Abstract
In this study, we show that the hyaluronan synthase 1 (HAS1) gene undergoes aberrant intronic splicing in multiple myeloma (MM). In addition to HAS1 full length (HAS1FL), we identify 3 novel splice variants of HAS1, HAS1Va, HAS1Vb, and HAS1Vc, detected in patients with MM or monoclonal gammopathy of undetermined significance (MGUS). HAS1Vb and HAS1Vc undergo intronic splicing with creation of a premature stop codon. MM cells expressing one or more HAS1 variants synthesize extracellular and/or intracellular hyaluronan (HA). Expression of the HAS1Vb splice variant was significantly correlated with reduced survival (P = .001). Together, alternative HAS1 gene splicing, the correlations between HAS1 splicing and HA synthesis, and the correlations between HAS1 splicing and reduced survival of MM patients support the hypothesis that the family of HAS1 protein plays a significant role in disease progression. Further, expression of HAS1Vb, in conjunction with HAS1FL and/or other HAS1 variants, may lead to accumulation of intracellular HA molecules and an impact on receptor for HA-mediated motility (RHAMM)-mediated mitotic abnormalities in MM. This study highlights the potential importance of HAS1 and its alternative splicing in pathophysiology of MGUS and MM. (Blood. 2005;105: 4836-4844)
Introduction
Multiple myeloma (MM) is an incurable malignancy of bone marrow (BM) plasma cells (PCs), with a median survival rate of 3 to 4 years after diagnosis. Molecular studies conducted by us and others have identified CD19+ late-stage B cells in the peripheral blood (PB) of patients with MM.1-4 They are characterized by a clonotypic immunoglobulin H (IgH) VDJ rearrangement identical to that of malignant PCs in the BM.1,3,4 MM B cells are drug resistant and express hyaluronan (HA)-specific receptors RHAMM (receptor for HA-mediated motility) and CD44, which are necessary for MM cell motility or adhesion.5-9
Previously, we have shown that the motility of circulating MM B cells is mediated by the extracellular matrix molecule HA.6 High or low levels of HA in the serum of patients with MM correlate with reduced median survival.10 In addition to extracellular HA, an intracellular HA has been detected in the nucleus, nucleolus, and rough endoplasmic reticulum.11-14 The source of the intracellular HA is unknown.
HA is synthesized by hyaluronan synthase (HAS).15 Three isoenzymes of HAS—HAS1 (hCh19), HAS2 (hCh8), and HAS3 (hCh16)—have been detected in humans thus far. Each isoenzyme synthesizes different sizes of HA molecules that exhibit different functions.16 Overexpression of HAS proteins promotes growth and/or metastatic development.17,18 Little is known about the role of HAS1 in cancer, reflecting a short lifetime of HAS1 transcripts, likely due to the presence of an adenylate/uridylate (AU)-rich element (ARE) in the 3′ untranslated region (UTR) gene controlling the half-life of mRNA (ARE database, http://rc.kfshrc.edu.sa/ared/).19,20
We have identified differential expression patterns of HAS genes in MM patients. HAS1 is expressed exclusively by circulating MM B cells and HAS2 by BM-localized MM PCs, perhaps reflecting different biologic imperatives of these 2 compartments of the MM clone. Both HAS1 and HAS2 are absent from B cells of healthy donors. Furthermore, we identify 3 novel splice variants of HAS1 designated as HAS1Va, HAS1Vb, and HAS1Vc. Expression of HAS1 splice variants is absent from B cells of healthy donors and in MM and monoclonal gammopathy of undetermined significance (MGUS) is restricted to the B-cell compartment. Expression of HAS1Vb, an intronic splice variant, correlates with poor survival in MM patients. This work indicates a potential role for alternative splicing of HAS1 in MM and MGUS.
Materials and methods
Patient samples
Blood and/or BM samples from 106 patients with MM and 62 patients with monoclonal gammopathy of MGUS were taken at diagnosis and subsequent clinical visits after approval from the Health Research Board (University of Alberta) and the Alberta Cancer Board and with informed consent per Declaration of Helsinki. Blood samples were also obtained from 10 healthy donors.
Tissue and cell preparation
Peripheral blood mononuclear cells (PBMCs) and bone marrow cells (BMCs) were stained and sorted as described by Szczepek et al.3 The sorted PB CD19+ B cells and BM PCs with more than 96% purity and PBMCs from 164 (106 MM and 58 MGUS) patients were collected and resuspended in Trizol (Invitrogen, Carlsbad, CA) or RNA lysis tissue buffer (RLT) (Qiagen, Mississauga, ON) for total RNA isolation, using either a standard Trizol isolation reaction or RNeasy kit (Qiagen) according to the manufacturer's instructions. Total RNA was obtained from cell lines in a similar manner.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Complementary DNA for the polymerase chain reaction (PCR) was reverse transcribed from total RNA isolated from sorted CD19+ B cells, CD38hiCD45lo PCs, non-B, non-PC, and T cells, or from total mononuclear cells isolated from blood or BM aspirates and cell lines. Total RNA (0.5 to 1 μg) was denatured for 10 minutes at 70°C followed by annealing with 100 ng dT15. Next, RNA was reverse transcribed as described by Keats et al.21 The HAS (HAS1, HAS2, and HAS3) gene-specific transcripts were amplified from cDNA obtained through a reverse transcriptase (RT) reaction using a reverse and forward primer set designed for each HAS transcript (Table 1). For the PCR reaction, 1.1 ng cDNA was added to 24 μL of PCR mix containing 10 × PCR buffer, 1 μL of 50 mM MgCl2, 0.5 μL of 10 mM deoxyribonucleoside triphosphate (dNTP), 1 μL each of 10 mM primers, and 5 units of platinum Taq (Invitrogen). The PCR cycling parameters were the following: primary denaturation, 5 minutes at 94°C; 35 cycles of denaturation for 30 seconds at 94°C; annealing, 30 seconds at 60°C; extension, 30 seconds at 72°C; and final extension, 7 minutes at 72°C. The samples were either stored at -20°C for later analysis or immediately processed for capillary electrophoresis on the ABI3100 DNA genetic analyzer (Applied Biosystems [ABI], Foster City, CA).
Nucleotide sequences of primer sets used in the study
. | |Primer sets| . | . | |
|---|---|---|---|
| Genes . | Reverse . | Forward . | |
| HAS1 | 5′VIC-GGGCTTGTCAGAGCTACTT | 5′AGGGCGTCTCTGAGTAGCAG | |
| HAS2 | 5′VIC-CCTCATCTGTGGAGATGGGT | 5′TCCCAGAGGTCCACTAATGC | |
| HAS3 | 5′VIC-CATCCAGGTGTGCGACTCTG | 5′CGCTGCTCAGGAAGGAAATC | |
| CD19 | 5′FAM-TACTATGGCACTGGCTGCTG | 5′CACGTTCCCGTACTGGTTCT | |
| RHAMM | 5′FAM-TGACAAAGATACTACCTTGCCTGCT | 5′CAGCATTTAGCCTTGCTTCCATC | |
| HAS1 cloning primers | 5′GCCTTCGCCCTGCTCATCCTG | 5′GTAGAACAGACGCAGCACA | |
. | |Primer sets| . | . | |
|---|---|---|---|
| Genes . | Reverse . | Forward . | |
| HAS1 | 5′VIC-GGGCTTGTCAGAGCTACTT | 5′AGGGCGTCTCTGAGTAGCAG | |
| HAS2 | 5′VIC-CCTCATCTGTGGAGATGGGT | 5′TCCCAGAGGTCCACTAATGC | |
| HAS3 | 5′VIC-CATCCAGGTGTGCGACTCTG | 5′CGCTGCTCAGGAAGGAAATC | |
| CD19 | 5′FAM-TACTATGGCACTGGCTGCTG | 5′CACGTTCCCGTACTGGTTCT | |
| RHAMM | 5′FAM-TGACAAAGATACTACCTTGCCTGCT | 5′CAGCATTTAGCCTTGCTTCCATC | |
| HAS1 cloning primers | 5′GCCTTCGCCCTGCTCATCCTG | 5′GTAGAACAGACGCAGCACA | |
For these primer sets, the reverse primers were labeled at their 5 ends with 6-carboxyflourescein (FAM) or VIC (chemical name not disclosed; ABI). Primers were designed using the “Primer 3” or “Gene Tool” programs based on the published cDNA sequences of the HASs, CD19, and RHAMM.
Capillary electrophoresis—DNA fragment analysis
For DNA fragment analysis, 1 μL of PCR product was mixed with 12 μL of HiDi formamide (ABI) and 1 μL of internal size standard GeneScan 500 (ABI). PCR products were then denatured for 4 minutes at 96°C and, after rapid centrifugation, samples were immediately placed on ice for 15 minutes. The electrophoresis conditions were as follows: run voltage, 15 kV; injection voltage, 1 kV; injection duration, 10 to 20 seconds; run temperature, 60°C; laser within power of 12 mW; and run current, 100 μA. The results were analyzed using GeneScan 3.7 software (ABI).
Cloning and sequencing
RNA for cloning was extracted using an RNeasy kit (Qiagen); 1 μg RNA was then reverse transcribed as described previously. HAS1 was amplified in 50 μL PCR reaction mix containing 8.8 ng cDNA, 5 μL of 1 × PCR buffer, 2 mM MgSO4, 0.2 mM dNTP, 0.4 mM HAS1 cloning primer set (Table 1), and 0.5 units of HiFi Platinum Taq (Invitrogen). The PCR cycling parameters were denaturation for 5 minutes at 94°C, followed by denaturation for 1 minute at 94°C, annealing for 2 minutes at 60°C, and extension at 72°C for 2 minutes for 35 cycles, with a final extension period of 7 minutes at 72°C. HAS1 PCR products were cloned into the TOPO TA cloning system and transformed into TOP 10 competent cells according to the manufacturer's instructions (Invitrogen). To identify colonies containing HAS1 plasmid, individual bacterial colonies were tested by PCR as described above. Positive HAS1/TOPO TA colonies were grown overnight in Luria-Bertani (LB) medium. Plasmids were prepared using the Qiagen Plasmid Purification Mini Kit (Qiagen) and sequenced with T7 and M13 primers with the ABI PRISM BigDye V3 Cycle Sequencing Ready Reaction DNA Sequencing Kit (ABI) on the ABI3100 DNA genetic analyzer.
Statistics
After obtaining informed consent, blood samples were collected from MM patients between December 1995 and March 2003. All patient records were reviewed retrospectively to verify the diagnosis of MGUS and MM.22 Information on diagnosis, patient demographics, baseline staging and clinical features, treatment, and survival were collected. Blood taken at time of diagnosis was available from 58 of the cases; only these cases were used to assess the correlation of HASs with baseline clinical features and survival. The primary analysis assessed the association between expression of the various HAS isoforms and survival. Secondary analyses explored correlations between HAS isoforms and other clinical or laboratory parameters. Categorical variables were compared between 2 groups using the Fisher exact test. Continuous variables were compared using the Student t test or the Wilcoxon rank sum test as appropriate. Survival distributions were determined using the Kaplan-Meier method and compared using the log-rank test. Multivariable analyses and hazard ratios were generated using Cox regression models. Statistical significance was set at a P = .05 using 2-sided analysis.
Particle exclusion assay (PEA)
The enzymatic activity of HAS proteins was determined using a particle exclusion assay (PEA) as described previously.23 Sorted CD19+ B cells (2 × 106), obtained from PBMCs of 7 MM patients and 3 healthy donors, and CD38hiCD45lo PCs (2 × 106), collected from the BM of 3 patients with MM, were cultured in a 35 mm culture dish coated with poly-l-lysine (PLL) (1 mg/mL; Sigma, St Louis, MO) and allowed to recover. After recovery, medium was removed at 4 hours, 12 hours, 24 hours, or 48 hours, and the cells were washed with calcium- and magnesium-free phosphate-buffered saline (CMF-PBS). Next, formalin- (3%) fixed sheep erythrocytes (50 × 105) in PBS/0.1% bovine serum albuin (BSA) were added to the cultured cells. The culture dish was placed on the microscope stage until the fixed red blood cells had settled (30 to 45 minutes). Imaging used an Axiovert 100M confocal laser scanning microscope (LSM 510; Zeiss, Oberkochen, Germany). As a negative control, the cells were treated with 100 μL (500 U/mL) hyaluronidase (HAase, type 4 from bovine testes; Sigma) for 1 hour at 37°C prior to adding fixed erythrocytes. Aliquots of the same samples were analyzed by RT-PCR for expression of HAS1 and its variants, as described.
Intracellular HA detection
PEA was modified and combined with indirect HA staining to verify that the detected pericellular matrix around the cell plasma membrane includes HA molecules. The sorted CD19+ B cells or CD38hiCD45lo PCs (2 × 105) were cultured and recovered as described. After the recovery time cells were fixed with 4% paraformaldehyde (PFA) for 15 minutes at 4°C. To localize HA, the cells were incubated with biotinylated HA binding protein (B-HABP) (Seikagaku America [BioLynx, San Antonio, TX]) (2 μg/mL in PBS/1% BSA overnight at room temperature). Alternatively, B-HABP was detected with streptavidin Alexa 594 (Molecular Probes, Eugene, OR) 1:500 dilution for 2 hours at room temperature. As a negative control, sorted cells were treated with 500 U/mL HAase for 1 hour at 37°C before and after fixation or B-HABP was preincubated with 100 μg HA (Seikagaku America). The specificity of streptavidin was determined by staining the cells with streptavidin Alexa 594 only. The cells were examined with an Axiovert 100M LSM 510.
To detect intracellular HA, CD19+ B cells were sorted and mobilized on a PLL-coated dish. Cells were treated with HAase for 2 hours at 37°C, washed with PBS, fixed/permeabilized with 4% PFA/0.03% saponin, and incubated with 2 μg/mL B-HABP overnight at 37°C. Intracellular HA was detected by streptavidin Alexa 594.
Cell lines and antibodies
The MM cell line RPMI 8226 was generously supplied by S. Treon (The Dana-Farber Cancer Institute). KMS-12-BM and KMS-12-PE were generous gifts from T. Otsuki (The Kawasaki Medical School). The rabbit polyclonal antibody recognizing HAS1 and variants was produced by Washington Biotechnology (Baltimore, MD) to the following peptide sequence: 135GNRAEDLYMVDMFRF150. The polyclonal serum was affinity purified, and the specificity was tested by enzyme-linked immunosorbent assay (ELISA) and Western blotting, the latter of which was performed on protein lysates obtained from the CCL 110 cell line expressing all isoenzymes of HAS (results not shown).
Western blotting
The cells RPMI 8226, KMS-12-BM, and KMS-12-PE were lysed at 5 × 106 to 107 cells per milliliter in RIPA buffer with protease inhibitors. Equal amounts, 2 μg of total proteins obtained from cell lysates, were separated by a 5% stacking/12% separating sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Immobilon-NC membranes (Millipore, Bedford, MA). Nonspecific binding was blocked by 5% BSA/0.1% Tween 20 PBS blocking buffer overnight at 4°C. The membranes were incubated with the anti-HAS1 serum (1:500), preimmune serum (1:500), and anti-HAS1 serum incubated with 5-fold excess blocking peptides overnight at 40°C. Next, membranes were incubated for 2 hours with an antirabbit horseradish peroxidase (HRP)-IgG (1:20 000) (Jackson ImmunoResearch Laboratories, Bar Harbor, ME). The immune complexes were visualized using ECL reagent according to the manufacturer's instructions (BioBar, Amersham Biosciences, Baie d'Urfe, Canada).
HAS genes are expressed in MM. Representative GeneScan electropherograms for DNA fragment analysis of HAS gene expression are shown. PCR products were obtained by RT-PCR amplification of fragments of HAS genes and aberrant variants. The x-axes represent molecular size (bp) of PCR product, and the y-axes indicate relative fluorescent units (RFU). The arrows indicate product peaks; faded peaks represent internal size standard peaks of LIZ 500 (ABI). (A) Expression of HAS1 and variants; (B) expression of HAS2; (C) expression of HAS3.
HAS genes are expressed in MM. Representative GeneScan electropherograms for DNA fragment analysis of HAS gene expression are shown. PCR products were obtained by RT-PCR amplification of fragments of HAS genes and aberrant variants. The x-axes represent molecular size (bp) of PCR product, and the y-axes indicate relative fluorescent units (RFU). The arrows indicate product peaks; faded peaks represent internal size standard peaks of LIZ 500 (ABI). (A) Expression of HAS1 and variants; (B) expression of HAS2; (C) expression of HAS3.
Results
Differential expression of HASs in MM or MGUS PBMC B cells and BM PCs
The MM clone includes both BM PCs and circulating CD19+ B cells previously shown to be clonotypic.3,4,7,24 The expression of HASs was examined in CD19+ B cells obtained from patients with MM, MGUS, and healthy donors as well as in CD38hiCD45lo PCs sorted from MM BM. Using RT-PCR and DNA fragment analysis, we detected ubiquitous expression of HAS3 in all analyzed patients and healthy donors (Figure 1C; Table 2). In contrast, cell type-specific expression patterns of HAS1 and HAS2 were detected in malignant B cells and PCs, respectively. CD19+ B cells from 7 of 13 MM patients expressed HAS1, while HAS2 was detected in CD38hiCD45lo PCs from 11 of 11 MM patients (Table 2; Figure 1B,C). No HAS2 was detectable in sorted PCs from 2 and unsorted BMCs from 3 lymphoma patients with uninvolved BM.
HAS1 gene expression is restricted to MM and MGUS B cells
Cell type and patient no. . | HAS1Va . | HAS1Vb . | HAS1Vc . | HAS1 . |
|---|---|---|---|---|
| MM CD38hiCD45loPCs* | ||||
| 1-8 | - | - | - | - |
| 9 | - | - | - | + |
| 10 | + | + | - | + |
| 11 | - | - | - | + |
| MM CD19+B cells† | ||||
| 12 | - | - | - | + |
| 13, 18 | - | + | - | + |
| 14, 16 | + | - | + | - |
| 15 | - | + | - | + |
| 17 | - | + | + | + |
| 19, 20 | + | - | - | + |
| 21 | + | + | - | - |
| 22, 23 | + | + | + | - |
| 24 | + | - | + | + |
| MGUS CD19+B cells‡ | ||||
| 25-27 | + | - | - | + |
| 28 | + | - | + | - |
| Healthy donors, CD19+B cells§ | ||||
| 1-10 | - | - | - | - |
Cell type and patient no. . | HAS1Va . | HAS1Vb . | HAS1Vc . | HAS1 . |
|---|---|---|---|---|
| MM CD38hiCD45loPCs* | ||||
| 1-8 | - | - | - | - |
| 9 | - | - | - | + |
| 10 | + | + | - | + |
| 11 | - | - | - | + |
| MM CD19+B cells† | ||||
| 12 | - | - | - | + |
| 13, 18 | - | + | - | + |
| 14, 16 | + | - | + | - |
| 15 | - | + | - | + |
| 17 | - | + | + | + |
| 19, 20 | + | - | - | + |
| 21 | + | + | - | - |
| 22, 23 | + | + | + | - |
| 24 | + | - | + | + |
| MGUS CD19+B cells‡ | ||||
| 25-27 | + | - | - | + |
| 28 | + | - | + | - |
| Healthy donors, CD19+B cells§ | ||||
| 1-10 | - | - | - | - |
Expression profiles of HAS genes and HAS1 novel variants in CD19+ B cells from PBMCs of MM and MGUS patients and healthy donors and from CD38hiCD45lo PCs from BM aspirates of MM patients. Expression profiles of HASs in MM PBMC non-B cells (CD19- fraction of PBMC cell populations obtained from the same samples from which CD19+ B cells were isolated), MM BM non-PCs (fraction of non-PC populations obtained from the same set of samples from which CD38hiCD45lo PCs were isolated), and MM T cells are not shown, because these samples express HAS3 only. No HAS1, HAS1 variants, or HAS2 was detected in these samples. The table indicates the number of patients analyzed for HAS gene expression; “+” and “-” indicate positive and negative expression of the indicated HAS transcripts. HAS2 expression was analyzed in sorted PCs obtained from 2 and unsorted BMCs from 3 lymphoma patients with uninvolved BM; no HAS2 was detected in any of these samples using RT-PCR and DNA fragment analysis/GeneScan software. Expression of CD19 in B cells obtained from patients and/or healthy donors and RHAMM expression in the BM PCs were used as positive control reactions to validate the integrity of RNA. All analyzed samples expressed CD19 (MM and healthy donor B cells) and RHAMM (MM PCs) transcripts (not shown). As a negative control for each PCR sample, reactions were always run in the absence of reverse transcriptase at the RT step (results not shown).
None of the 13 patients expressed HAS2; all 13 expressed HAS3.
Eleven of the 11 patients expressed HAS2 and HAS3.
None of the 4 patients expressed HAS2; all 4 expressed HAS3
None of the 10 healthy donors expressed HAS2; all 10 expressed HAS3.
Expression of HAS2 was absent in MM CD19+ B cells even after the precipitation of 25 μL of PCR products, a method to increase the sensitivity of detection for templates with low abundance. Similar to MM B cells, CD19+ B cells obtained from the PBMCs of 4 patients with MGUS expressed HAS1 and HAS3 but not HAS2 (Figure 1A,B; Table 2). Precipitation of PCR products revealed weakly detectable expression of HAS1 in CD38hiCD45lo PCs from only 3 of 11 MM patients (Table 2). No HAS1 or HAS2 expression was detectable in non-PCs obtained from the same set of samples from which CD38hiCD45lo PCs were isolated. Furthermore, no HAS1 was detectable in non-B cells (CD19- PBMCs) in the same samples from which MM and MGUS CD19+ B cells were isolated or from sorted T-cell populations obtained from PBMCs of 4 MM patients. Overall, strong HAS1 expression in MM and MGUS was restricted to CD19+ B cells, while HAS2 is restricted to MM PCs.
To determine whether or not expression of HAS1 is unique to MM and MGUS CD19+ B cells, we analyzed CD19+ B cells from the PBMCs of 10 healthy donors. B cells from healthy donors expressed only HAS3 transcripts (Table 2; Figure 1C). HAS1 and HAS2 were undetectable in these cells, even after precipitation/analysis of 25 μL of PCR products.
Furthermore, HAS1 expression was assessed for 82 MM and 58 MGUS patients. Archived PBMCs were used, justified by our demonstration that in MM and MGUS, expression of HAS1 is restricted to the CD19+ B cells in PBMCs and was undetectable in non-CD19+ PBMCs (Table 2). Table 3 shows HAS1 expression for PBMCs from 58% of MM and 74% of MGUS patients.
HAS1 and aberrant novel variants are expressed in PBMCs obtained from most MM and MGUS patients
Sample type . | No. patients . | HAS1Va, % . | HAS1Vb, % . | HAS1Vc, % . | HAS1, % . |
|---|---|---|---|---|---|
| MM PBMCs | 82 | 68 | 46 | 33 | 58 |
| MGUS PBMCs | 58 | 74 | 41 | 9 | 74 |
Sample type . | No. patients . | HAS1Va, % . | HAS1Vb, % . | HAS1Vc, % . | HAS1, % . |
|---|---|---|---|---|---|
| MM PBMCs | 82 | 68 | 46 | 33 | 58 |
| MGUS PBMCs | 58 | 74 | 41 | 9 | 74 |
Numbers indicate percentage of patients whose PBMCs expressed HAS1 and its novel variant. Results were obtained by RT-PCR/DNA fragment analysis, and data analyses were conducted using GeneScan software. HAS1Vc expression is not significantly different between MM and MGUS.
Novel aberrant splice variants of HAS1 in sorted MM and MGUS B cells
Using RT-PCR and DNA fragment analysis, we identified 3 splice variants of HAS1, designated as HAS1Va, HAS1Vb, and HAS1Vc, in MM and MGUS CD19+ B cells (Figure 1A). We analyzed the expression of these variants in sorted CD19+ B cells isolated from PBMCs of 13 MM patients and CD38hiCD45lo PCs obtained from BM of 11 MM patients. HAS1Va was expressed in CD19+ B cells from 8 of 13 MM and 4 of 4 of MGUS patients (Table 2). From the same set of samples HAS1Vb was expressed in 7 of 13 MM patients, while no expression of these transcripts was detected in CD19+ B cells obtained from 4 patients with MGUS (Table 2). HAS1Vc was expressed in 6 of 13 MM and 1 of 4 MGUS patients (Table 2). MM PCs from only 1 of 11 patients expressed HAS1Va and HAS1Vb, and none expressed detectable HAS1Vc (Table 2). Additionally, no splice variants of HAS1 were detectable in CD19- fractions of PBMCs (non-B cells) from 9 MM patients or in non-PC populations (BMCs remaining after gating for CD38hiCD45lo PCs) obtained from 3 MM BM samples. No HAS1 variants were detectable in CD19+ B cells from 10 healthy donors or in sorted T cells of 4 MM patients. Thus, the expression of one or more HAS1 variants was restricted to MM and MGUS CD19+ B cells. This allowed us to examine recurrent expression of HAS1 splice variants in unfractionated PBMCs obtained from 82 MM and 58 MGUS patients. HAS1Va transcripts were detected in 68% of MM and 74% of MGUS patients, while HAS1Vb was identified in 46% of MM and 41% of MGUS PBMCs (Table 3). HAS1Vc was detected in 33% of MM and 9% of MGUS PBMCs (Table 3). HAS3 was ubiquitously expressed in all samples tested, providing a control for RNA integrity. This analysis demonstrates that most MM and MGUS patients express one or more HAS1 variants in various combinations.
Longitudinal analysis of HAS1 and variants in MM patients
Longitudinal monitoring of HAS1 and its variants was performed for PBMCs from 18 unselected MM patients. At the time of diagnosis, 65% (11 of 18) of MM patients expressed HAS1 alone or in combination with one or more novel variants, while 71% (10 of 14) of MM patients expressed these genes at the time of relapse. No distinct expression pattern was observed in patient samples obtained during disease progression or remission, possibly reflecting treatment of many patients with prednisone and/or dexamethasone, which are known to play a significant role in tissue-specific regulation, activation, and/or suppression of HAS genes and production of HA.25,26
Cloning and sequencing of HAS1 variants
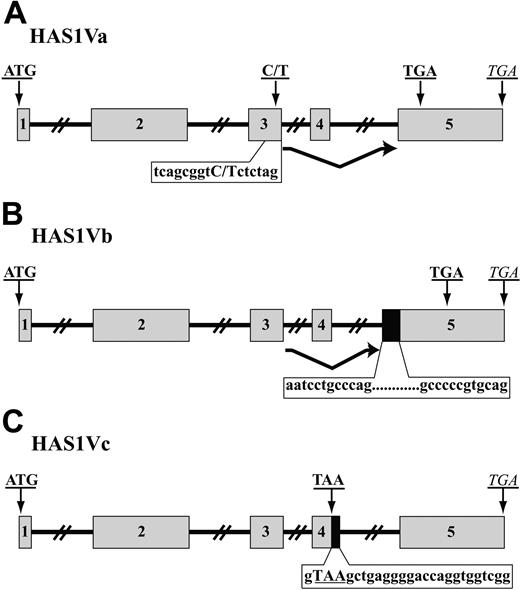
Using RT-PCR, we amplified HAS1Va and HAS1Vb cDNA fragments from CD19+ B cells of 3 MM patients and HAS1Vc cDNA fragments from 1 MM patient. The variants of HAS1 were amplified using hot-start PCR and an exon-intron spanning primer set. The PCR products were cloned into the TOPO TA vector, and positive subclones were identified by PCR with HAS1 gene-specific primers. Plasmids isolated from positive clones were sequenced. Sequences were identified through alignment with the published sequence of human HAS mRNA (gi: 4504338; National Center for Biotechnology Information [NCBI]). This analysis identified HAS1Va as a result of complete deletion of exon 4 leading to a frameshift after the deletion and “insertion” of a premature termination codon (PTC) 56 nucleotides downstream of the deletion (Figure 2A). HAS1Vb is the result of partial retention of intron 4 (59 bp) at the 5′ end of exon 5 and deletion of the entire exon 4. These aberrations lead to a frameshift after deletion of exon 4 that creates a PTC 93 nucleotides downstream of retained intron 4, at the beginning of the exon 5 (Figure 2B). HAS1Vc results from the retention of 26 nucleotides of intron 4 at the 3′ end of exon 4, causing truncation of the HAS1 transcripts and insertion of PTC at the 3′ end of exon 4 (Figure 2C). For all 3 variants, the start codon and the conserved sequence of the glycosyltransferase motif was present in the aligned cDNA sequences, consistent with the evidence below (“Expression of HAS1 variant protein and HA synthesis in MM cells”) that HAS1 variants retain the ability to synthesize HA.
MM B cells express aberrant splice variants of HAS1. A schematic representation of HAS1Va (A), HAS1Vb (B), and HAS1Vc (C). ▦ represents exons, while ▪ indicates retained fragments of intron 4. Introns are shown with solid lines. Original stop codons of each novel variant are marked with italic letters, while bold uppercase letters indicate start codons and PTC. (A) HAS1Va: Point mutation 7760C>T detected in this novel variant transcript is shown in bold, uppercase letters; PTC is located 56 nucleotides downstream of deleted exon 4. (B) HAS1Vb is the result of deletion of the entire exon 4 and partial retention of intron 4 (59 bp; 12 first and last nucleotides of retained introns are shown) at the 5′ end of exon 5. These aberrations harbored PTC 93 nucleotides downstream of the retained intron 4. (C) HAS1Vc intronic splice variant is similar to HAS1Vb and is the result of the retention of 26 nucleotides of intron 4, causing insertion of a PTC, TAA, at the 3′ end of exon 4. The 26 nucleotides of retained intron 4 are shown on the figure. PTC is shown in bold uppercase letters.
MM B cells express aberrant splice variants of HAS1. A schematic representation of HAS1Va (A), HAS1Vb (B), and HAS1Vc (C). ▦ represents exons, while ▪ indicates retained fragments of intron 4. Introns are shown with solid lines. Original stop codons of each novel variant are marked with italic letters, while bold uppercase letters indicate start codons and PTC. (A) HAS1Va: Point mutation 7760C>T detected in this novel variant transcript is shown in bold, uppercase letters; PTC is located 56 nucleotides downstream of deleted exon 4. (B) HAS1Vb is the result of deletion of the entire exon 4 and partial retention of intron 4 (59 bp; 12 first and last nucleotides of retained introns are shown) at the 5′ end of exon 5. These aberrations harbored PTC 93 nucleotides downstream of the retained intron 4. (C) HAS1Vc intronic splice variant is similar to HAS1Vb and is the result of the retention of 26 nucleotides of intron 4, causing insertion of a PTC, TAA, at the 3′ end of exon 4. The 26 nucleotides of retained intron 4 are shown on the figure. PTC is shown in bold uppercase letters.
Cloning, sequencing, and alignment analysis of HAS1 variants from CD19+ B cells of one MM patient revealed a point mutation (a possible single nucleotide polymorphism [SNP]) on exon 3 of the HAS1Va transcripts (Figure 2A). HAS1Va from this MM patient had the nucleotide T instead of C in this position (7760C>T). The detected point mutation was confirmed through twice sequencing both strands (plus and minus strands) of HAS1Va cDNA in a triplicate sequencing reaction by triplicate runs. Transcripts also were amplified using HiFi Platinum Taq, which has proofreading ability. The point mutation 7760C>T was absent in the HAS1 full length (HAS1FL), HAS1Vb, and HAS1Vc transcripts obtained from the same B-cell population of the same patient, suggesting that the HAS1Va and the HAS1/HAS1Vb/HAS1Vc group were derived from different alleles.
Correlated expression of intronic variants HAS1Vb and HAS1Vc
This analysis incorporated 74 randomly selected MM cases. Pairwise relationships between various HASs were assessed with the Fisher exact test. This analysis showed that expression of HAS1Vb and HAS1Vc intronic splice variant transcripts in MM patients was significantly correlated (P = .01). Sixteen MM patients coexpressed both variants, 33 expressed neither variant, 16 expressed only HAS1Vb, and 9 expressed only HAS1Vc. However, the correlation between HAS1Vb and HAS1Vc is only moderate; the population of HAS1Vb expressors only partly overlaps the population of HAS1Vc expressors. This result was similar when only 58 diagnosis samples were considered (P = .03). There was no statistically significant relationship in expression among the other HAS1 variants.
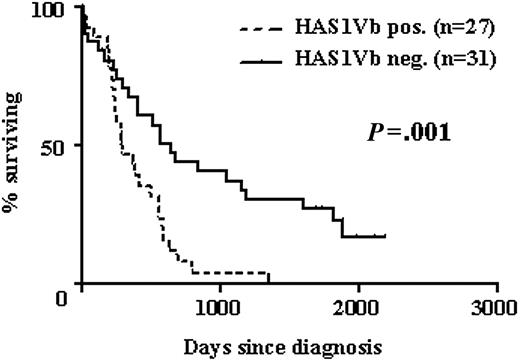
HAS1Vb expression by MM B cells correlates with poor survival. Kaplan-Meier survival distributions of MM patients with (-) or without (—) detectable HAS1Vb in the PB at time of diagnosis. HR, 2.6; 95% CI, 1.4 to 4.8; P = .001 using the log-rank rest.
HAS1Vb expression by MM B cells correlates with poor survival. Kaplan-Meier survival distributions of MM patients with (-) or without (—) detectable HAS1Vb in the PB at time of diagnosis. HR, 2.6; 95% CI, 1.4 to 4.8; P = .001 using the log-rank rest.
HAS1Vb expression by MM B cells correlates with reduced survival
Expression of HASs was analyzed in PBMCs obtained from 58 MM patients at time of diagnosis. The univariate analysis of these cases showed that expression of HAS1Vb in MM patients was most strongly correlated with a shorter survival (hazard ratio [HR], 2.6; 95% confidence interval [CI], 1.4 to 4.8; P = .001; Figure 3). Associations between HAS1Va (P = .048) and HAS1FL (P = .12) and poor survival were of borderline or nonstatistical significance. There was no association between expression of HAS1Vc and poor survival (P = .66).
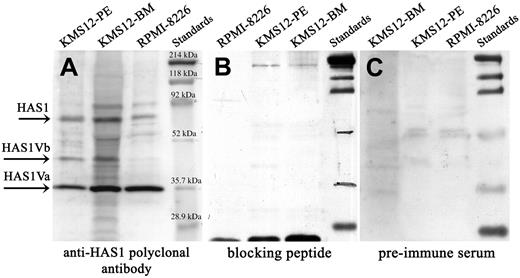
Expression of HAS1 and novel variant genes and proteins in MM cell lines. Western blot analysis. Protein lysates of MM cell lines were separated on SDS-PAGE, blotted onto nitrocellulose, and probed with (A) anti-HAS1 antibodies, (B) preincubated anti-HAS1 with blocking peptide, and (C) before bleed (preimmume serum). Arrows identify HAS1Va, about 35.9 kDa; HAS1Vb, about 39.5 kDa; and HAS1, about 65 kDa bands. The size of HAS1Va (about 35.9 kDa) and HAS1Vb (about 39.5 kDa) proteins were predicted using the ExPASy Molecular Biology Server. The anti-HAS1 specificity was evaluated by preincubating anti-HAS1 serum with the blocking peptide overnight before probing the membrane. The extra bands presented on the blot most likely are bands corresponding to HAS1 proteins encoded by other yet to be identified variants, and/or these bands represent the HAS proteins subjected to posttranslation modifications and glycosylation.
Expression of HAS1 and novel variant genes and proteins in MM cell lines. Western blot analysis. Protein lysates of MM cell lines were separated on SDS-PAGE, blotted onto nitrocellulose, and probed with (A) anti-HAS1 antibodies, (B) preincubated anti-HAS1 with blocking peptide, and (C) before bleed (preimmume serum). Arrows identify HAS1Va, about 35.9 kDa; HAS1Vb, about 39.5 kDa; and HAS1, about 65 kDa bands. The size of HAS1Va (about 35.9 kDa) and HAS1Vb (about 39.5 kDa) proteins were predicted using the ExPASy Molecular Biology Server. The anti-HAS1 specificity was evaluated by preincubating anti-HAS1 serum with the blocking peptide overnight before probing the membrane. The extra bands presented on the blot most likely are bands corresponding to HAS1 proteins encoded by other yet to be identified variants, and/or these bands represent the HAS proteins subjected to posttranslation modifications and glycosylation.
Expression of HAS1 variant protein and HA synthesis in MM cells
Protein expression was evaluated by Western blotting in MM cell lines (RPMI 8226, KMS-12-BM, KMS-12-PE) (Figure 4 and Table 4). Bands corresponding to the HAS1Va (35.9 kDa) and HAS1Vb (39.5 kDa) were detected in all 3 cell lines although expression of HAS1Vb was weak in RPMI 8226. Overall, expression levels of HAS1 proteins were low. In parallel, expression of HAS1, HAS1Va, and HAS1Vb was consistent with the protein expression in aliquots of the same cells (Table 4); HAS1Vc was undetectable.
RT-PCR DNA fragment analysis
Cell lines . | HAS1Va . | HAS1Vb . | HAS1 . |
|---|---|---|---|
| KMS-12-PE | + | + | + |
| KMS-12-BM | + | + | + |
| RPMI 8226 | + | - | + |
Cell lines . | HAS1Va . | HAS1Vb . | HAS1 . |
|---|---|---|---|
| KMS-12-PE | + | + | + |
| KMS-12-BM | + | + | + |
| RPMI 8226 | + | - | + |
Transcript levels of HAS1 and novel variants were measured in 3 MM cell lines, RPMI 8226, KMS-12-PE, and KMS-12-BM.
Total RNA for RT-PCR and cell lysate for Western blot analysis were obtained from same cell culture for each cell line.
HAS1-expressing MM B cells synthesize a pericellular HA matrix. The HA pericellular matrix around the cell was visualized by the addition of fixed erythrocytes to sorted CD19+ B cells from MM (n = 15) and healthy donors (n = 3) and from MM CD38hiCD45lo PCs (n = 3) in short-term culture. Fixed erythrocytes were physically excluded from the areas surrounding a cell that had synthesized an HA pericellular matrix. HA matrix was detected only around MM CD19+ B cells. MM BM CD38hiCD45lo PCs and CD19+ B cells obtained from healthy donors did not exhibit an HA matrix around their plasma membranes (results not shown). Furthermore, no HA matrix was detected around the MM CD19+ B cells at 4 hours and 12 hours of culturing; however, 24 hours later, a small amount of HA matrix was detected around some MM CD19+ B cells, while other B cells in the culture did not exhibit an HA matrix. The size of an HA matrix significantly increased around some MM CD19+ B cells 48 hours after culture (A, arrows; B), while other CD19+ B cells did not develop an HA matrix (A, stars). In the culture some cells were characterized by a prominent coat of HA at one edge of the cell, while the opposite edge of the same cell exhibited lesser amounts of HA matrix (B). This type of distribution of HA matrix around the cells results from cell motility. Motile cells exhibit a prominent HA halo at their trailing edge and a lesser pronounced halo at their leading edge. After HAase treatment no HA matrix was detected around MM CD19+ B cells (C). Scale bar = 10 μm. Magnification 40×, numerical aperture 1.3. Images were processed using LSM 510 software.
HAS1-expressing MM B cells synthesize a pericellular HA matrix. The HA pericellular matrix around the cell was visualized by the addition of fixed erythrocytes to sorted CD19+ B cells from MM (n = 15) and healthy donors (n = 3) and from MM CD38hiCD45lo PCs (n = 3) in short-term culture. Fixed erythrocytes were physically excluded from the areas surrounding a cell that had synthesized an HA pericellular matrix. HA matrix was detected only around MM CD19+ B cells. MM BM CD38hiCD45lo PCs and CD19+ B cells obtained from healthy donors did not exhibit an HA matrix around their plasma membranes (results not shown). Furthermore, no HA matrix was detected around the MM CD19+ B cells at 4 hours and 12 hours of culturing; however, 24 hours later, a small amount of HA matrix was detected around some MM CD19+ B cells, while other B cells in the culture did not exhibit an HA matrix. The size of an HA matrix significantly increased around some MM CD19+ B cells 48 hours after culture (A, arrows; B), while other CD19+ B cells did not develop an HA matrix (A, stars). In the culture some cells were characterized by a prominent coat of HA at one edge of the cell, while the opposite edge of the same cell exhibited lesser amounts of HA matrix (B). This type of distribution of HA matrix around the cells results from cell motility. Motile cells exhibit a prominent HA halo at their trailing edge and a lesser pronounced halo at their leading edge. After HAase treatment no HA matrix was detected around MM CD19+ B cells (C). Scale bar = 10 μm. Magnification 40×, numerical aperture 1.3. Images were processed using LSM 510 software.
Enzymatic activity of the HASs was detected using a PEA (Figure 5A,B). HA matrix was detected around MM CD19+ B cells after 48 hours of culturing. No matrix accumulation was observed around MM BM PCs or B cells obtained from healthy donors (not shown). HAase treatment removed the deposited matrix around cell plasma membrane of MM CD19+ B cells (Figure 5C), confirming the presence of HA (Figure 5A,B). The existence of HA molecules in the pericellular matrix detected by PEA was verified by incubating cells with B-HABP (Figure 6). Using B-HABP, we also detected intracellular HA in permeabilized MM CD19+ B cells (Table 5). Intracellular HA in these cells is abundantly distributed around the perinuclear compartment of the cells as well as along the cell cytoskeleton (Figure 6F).
Expression of HAS1 variants correlates with production of extracellular and/or intracellular HA
HAS RNA expression pattern . | HA production . | No. patients . |
|---|---|---|
| HAS1Va, HAS3 | Extracellular HA matrix | 2 |
| HAS1Va, HAS1, HAS3 | Extracellular HA matrix | 3 |
| HAS1Va, HAS1Vb, HAS1, HAS3 | Extracellular HA matrix & intracellular HA | 3 |
| HAS1Vb, HAS1, HAS3 | Intracellular HA and very weak extracellular HA | 2 |
| HAS1, HAS3 | No HA production | 3 |
| HAS3 | No HA production | 2 |
HAS RNA expression pattern . | HA production . | No. patients . |
|---|---|---|
| HAS1Va, HAS3 | Extracellular HA matrix | 2 |
| HAS1Va, HAS1, HAS3 | Extracellular HA matrix | 3 |
| HAS1Va, HAS1Vb, HAS1, HAS3 | Extracellular HA matrix & intracellular HA | 3 |
| HAS1Vb, HAS1, HAS3 | Intracellular HA and very weak extracellular HA | 2 |
| HAS1, HAS3 | No HA production | 3 |
| HAS3 | No HA production | 2 |
Summary of the expression of HAS1, its novel variants, and HAS3 genes in PBMCs obtained from 15 MM patients. In conjunction with gene expression analysis, we conducted PEA and HA staining on PBMC B cells obtained from aliquots of the same populations of cells from MM patients to evaluate the potential role of HAS1 variants in the enhanced production of extracellular and intracellular HA.
To clarify the role of HAS1 and its variants in the enhanced extracellular and intracellular production of HA molecules by the CD19+ B cells of patients with MM, we have correlated the expression of HASs with the production of extracellular and/or intracellular HA in these cells using PEA, HA staining, and RT-PCR. Although the patterns are complex, analysis of B cells from 15 MM patients (Table 5) shows that synthesis of HA is detectable only by those populations of B cells expressing HAS1 variants. The potential contribution of HAase has not yet been assessed. Overall, HAS1Va appeared associated with synthesis of extracellular HA and HAS1Vb with synthesis of intracellular HA. Expression of HAS1 with HAS3 appeared insufficient for synthesis of either form of HA.
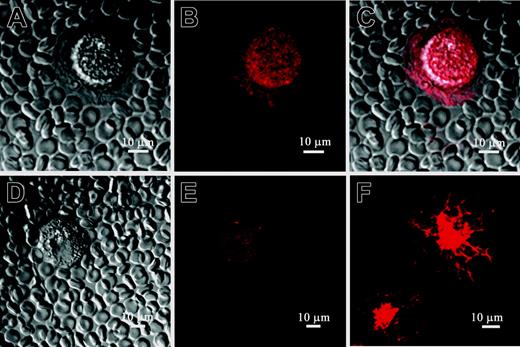
Pericellular matrix synthesized by MM B cells includes HA: MM B cells express intracellular HA. PEA in combination with indirect HA staining was used to verify the existence of HA molecules in the pericellular matrix detected by PEA on Figure 2. MM CD19+ B cells were cultured for 48 hours and then were incubated with B-HABP. HA binding to B-HABP was visualized using streptavidin Alexa 594. (A) CD19+ B cell without HA staining. (B) The cell and pericellular matrix around MM CD19+ B cells, which excluded fixed erythrocytes, were stained with streptavidin Alexa 594, indicating the presence of HA in the pericellular matrix. (C) Merged image of PEA and HA staining. (D) The cells were treated with HAase, which degraded the HA pericellular matrix. (E) HAase treatment also diminished cell surface HA staining. (F) Staining cells with B-HABP also detected intracellular HA in permeabilized MM CD19+ B cells. The staining pattern suggests that intracellular HA is distributed along the cell cytoskeleton and perinuclear compartment of MM B cells. Additionally, for one MM patient, weak nuclear staining of HA was observed in CD19+ B cells (not shown). Scale bar = 10 μm. Magnification 40×, numerical aperture 1.3. Images were processed using LSM 510 software.
Pericellular matrix synthesized by MM B cells includes HA: MM B cells express intracellular HA. PEA in combination with indirect HA staining was used to verify the existence of HA molecules in the pericellular matrix detected by PEA on Figure 2. MM CD19+ B cells were cultured for 48 hours and then were incubated with B-HABP. HA binding to B-HABP was visualized using streptavidin Alexa 594. (A) CD19+ B cell without HA staining. (B) The cell and pericellular matrix around MM CD19+ B cells, which excluded fixed erythrocytes, were stained with streptavidin Alexa 594, indicating the presence of HA in the pericellular matrix. (C) Merged image of PEA and HA staining. (D) The cells were treated with HAase, which degraded the HA pericellular matrix. (E) HAase treatment also diminished cell surface HA staining. (F) Staining cells with B-HABP also detected intracellular HA in permeabilized MM CD19+ B cells. The staining pattern suggests that intracellular HA is distributed along the cell cytoskeleton and perinuclear compartment of MM B cells. Additionally, for one MM patient, weak nuclear staining of HA was observed in CD19+ B cells (not shown). Scale bar = 10 μm. Magnification 40×, numerical aperture 1.3. Images were processed using LSM 510 software.
Discussion
This study shows that circulating MM and MGUS B cells express HAS1 and a family of alternative splice variants and that alternative splicing of the HAS1 gene in MM B cells predicts for reduced survival. The HAS1 gene and its variants are largely absent from non-B cells as well as from BM-localized MM PCs, perhaps reflecting differentiation events within the MM clone. HAS1Vb is the result of abnormal intronic splicing events, which appear to involve the activation cryptic splice sites within HAS1. Similar splicing patterns have been observed for other genes that are associated with malignant phenotypes.27-30 HAS1 and its variants are absent from B cells of healthy donors and from non-B or T cells from blood and BM of MM patients. Analysis of normal human tissues showed up-regulation of HAS1 only in lung tissue (NCBI GEO profiles), consistent with the view that it may be predominantly expressed in malignant cells.31 No HAS1 variants have been reported in the GEO database. Expression of HAS1 variants appears to be exclusive to malignant B lineage cells in MM, MGUS, and Waldenstrom macroglobulinemia.32
In MM, expression of HAS1 variants is restricted to MM CD19+ B cells. BM CD38hiCD45lo PCs expressed HAS1Va and HAS1Vb, at very low levels, in only 1 of 11 MM patients. HAS1Vc transcripts were undetectable in MM PCs. HAS1 variants were absent from T cells obtained from MM PBMCs of 4 of 4 patients and non-B cells (CD19- populations) from the PBMCs of 9 of 9 MM patients. Thus, HAS1 variants are consistently detected in sorted MM CD19+ B cells but not in BM CD38hiCD45lo PCs or in other cell populations comprising PBMCs of MM and MGUS patients or BMCs of MM patients. This suggests biologically important changes in MM gene expression profiles as malignant B cells differentiate to PCs.
Recently, it was reported that expression of HAS1 and HAS2 in HR-3Y1 cells corresponds to the degree of malignant cell transformation.33 Up-regulation of the HAS1 gene was observed in highly malignant cells transformed with v-src and/or with v-fos.33 Although alternative splicing is a normal event contributing to protein diversity in humans, more than a dozen human cancers are associated with abnormalities in alternative splicing, including intronic splicing. One cause of aberrant splicing is mutation, the consequences of which are exon skipping and/or intron retention.34-36 Our study suggests that HAS1 and its variants, particularly HAS1Vb, may contribute to early myelomagenesis because these transcripts are detected individually or in combination with other variants in PBMCs obtained from most MM and MGUS patients at the time of diagnosis.
Longitudinal expression analysis of HAS1 and variants in PBMCs of 18 unselected MM patients showed sporadic expression of HAS1 and its variants throughout disease, with expression in most MM patients at diagnosis (65% of patients) and in relapse (71.4% of patients). This may reflect treatment of many patients with corticosteroids, known to inhibit some HASs and production of HA.25,26,33 Thus, expression of the HAS1 family genes, mainly HAS1Vb, appears to characterize circulating MM cells in the blood of patients at both early and late stages of the disease. Furthermore, the observation that HAS1 and variants are found in PBMCs of MM patients but not healthy donors, together with the association between HAS1Vb and poor survival, suggests that these alternatively spliced HASs are up-regulated at early stages of malignant transformation and may contribute to the spread of malignancy.
Alignment analysis of HAS1 variants with HAS1FL demonstrated that the complete motif of glycosyltransferase is retained, consistent with demonstration that expression of HAS1 and/or HAS1 variants is required for the production of HA. Expression of HAS1Va, which was detected in circulating B cells of 8 of 13 MM patients, correlated with poor survival (P = .048) and may be required for synthesis of extracellular HA. A point mutation 7760C>T detected on the highly conserved exon 3 of HAS1Va could promote activation of cryptic splice sites and consequently mediate the splicing and truncation of HAS1, because this mutation is located in the vicinity of splicing signals of the gene. The occurrence of the point mutation 7760C>T in HAS1Va transcripts and its absence in HAS1FL, HAS1Vb, and HAS1Vc obtained from the same patient suggest the presence of a new variant allele of HAS1 in MM patients. Currently, we are cloning and sequencing the HAS1 gene from genomic DNA of MM patients to clarify whether or not the changes detected on HAS1Va transcript represent mutation or polymorphism.
HAS1Vb, which correlates strongly with poor survival (P = .001), partially retains intron 4 most likely through the activation of cryptic 5′ and/or 3′ splicing sites. Previous analyses of other genes have shown that this type of aberration, the retention of introns during splicing, is uncommon (6%), and it is often associated with short introns.36 In most cases, partial retention of introns appears to be characteristic of genes associated with a malignant phenotype.27-29,37 As shown here, intronic splicing of HAS1 also correlates with the malignant phenotype. The alternatively spliced HAS1Vb transcripts are expressed in circulating B cells from most MM patients at diagnosis and relapse. Alternative splicing of HAS1Vb causes a frameshift and insertion of PTC downstream of the deleted exon and after the retained part of intron, leading to a truncated protein. Moreover, HAS1Vb may be required for synthesis of intracellular HA; only MM B-cell populations expressing HAS1Vb produced intracellular HA. In addition, protein encoded by HAS1Vb, similar to the MDM2 splice variant, may form dimers with wild-type HAS1 and thus alter normal functioning of the full-length HAS1 protein.37,38 HAS1Vc, another intronic splice variant of HAS1 that retains exon 4, shares similar but not identical splicing and expression patterns with HA1Vb. No correlation was found between HAS1Vc and patient survival. HAS1Vc may act in a dominant-negative manner to compromise normal functioning of HAS1.
All 3 HAS1 splice variants are truncated. However, alignment and protein motif screening analysis (Prosite-MotifScan) showed that all 3 variants of HAS1 retain the complete motif of glycosyltransferase, which carries out the synthesis of HA molecules. Protein expression of HAS1 variants is supported by the Western blot analysis conducted on MM cell lines. Furthermore, HAS1Va and/or HAS1Vb, perhaps in concert with HAS3, appear to be required for, respectively, synthesis of extracellular HA matrix or intracellular HA by MM B cells; B cells obtained from healthy donors expressing only HAS3 and MM PCs, which express HAS2 together with HAS3 but lack HAS1, are unable to synthesize extracellular/intracellular HA. Further, expression of HAS1, HAS1Va, and HAS1Vb and detection of an HA matrix correlates with cell motility. Among the B lineage cells in MM, only MM B cells include a motile subset. Up-regulation of HAS1 and variants may contribute to the spread of MM independently or in concert with down-regulation of HAase.
Recently, the interaction between endogenous HA synthesis and multidrug resistance has been documented.39 Ex vivo and in vivo MM B cells, the only components of the MM clone to express the HAS1 family, are highly drug resistant.7 Baumgartner et al showed that HAase treatment improved the effects of various chemotherapeutic agents.40 Thus, synthesis of extracellular HA by MM B cells may impact disease biology by contributing to drug resistance.
Based on alignment and Western blot analysis, transcripts of HAS1 and its variants appear to be translated to form functional proteins. However, for the HAS1 variants, deletion of the entire exon 4 and most of the exon 5 could alter their proper membrane folding, compromising translocation of HA into the extracellular matrix (ECM) and potentially distributing HA molecules into an interior cellular compartment, as detected here (Figure 6F). A similar localization of intracellular HA in Ras-transformed cells has been reported by others.12 We believe that intracellular HA detected in MM B cells is produced by one or more HAS1 variants, particularly HAS1Vb, which is associated with poor survival. Strong perinuclear localization of intracellular HA branches out from the perinuclear compartment toward the cell plasma membrane. Identification of this type of intracellular HA staining pattern suggests that these molecules may also contribute to the process through which malignant cells maintain cellular architecture. Finally, it is intriguing to speculate that the intracellular HA molecules produced by HAS1Vb may through HA binding modulate the function of RHAMM, which is overexpressed and contributes to mitotic abnormalities in MM.41-43
Prepublished online as Blood First Edition Paper, February 24, 2005; DOI 10.1182/blood-2004-10-3825.
Supported by the Canadian Institutes of Health Research and CA80963 from the National Cancer Institute. S.A. supported by the Alberta Heritage Foundation for Medical Research (AHFMR) and National Research Council (NRC); L.M.P. is the Canada Research Chair in Biomedical Nanotechnology.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the many patients at the Cross Cancer Institute and University of Alberta for their generous donations and their participation in this study. The skilled assistance of Viet Hoang, Juanita Wizniak, and Tara Tiffinger is gratefully acknowledged.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal