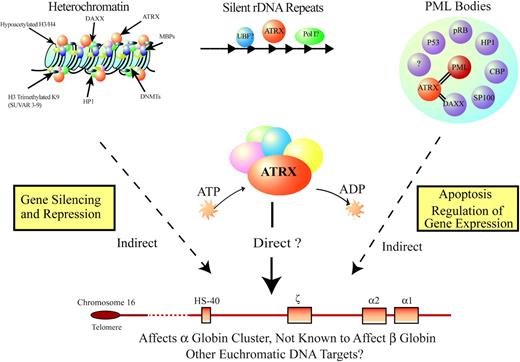
Model for how ATRX might influence gene expression. ATRX is part of a multiprotein complex that uses the energy of ATP to remodel chromatin or its associated DNA in a way that affects transcriptional activity at euchromatic loci, including the α-globin gene cluster. This interaction may be a result of alteration of the regional distribution of heterochromatin by the complex or of recruitment of transcription factors (directly or indirectly) that alter gene expression. ATRX function may be associated with its presence at one of its target nuclear locations (eg, heterochromatin, ribosomal DNA repeats, PML bodies) in addition to euchromatic sites. Potential points for interaction between ATRX and MDS, leading to a more severe hematologic phenotype than in ATR-X syndrome, include the multiprotein complex, of which ATRX is a part; transcription factors, whose binding to DNA is affected by the ATRX complex; and epigenetic modifications of histone-associated DNA, leading to alterations in local chromatin conformation.
Model for how ATRX might influence gene expression. ATRX is part of a multiprotein complex that uses the energy of ATP to remodel chromatin or its associated DNA in a way that affects transcriptional activity at euchromatic loci, including the α-globin gene cluster. This interaction may be a result of alteration of the regional distribution of heterochromatin by the complex or of recruitment of transcription factors (directly or indirectly) that alter gene expression. ATRX function may be associated with its presence at one of its target nuclear locations (eg, heterochromatin, ribosomal DNA repeats, PML bodies) in addition to euchromatic sites. Potential points for interaction between ATRX and MDS, leading to a more severe hematologic phenotype than in ATR-X syndrome, include the multiprotein complex, of which ATRX is a part; transcription factors, whose binding to DNA is affected by the ATRX complex; and epigenetic modifications of histone-associated DNA, leading to alterations in local chromatin conformation.
Another potential clue to the effect of ATRX mutations in ATMDS is the dramatic male predominance of this condition, which is so striking that it demands an explanation. Sex imbalance in human disease can occur as a result of the differences between males and females at many different levels.12 A common occupational exposure underlying ATMDS seems improbable, and though the hormonal milieu certainly affects hematopoiesis (eg, the moderate contribution of androgens to normal erythropoiesis), a hormonal explanation for the dramatic sex imbalance in ATMDS also seems unlikely. At the molecular level, an excess of male patients might imply involvement of an X-linked gene with a “gene dosage” effect, and the fact that ATRX is X-linked superficially seems to provide support for this hypothesis. However, male and female somatic cells contain only one fully active X chromosome. Males have one X and a Y chromosome that encode few genes, but in females (XX genotype), one X chromosome is randomly inactivated (though some genes escape X inactivation). Therefore, unlike inherited X-linked mutations, there is no a priori reason why acquired X-encoded mutations should manifest more frequently in males than in females. An instructive parallel condition is paroxysmal nocturnal hemoglobinuria (PNH), which is also associated with acquired, somatic mutations in an X-linked gene (PIGA); in PNH, there is actually a slight female excess (approximately 1.2:1) in disease incidence.92 This difference in ATMDS sex distribution is a major issue yet to be resolved.
Conclusion
Although ATMDS and ATR-X syndrome are rare conditions, there have been many examples in which elucidating the molecular basis of uncommon disorders sheds important light on the general principles underlying more common, clinically important diseases. It is likely that acquired ATRX mutations arise as part of the widespread background of genetic mutations that occur in malignant cells. We identify ATRX mutations not because they give the MDS clone a selective advantage but because such mutations cause a dramatic red blood cell phenotype. If the acquired mutations simply mimicked the hematologic effect of the inherited mutation, this would be of limited interest. However, it appears that ATRX mutations in patients with MDS produce a remarkably severe phenotype, suggesting that there are other genetic or epigenetic abnormalities in MDS that interact with the ATRX mutations to enhance their effect in silencing α-globin expression. It is presumed that ATRX mutations rarely occur in MDS, yet they are consistently associated with the ATMDS phenotype, suggesting that the “enhancing” mutation(s) in MDS is a common defect in MDS and, therefore, may be important to discover.
Prepublished online as Blood First Edition Paper, September 9, 2004; DOI 10.1182/blood-2004-07-2792.
Supported by the Medical Research Council in the United Kingdom (D.R.H., R.J.G.). D.P.S. is a Mayo Foundation Research Scholar supported by the Mayo Clinic.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Sir David J. Weatherall and William G. Wood for their helpful comments on the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal