Abstract
To develop a tool to obtain a high level of gene expression specifically in endothelial cells (ECs), we assessed enhancer activity of fragments in the first intron of the VE-cadherin gene using 3 different experimental systems: luciferase assay in the F2 EC line, green fluorescent protein (GFP) expression in ECs generated in embryonic stem (ES) cell differentiation culture, and GFP expression in transgenic mice. Although the 2.5-kbp (kilobase pair) 5′ flanking sequence of the VE-cadherin gene is EC specific, adding 4 kbp of the 5′ half of the first intron affected an enhancement of the gene expression level in all 3 assay systems. No other fragments tested in this study could confer such effects. Compared with other gene expression units, the unit described in this study would be the most optimum one available to date for EC-specific gene expression. Because this unit can express genes in VE-cadherin+ progenitors of hematopoietic cells but not in fully committed hematopoietic cells, it will be useful to manipulate specifically the uncommitted progenitor stage during hematopoietic cell differentiation. (Blood. 2005; 105:4657-4663)
Introduction
Embryonic stem (ES) cells are now established as a useful source of normal differentiated cells of various lineages, required not only for cell therapies of various degenerative diseases but also for preparing a panel of normal cells for drug screening. Although the advantage of this system lies in its ability to provide essentially unlimited amounts of cells, difficulties exist in steering the differentiation process to produce pure cell populations in culture. To overcome this problem, methods to purify the cells of interest from other unwanted populations have been developed. One such method uses cell sorting technology. We1,2 and others3 demonstrated that a set of surface markers can be used for defining and purifying various cell populations in the ES cell culture. Moreover, beta-galactosidase and green fluorescent protein (GFP) genes driven by specific promoters are also useful for purifying a subset of cells, particularly when applied in combination with surface markers. Another method is the drug-mediated selection of a particular lineage by which drug-resistant genes, driven by cell lineage-specific promoter units, are introduced into ES cells, thereby allowing the depletion of unwanted cell lineages during culture.4 In both cases, the success of purification depends entirely on the availability of the lineage-specific promoter/enhancer unit. Moreover, such promoters are essential for expressing exogenous genes specifically in the cell lineage.
In endothelial cell (EC) lineages, many promoters have been developed and shown useful for EC-specific gene expression. To our knowledge, however, most available promoters have a problem of expression in non-EC lineages. For instance, promoters of P-selectin and von Willebrand factor are active in megakaryocytes,5,6 preproendothelin-1 is active in arterial smooth muscle cells,7 and intercellular adhesion molecule-2 (ICAM-2) and platelet endothelial cell adhesion molecule-1 (PECAM-1) are active in hematopoietic cells.8,9 Promoters for Tie-1 and Tie-2, which are most commonly used as endothelial-specific promoters, are also expressed in hematopoietic cells in the embryo and the adult.10 The promoter of vascular endothelial growth factor receptor 2 (VEGFR2) has also been reported to be specific to ECs, but we have shown that this promoter is active in undifferentiated ES cells.11
In contrast to the molecules just described, VE-cadherin (VECD) is known to be expressed specifically and continuously by ECs.12 Thus, Gory et al13 attempted to isolate endothelial-specific regulatory elements from the VECD gene and found that a 2.5-kbp (kilobase pair) upstream region of the coding sequence can drive the EC-specific gene expression in vivo. Although the endothelial specificity of this regulatory unit appears to be sufficient, our preliminary data suggest that the overall activity of this unit is weak and, in some ECs, such as those in brain capillaries, completely inactive.
The aim of the present study is to investigate the possibility of modifying this unit to obtain a stronger yet endothelial-specific regulatory unit that will be useful for vascular biology. We will show the presence of an enhancer region in the first intron of the VECD gene, which can augment the activity of the 2.5-kbp 5′ promoter region while maintaining endothelial specificity. We will also show that this unit can be used in ES cell differentiation culture and in transgenic mice.
Materials and methods
Monoclonal antibodies
The monoclonal antibodies (mAbs) AVAS12 (anti-VEGFR2)14 and VECD1 (anti-VECD)15 were purified from hybridoma culture supernatants by protein G-Sepharose columns (Pharmacia, Uppsala, Sweden) and were labeled with allophycocyanin (APC) by standard methods. Unconjugated or phycoerythrin (PE)-conjugated anti-Gr1, anti-CD31, anti-B220, anti-CD45, and Mac1 mAbs were purchased from PharMingen (San Diego, CA).
Isolation and analysis of genomic clones
A genomic clone that contains exon 1 of the VECD gene was isolated from a 129/svj mouse genomic library in the FIX II vector (Stratagene, La Jolla, CA) by screening with a 219-bp fragment spanning the exon 1 sequence of the VECD gene. This probe was generated from mouse genomic DNA by reverse transcription-polymerase chain reaction (PCR) using the following primers: sense, 5′-ACA AAG GAA CAA TAA CAG GAA ACC-3′; antisense, 5′-CAG GAG TAG AGA GGA ATT TGG GAG-3′). The restriction enzyme map of this clone was consistent with that reported previously by Huber et al.16
According to the restriction map, the 6.8-kbp EcoRI-EcoRI fragment that contains exon 1 and the 5′ flanking sequence of the VECD gene was excised and subcloned to pBluescript II SK (pBS SK; Stratagene). A 9-kbp PstI-NotI (a site derived from FIXII vector) fragment, which contains most of the first intron, was also subcloned to PstI-NotI sites of pBS SK (pBS-VECD Int1). Either 4.5 kbp of the BglII-NotI fragment containing the 3′ half of the first intron or 4 kbp of the PstI-BglII fragment containing the 5′ half of the first intron was excised from pBS-VECD-Int1, thereby making a new construct, pBS-VECD-5′ Int1 or pBS-VECD-3′ Int1.
Enhanced GFP reporter constructs
pEGFP-N3 vector (Clontech, Palo Alto, CA) was used to measure promoter/enhancer activity in terms of the intensity of GFP expression. To make a construct containing the promoter of the VECD gene, 2.8-kbp SacI-PstI of the VECD 5′ flanking fragment was first inserted in SacI-PstI sites of the pEGFP-N3 vector, and then the150-bp PstI-PstI fragment was added to the PstI site of the resultant construct. The orientation of inserted fragments was confirmed by sequencing. The SpeI-NotI fragment, which contains the promoter fragment of the VECD and GFP genes, was excised from this construct and was inserted in the pBSII KS (+) vector containing the SV40 intron/polyadenylation site derived from pCDM8 (Invitrogen, Carlsbad, CA). The resultant plasmid was designated pVECDp-GFP. The ClaI-BssHII fragment containing VECDp-GFP was prepared and inserted in the ClaI site of either pBS-VECD-5′ Int1 or pBS-VECD-3′ Int1, and the resultant constructs were designated pVECDp-GFP-5′ Int1 and pVECDp-GFP-3′ Int1, respectively. These constructs were linearized by BssHII digestion before transfection to ES cells. The ES cells were electroporated with linearized plasmids and pPGKpuro, which carries a puromycin-resistant gene for selecting gene-transduced ES cells.
Luciferase reporter constructs
A 2.5-kbp EcoRV-SalI fragment was prepared from pVECDp-GFP and was inserted in the multiple cloning site of PGL3 basic containing the firefly luciferase gene (Promega, Madison, WI). The blunted SalI site of this construct was inserted with 0.8 kbp of Pst1-SacI, 1.5 kbp of the PstI-HindIII fragment, 4 kbp of the EcoRV-BglII fragment, or 4.5 kbp of the BglII-NotI fragment from pVECD-Int1 (Figure 1).
Transient transfection was performed using Effectene (Qiagen, Valencia, CA) according to the protocol recommended by the manufacturer. PRL-TK (Promega), which contains a cDNA encoding Renilla luciferase, was cotransfected with pGL3 vectors to normalize the firefly luciferase activity. Luciferase activity was measured using Dual-Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions.
Enhancer activity in the first intron of the vecd gene. Four different fragments indicated by thick lines in this figure were prepared from the first intron of the VECD gene (a restriction map of the first intron was given at the top). Enhancer activity of each fragment was assessed by using the luciferase gene driven by 2.5-kbp 5′ promoter region of the VECD gene. Luciferase activity was measured either in NIH3T3 (□) or F2 (▪) after transient transfection of each construct. Each bar is the mean ± SD of 5 independent experiments.
Enhancer activity in the first intron of the vecd gene. Four different fragments indicated by thick lines in this figure were prepared from the first intron of the VECD gene (a restriction map of the first intron was given at the top). Enhancer activity of each fragment was assessed by using the luciferase gene driven by 2.5-kbp 5′ promoter region of the VECD gene. Luciferase activity was measured either in NIH3T3 (□) or F2 (▪) after transient transfection of each construct. Each bar is the mean ± SD of 5 independent experiments.
NIH3T3 and F2 mouse EC lines were cultured in Dulbecco modified Eagle medium (DMEM; Gibco BRL, Grand Island, NY) supplemented with 10% fetal calf serum.
GFP reporter assay in ES cell differentiation cultures
EB5, an ES cell line derived from the E14tg2a ES cell line, was a kind gift from Dr H Niwa (Riken Center for Developmental Biology, Kobe, Japan). In EB5, the Aspergillus blastcidin-S deamininase gene was inserted into an Oct-3/4 allele by homologous recombination. This cell line allows us to select immature cells expressing Oct-3/4 by blastcidine-S hydrochloride (Blast S; Funakoshi, Tokyo, Japan) selection. EB5 was maintained with Glasgow MEM (G-MEM; Gibco BRL) supplemented with 10% knockout serum (Gibco BRL), 1% fetal calf serum (FCS; Whittaker Bioproducts, Walkersville, MD), 100 μM 2-mercaptoethanol (2 ME; Merck, Darmstadt, Germany), 10 mM nonessential amino acids (Gibco BRL), 1 mM sodium pyruvate (Sigma), 20 μg/mL Blast S, and 1000 U/mL leukemia inhibitory factor (LIF) in culture dishes coated with gelatin (type A from porcine skin; Sigma, St Louis, MO). The OP9 stromal cell line was maintained in αMEM (Gibco BRL) supplemented with 20% FCS (HyClone Laboratories, Logan, UT).17
Gene transfection and selection of transduced ES cells were performed as described previously.11 GFP expression was assessed by FACS Vantage or FACS Calibur (Becton Dickinson Labware, Bedford, MA).
Two-step culture for the induction of ES cell differentiation to ECs was carried out as described previously.18 Briefly, ES cells were cultured on a type 4 collagen-coated plate (Biocoat; Becton Dickinson) and incubated for 4 days in αMEM (Gibco BRL) supplemented with 10% FCS in the absence of LIF. Cultured cells were harvested with cell dissociation buffer (Gibco BRL), and VEGFR2+ cells were sorted and recultured on OP9. Cells were recovered 1 to 5 days later and analyzed for expression of GFP.
GFP expression during the differentiation of hemogenic ECs to hematopoietic cells was also examined. VECD+ CD31+CD45- ECs were obtained by incubating ES cells for 7 days on OP9. GFP+ cells were further selected and cultured under conditions to induce various hematopoietic cells. For the induction of myeloid cells, sorted cells were transferred to fresh OP9 cells in the medium supplemented with a mixture of recombinant growth factors containing 200 U/mL murine interleukin-3 (IL-3), 2 U/mL human erythropoietin (EPO), 100 ng/mL human granulocyte colony-stimulating factor (G-CSF), and 100 ng/mL murine c-Kit ligand (SCF). For the induction of B-lymphoid cells, sorted cells were cultured with OP9 stromal cells in the presence of 100 ng/mL SCF, 100 ng/mL human Flt3/Flk-2 ligand (FL), and 200 U/mL murine IL-7. Recombinant EPO, G-CSF, and FL were purchased from R&D Systems (Minneapolis, MN). Recombinant murine IL-3, SCF, and IL-7 were prepared as described.19 Cultured cells were analyzed for expression of surface markers, and GFP was analyzed by flow cytometry.
Generation and analysis of transgenic mice
Linearized plasmids were purified by NACS PREPAC (Gibco BRL), suspended at 5 ng/mL, and injected into mouse oocytes as described.20 Transgenic integration was confirmed by polymerase chain reaction (PCR) of genomic DNA obtained from mouse ears. A primer set, 5′-ACAAAGGAACAATAACAGGAAACC-3′ and 5′-GAACTCCAGCAGGACCATGT-3′, was derived from the VECD promoter region (5′) and the egfp gene (3′), and another primer set, 5′-ACATACGGAAAACTTACCCTT-3′ and 5′-GGAATCACAGAGCCGTTATT-3′, was derived from the EGFP gene (5′) and the VECD 5′ Intron 1 (3′), respectively. Four independent transgenic lines were established and analyzed for the expression of GFP. In some experiments, embryos were harvested, dissociated to single-cell suspensions, and analyzed by flow cytometry. For immunofluorescence staining, tissues were fixed in 4% paraformaldehyde, embedded in optimum cutting temperature (OCT) compound, and cryosectioned. Sections (9-12 μm) were stained with rabbit anti-GFP antibody and rat anti-PECAM-1 mAb and were developed by Alexa 488-conjugated anti-rabbit immunoglobulin G (IgG) (H+L) and Alexa 564-conjugated anti-rat IgG (H+L) antibodies (Molecular Probes, Eugene, OR). Samples were mounted with ProLong Antifade kit (Molecular Probes). Immunostained specimens of the spleen and embryo were detected using a DMIR/E2 microscope (LEICA-Microsystems, Wetzlar, Germany) equipped with HC PL APO 40/1.25-0.75 oil (Figure 3A spleen) and HC PL APO 20/0.70 objective lens (Figure 3A embryo) with a TCS SP2 AOBS spectral confocal scanning system (LEICA-Microsystems). Acquisition software was Leica Confocal Software (LEICA-Microsystems). Other specimens in Figure 3A and Figure 4 were visualized using Axiovert 200 microscope (Carl Zeiss, Oberkochen, Germany) equipped with Plan-Apochromat 63/1.4 oil (Figure 3), Plan-Apochromat 10/0.45 (Figure 4 A, B, and C), and Plan-Apochromat 20/0.60 objective lens (Figure 4 D, E, and F) with a Radiance 2100 (Bio-Rad, Hercules, CA). Acquisition software was LaserSharp 2000 (Bio-Rad). The images were further processed using Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Inflammatory neovascularization was induced by topical application of 2 μL 0.15 M NaOH to the corneal surface and subsequent scraping of the corneal and limbal epithelia with the blunt end of a razor blade and a cotton-wool swab in a rotary motion parallel to the limbus. To prevent infection, erythromycin ophthalmic ointment was instilled immediately after epithelial denudation. All procedures were carried out under an anesthesia with sodium-pentobarbital under the guidelines of the Stem Cell Biology Laboratory, Center for Developmental Biology (Kobe, Japan), and this study protocol was approved by the review board of the Center for Developmental Biology.
Results
Enhancer activity of the first intron of the vecd gene on EC-specific gene expression
Gory et al13 previously reported that the 2.5-kbp 5′ flanking region of the VECD gene (VECDp) could allow EC-specific gene expression in EC lines and in ECs of the transgenic mouse. However, we had difficulty using VECDp to obtain a high expression level of GFP in living ECs generated in the differentiation culture of ES cells (data shown in Figure 2). The fact that cells expressing endogenous VECD are readily detected by flow cytometry in the same culture indicates a requirement of an appropriate enhancer region for facilitating the usefulness of VECDp.
According to a previous analysis of the enhancer in the first intron of the VEGFR2 gene, Ets-1, GATA, and SCL/TAL-1 binding sites in this region were implicated as essential for the endothelial-specific enhancement of gene expression in the transgenic mouse.21-23 To identify similar enhancer activity in the VECD gene, we screened the Ensemble genome database and found multiple binding sites for those transcriptional regulators in the first intron. We consequently examined the possibility that this first intron of the VECD gene enhances the promoter activity of VECDp in ECs.
The fragments indicated by thick lines in Figure 1 were conjugated to the luciferase gene driven by VECDp, and then luciferase activity was measured in NIH3T3 fibroblast cell and F2 EC lines. Addition of the 4-kbp 5′ fragment of the first intron (5′ Int1) effected significant enhancement of the promoter activity of VECDp, whereas no enhancement was attained by the other fragments of the first intron, including a 4.5-kbp 3′ fragment (3′ Int1) (Figure 1). This result suggests the presence of an enhancer element in the 5′ Int1.
We next examined the enhancer activity of 5′ Int1 on VECDp-driven GFP expression of ECs generated in the ES cell differentiation culture. In ES cell cultures, multiple cell lineages were generated at the same time, enabling us to evaluate the specificity of promoter activity by comparison with EC and non-EC lineages distinguished by the expression of endogenous VECD. Either VECDp-EGFP, pVECDp-5′ Int1-EGFP, or pVECDp-3′ Int1-EGFP was stably transfected to ES cells. The gene-transduced ES cell clones were then induced to differentiate to ECs. GFP expression was measured by FACS Calibur 5 days after culturing VEGFR2+ cells on OP9 (Figure 2). Under this culture condition, multiple mesendodermal cells, including hematopoietic cells, are generated. In this experimental setting, all non-EC lineages are expected to be included in the VECD- population, and the expression level of endogenous VECD on the cell surface serves as a reference for the specificity and quantitative intensity of the enhancer activity.
Enhancer activity of the 5′ half of the first intron of the vecd gene in ES cell differentiation cultures. The first intron of the VECD gene was split into 4-kbp 5′ (5′ Int1) and 4.5-kbp 3′ (3′ Int1) fragments, and their enhancer activity was assessed by using the EGFP gene driven by 2.8-kbp 5′ flanking region of the VECD gene (VECDp). ES cell clones harboring VECDp-egfp, VECDp-5′ Int1-EGFP, or VECDp-3′ Int1-egfp were established and induced to differentiate into ECs. GFP expression in the cells in each culture were assessed after staining with anti-VECD mAb and were analyzed using FACS Vantage. Each panel represents the result of clones that showed the highest GFP expression level among each group.
Enhancer activity of the 5′ half of the first intron of the vecd gene in ES cell differentiation cultures. The first intron of the VECD gene was split into 4-kbp 5′ (5′ Int1) and 4.5-kbp 3′ (3′ Int1) fragments, and their enhancer activity was assessed by using the EGFP gene driven by 2.8-kbp 5′ flanking region of the VECD gene (VECDp). ES cell clones harboring VECDp-egfp, VECDp-5′ Int1-EGFP, or VECDp-3′ Int1-egfp were established and induced to differentiate into ECs. GFP expression in the cells in each culture were assessed after staining with anti-VECD mAb and were analyzed using FACS Vantage. Each panel represents the result of clones that showed the highest GFP expression level among each group.
Activity of VECDp-5′ Int1 in transgenic mice
It was previously shown that VECDp is sufficient for EC-specific gene expression in transgenic mice.13 In our transgenic mice harboring VECDp-EGFP, we could detect EC-specific GFP expression in the small intestine, spleen, kidney, and heart to varying extents (Figure 3A). However, in all VECDp-EGFP transgenic mice produced in this study, it was difficult to detect GFP expression in the brain and retina. In contrast, GFP expression was detected in the ECs of retina and brain of transgenic mice bearing VECDp-5′ Int1-EGFP (Figure 3A). In addition, in organs in which GFP expression is detectable in VECDp-EGFP transgenic mice, more ECs express GFP in VECDp-EGFP-5′ Int1 transgenic mice than in VECDp-EGFP mice.
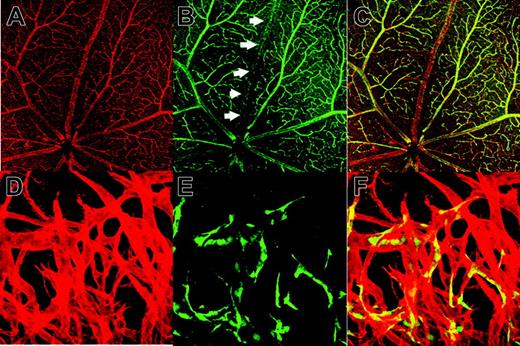
Enhancer activity of 5′ Int1 of the VECD gene in transgenic mice. Transgenic mouse strains harboring VECDp-EGFP or VECDp-5′ Int1-EGFP were established. (A) Histologic sections of the small intestine, retina, spleen, brain cortex, kidney, or heart were prepared from 8- to 10-week-old transgenic mice and were stained by anti-GFP antibody (green) and anti-PECAM-1 mAb (red). Two pictures are merged to show the specificity of expression. GFP expression was detected at varying extent in the small intestine, spleen, kidney, and heart of the VECDp-EGFP transgenic mouse, but not in the brain and retina of the same mouse. On the other hand, a high level of GFP expression was detected in all these organs of VECDp-5′ Int1-EGFP transgenic mice. Note, however, the presence of GFP- PCAM-1+ cells in these tissues. Sagittal sections of E12.5 embryos were prepared from 2 stains of mouse, and expression of GFP and PCAM-1 was examined immunohistochemically. Arrows indicate the dorsal aorta. (B) E12.5 embryos of VECDp-EGFP or VECDp-5′ Int1-EGFP transgenic strains were collected, dissociated to single-cell suspension, stained by anti-VECD CD31 mAbs, and analyzed by FACS Vantage. Expression of VECD and GFP of the CD31+ population is presented.
Enhancer activity of 5′ Int1 of the VECD gene in transgenic mice. Transgenic mouse strains harboring VECDp-EGFP or VECDp-5′ Int1-EGFP were established. (A) Histologic sections of the small intestine, retina, spleen, brain cortex, kidney, or heart were prepared from 8- to 10-week-old transgenic mice and were stained by anti-GFP antibody (green) and anti-PECAM-1 mAb (red). Two pictures are merged to show the specificity of expression. GFP expression was detected at varying extent in the small intestine, spleen, kidney, and heart of the VECDp-EGFP transgenic mouse, but not in the brain and retina of the same mouse. On the other hand, a high level of GFP expression was detected in all these organs of VECDp-5′ Int1-EGFP transgenic mice. Note, however, the presence of GFP- PCAM-1+ cells in these tissues. Sagittal sections of E12.5 embryos were prepared from 2 stains of mouse, and expression of GFP and PCAM-1 was examined immunohistochemically. Arrows indicate the dorsal aorta. (B) E12.5 embryos of VECDp-EGFP or VECDp-5′ Int1-EGFP transgenic strains were collected, dissociated to single-cell suspension, stained by anti-VECD CD31 mAbs, and analyzed by FACS Vantage. Expression of VECD and GFP of the CD31+ population is presented.
The specificity of this enhancer region was also evaluated in embryos. In embryonic day 12.5 (E12.5) embryos of VECDp-5′ Int1-EGFP transgenic mice, PECAM-1 expression correlates well with that of GFP expression (Figure 3A), whereas only a low level of GFP expression was detected in the ECs of VECDp-EGFP transgenic embryos at the same age.
Because of a technical difficulty in fixation procedure, we could not find the optimum condition for dual-color immunohistochemistry with anti-VECD mAb and the anti-GFP antibody. To evaluate the expressions of GFP and endogenous VECD at the same time, we used flow cytometry and compared gene expression activity of VECDp and VECDp-5′ Int1 in ECs dissociated from embryos (Figure 3B). Although VECDp-5′ Int1-EGFP transgenic mice expressed a significant level of GFP, the expression in VECDp-EGFP transgenic mice remained at a low level. Moreover, expression levels of GFP and endogenous VECD showed a good correlation quantitatively. Thus, the 5′ Int1 enhancer is useful for high-level, EC-specific gene expression in transgenic mice.
Although GFP expression was essentially confined to ECs in these transgenic mice, we recognized that GFP expression is not detected in all ECs even with VECDp-5′ Int1. In the kidney of this transgenic strain, for example, a considerable proportion of PECAM-1+ cells were GFP-. This problem was further investigated in the adult retina, in which the vascular architecture is easily visualized (Figure 4A-C). In the adult retina of mouse, PECAM-1 is expressed in most ECs. In contrast, GFP expression appears to be suppressed specifically in ECs of venous trunks. Nonetheless, the suppression of VECDp-5′ Int1 activity is not necessarily specific to venous vessels because only a small proportion of ECs in the neo-angiogenesis site of cornea express GFP and GFP+ ECs are distributed randomly (Figure 4D-F). In this experimental setting, vascular sprouting into cornea occurs only inducibly; thus, all ECs in sprouted vasculatures corresponded to newly formed ECs. In all whole-mount specimens analyzed in Figure 4, no GFP expression was detected in round cells in vascular lumen. All histologic analyses indicated that VECDp-5′ Int1 is highly specific to ECs during embryogenesis and postnatal life but that its activity is variable among ECs.
GFP- ECs in the VECDp-5′ Int1- egfp. Flat-mount specimen of a retina of an 8-week-old VECDp-5′ Int1-egfp transgenic mouse was stained with (A) anti-PECAM-1 mAb (red) (B) anti-GFP antibody (green). Arrows indicate a venous trunk in this retina, in which GFP expression was not detected. (C) Overlay. In one experiment (C-D), neo-angiogenesis was induced in the cornea of this transgenic strain. Whole-mount specimen of a newly formed vascular plexus was stained by (D) anti-PECAM-1 mAb (red) and (E) anti-GFP antibody (green). (F) Overlay. Only portions of ECs in this region are GFP+, and they are distributed randomly.
GFP- ECs in the VECDp-5′ Int1- egfp. Flat-mount specimen of a retina of an 8-week-old VECDp-5′ Int1-egfp transgenic mouse was stained with (A) anti-PECAM-1 mAb (red) (B) anti-GFP antibody (green). Arrows indicate a venous trunk in this retina, in which GFP expression was not detected. (C) Overlay. In one experiment (C-D), neo-angiogenesis was induced in the cornea of this transgenic strain. Whole-mount specimen of a newly formed vascular plexus was stained by (D) anti-PECAM-1 mAb (red) and (E) anti-GFP antibody (green). (F) Overlay. Only portions of ECs in this region are GFP+, and they are distributed randomly.
Activity of VECDp-5′ Int1 in hematopoietic cell lineages
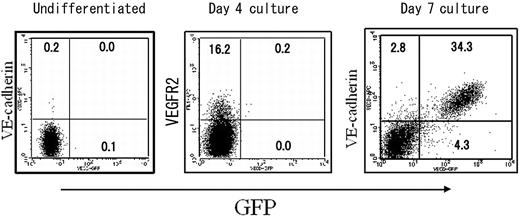
We next investigated the detailed specificity of this VECDp-5′ Int1 in ES cell differentiation culture on OP9. In this culture, VEGFR2+VECD- mesoderm cells appeared approximately 4 days after the incubation of ES cells on OP9, and VECD+ ECs started to appear one day later. To examine the specificity and activity of VECDp-5′ Int1 in ES cell differentiation cultures, we analyzed the GFP expression of undifferentiated ES cells and that of cells harvested after 4- or 7-day incubation of ES cells on OP9. As shown in Figure 5, GFP is not expressed in undifferentiated ES cells or in cells from 4-day-old ES cell cultures, including VEGFR2+ mesoderm cells. On the other hand, strong expression was detected in most VECD+ ECs generated in 7-day-old ES cell cultures. Consistent with results from the previous sections, nearly all GFP+ cells coexpressed VECD, and expression levels of GFP and VECD on the surfaces of ECs correlated well. In this experimental condition, not only were ECs and hematopoietic cells induced, endodermal and somitic cells were induced at the same time. Thus, at least in ES cell differentiation cultures, VECDp-5′ Int1 is highly specific to ECs.
EC-specific gene expression by VECDp-5′ Int1 in ES cell differentiation cultures. The same VECDp-5′ Int1-EGFP ES cell clone as that presented in Figure 2 was cultured on OP9 for inducing differentiation. Undifferentiated ES cells and cells harvested 4 or 7 days after the induction of differentiation were stained with anti-VECD mAb and analyzed by FACS Calibur.
EC-specific gene expression by VECDp-5′ Int1 in ES cell differentiation cultures. The same VECDp-5′ Int1-EGFP ES cell clone as that presented in Figure 2 was cultured on OP9 for inducing differentiation. Undifferentiated ES cells and cells harvested 4 or 7 days after the induction of differentiation were stained with anti-VECD mAb and analyzed by FACS Calibur.
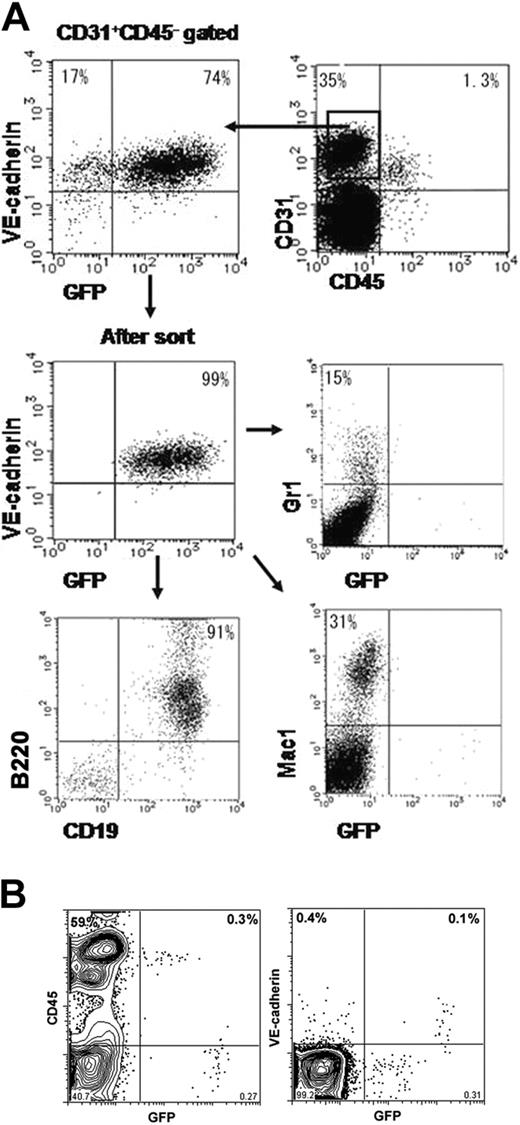
Our previous study showed that a subset of VECD+ ECs could give rise to various hematopoietic cells.2 It was difficult to obtain cis-regulatory regions that enabled us to express the gene in this VECD+ hematopoietic progenitor but not in CD45+ hematopoietic cells. For instance, the promoter/enhancer region of the VEGFR2 gene that appeared to be the most EC specific24 failed to express the gene in this population.11 Thus, we investigated whether GFP+ ECs generated from VECDp-5′ Int1-EGFP ES cells contained progenitors of hematopoietic cells. ES cells were cultured on OP9 for 7 days, and VECD+CD31+CD45-GFP+ cells were sorted and recultured in OP9 for another 1 to 2 weeks.
As shown in Figure 6A, Gr-1+ granulocyte, Mac1+ monocyte, and B220+CD19+ B-lymphocyte lineages were generated from GFP+ cells. In contrast, these 3 hematopoietic cell lineages generated from GFP+ cells are GFP-. This result indicates that VECDp-5′ Int1 can dictate the expression of endogenous VECD during differentiation of hemogenic ECs to hematopoietic cells.
We also attempted to assess GFP expression in hematopoietic tissues of VECDp-5′ Int1 transgenic mice. As shown in Figure 6B, all ECs that expressed endogenous VECD expressed a high level of GFP, though the proportion was only 0.1% of total dissociated cells. In addition, approximately 0.3% of dissociated cells expressed GFP at the level 5- to 10-fold lower than that of VECD+ ECs. All GFPdull cells were CD45+ but did not express endogenous VECD. In the same sample, nearly 60% of cells were CD45+ so that the GFPdull population comprised approximately 0.5% of total hematopoietic cells in neonatal spleens. This result suggests that 5′ Int1 may direct an ectopic expression of gene in some hematopoietic cells, though its expression level may be low.
Discussion
The aim of this study was to develop a tool that directs gene expression specifically in ECs in the differentiation culture of ES cells. In surveying the literature, we found that 2 units would be most promising in terms of EC-specific gene expression: the 5′ promoter/first intron enhancer combination of VEGFR2 gene24 and the 5′ promoter of the VECD gene.13 Although other gene expression units have been reported and have been used to express exogenous genes in ECs, all appeared to be active in other cell lineages such as hematopoietic cells. Thus, we chose these 2 units and investigated the possibility of using EC-specific regulatory units in ES cell differentiation culture and transgenic mice.
First, we analyzed the activity of the promoter/first intron of VEGFR2 that had been originally reported by Kappel et al.24 Contrary to our expectation, we found that it is ectopically active in undifferentiated ES cells, though its activity is strictly confined to ECs in later stages.11 Another problem of this unit was that it is not active in ECs with hemogenic activity, which is an important stage during the development of hematopoietic stem cells.2 Thus, this unit may not be ideal for EC-specific gene expression in the ES cell differentiation culture, particularly to follow the differentiation course of VECD+ cells to hematopoietic cells.
Activity of VECDp-5′ Int1 in hematopoietic cell differentiation. (A) ES cells were cultured on OP9 for 7 days to induce ECs. Activity of VECDp-5′ Int1 in hemogenic ECs was tested by culturing GFP+CD31+VECD+CD45- cells on OP9 under appropriate culture conditions for inducing hematopoietic cell lineages. Gr1+ granulocytes, Mac1+ monocytes, and CD19+B220+ B lymphocytes were induced from GFP+ population, indicating that VECDp-5′ Int1 was active also in hemogenic ECs. GFP expression could not be detected in hematopoietic cell lineages in this culture. (B) GFP expression was measured in the neonatal spleen cells from VECDp-5′ Int1-EGFP transgenic mice after staining with anti-CD45 mAb or anti-VECD mAb. All VECD+ cells expressed a high level of GFP. A dull GFP expression was also detected in a low proportion of CD45+ cells.
Activity of VECDp-5′ Int1 in hematopoietic cell differentiation. (A) ES cells were cultured on OP9 for 7 days to induce ECs. Activity of VECDp-5′ Int1 in hemogenic ECs was tested by culturing GFP+CD31+VECD+CD45- cells on OP9 under appropriate culture conditions for inducing hematopoietic cell lineages. Gr1+ granulocytes, Mac1+ monocytes, and CD19+B220+ B lymphocytes were induced from GFP+ population, indicating that VECDp-5′ Int1 was active also in hemogenic ECs. GFP expression could not be detected in hematopoietic cell lineages in this culture. (B) GFP expression was measured in the neonatal spleen cells from VECDp-5′ Int1-EGFP transgenic mice after staining with anti-CD45 mAb or anti-VECD mAb. All VECD+ cells expressed a high level of GFP. A dull GFP expression was also detected in a low proportion of CD45+ cells.
The next candidate was the 2.5-kbp 5′ promoter region of the VECD gene because Gory et al13 indicated that it could dictate the endogenous expression of VECD by multiple criteria. Our present study confirmed their results. As shown in Figure 2, the expression of GFP driven by VECDp and endogenous VECD is well correlated in cells generated from ES cells. However, the GFP expression level was not yet high enough to be used as a convenient unit for EC-specific gene expression. Moreover, in our transgenic experiments, it was difficult to detect GFP expression in ECs of several organs, such as the adult brain and retina of VECDp-EGFP transgenic mice. We established 4 transgenic strains, and none of them showed detectable levels of GFP expression in those organs. Our failure could not rule out the possibility that an appropriate transgenic strain can be obtained by repeated trials. In fact, the initial report by Gory et al13 demonstrated that this unit could dictate the expression of the endogenous VECD gene when used to express the luciferase gene. Nonetheless, it is clear that VECDp alone is unable to attain consistent EC-specific expression in transgenic mice and in ES cell differentiation cultures.
To attain higher EC-specific gene expression, we next explored cis-regulatory regions that could enhance the activity of VECDp. Although multiple cis regions may regulate the expression of endogenous VECD, we began by assessing the activity of the first intron because it contains multiple binding regions for Ets-1, Tal-1/SCL, and GATA, which have been shown to be essential for the enhancer activity of the first intron in the VEGFR2 gene. We found enhancer activity in the first intron of the VECD gene. Hence, through all 3 methods used in this study, 5′ Int1 of the VECD gene enhanced the activity of VECDp without altering specificity.
To assess the intensity and specificity of the enhancer activity of 5′ Int1 in ES cell cultures, we used flow cytometry analysis. It should be noted that flow cytometry analysis of living cells enables a more quantitative measurement of intensity and specificity of gene expression than immunohistochemical analysis of tissues. Flow cytometry analysis is particularly useful in such a molecule as VECD because it allows comparison of the expression levels of the marker gene with those of the endogenous molecules on the cell surface, which are readily detectable by cell surface staining.
In this study, we established at least 3 independent ES cell clones for VECDp-EGFP with or without 5′ Int1 and selected the cell lines expressing the highest level of GFP within each group. Although there were variations in the level of GFP expression among gene-transduced ES cell clones, none of the clones transduced with VECDp-5′ Int1-EGFP showed a lower level of GFP expression than clones harboring VECDp-EGFP. As shown in Figure 2, we compared the 2 best clones representing the groups transduced with either VECDp-EGFP or VECDp-5′ Int1-EGFP and could see that an enhancement of GFP expression is attained by adding the 5′ Int1 enhancer to VECDp. Moreover, adding this enhancer fragment has an effect in reducing the variation in the level of gene expression driven by VECDp. Indeed, all 3 ES clones transduced independently with VECDp-5′ Int1-EGFP could generate GFP+VECD+ ECs, whereas only 1 of 4 clones bearing VECDp-EGFP could do so. Finally, the expression levels of GFP and endogenous VECD showed a correlation in specificity and intensity. Hence, this unit provided a useful tool for EC-specific gene expression in ES cell cultures.
Our results showed that this unit is also useful for EC-specific gene expression in transgenic mice. As shown in Figure 3A, this unit is active in ECs of many tissues in embryos and adults. The addition of this enhancer eventually conferred the ability to express genes in ECs of the brain and retina, which had been difficult with VECDp alone. Analysis of dissociated cells from transgenic embryos also demonstrated that VECDp-5′ Int1 has activity that was stronger than that of VECDp and that was specific to ECs. However, we also noticed the presence of ECs in which VECDp-5′ Int1 was not active. For instance, ECs of venous trunks in the retina of VECDp-5′ Int1-EGFP transgenic mice did not express GFP. Moreover, though this unit was active in most ECs during early embryogenesis, a large proportion of ECs in the site of neoangiogenesis of the cornea failed to express GFP. Thus, there was variation in the expression level of VECDp-5′ Int1 among ECs. At present, it is difficult to find a specific reason for this variability, but it should be noted that even the expression level of endogenous VECD, as measured by fluorescence-activated cell sorter (FACS), sometimes varies by a magnitude of 10-fold (Figure 3B). Thus, some of variability in the activity of VECDp-5′ Int1 may reflect that of endogenous VECD. Nonetheless, it is likely that more cis-regulatory regions are involved in the regulation of the endogenous VECD gene. In summary, this unit may be superior to the previous units developed for EC-specific gene expression, but it may not be complete.
It is now widely accepted that at least a part of hematopoietic cells is derived from VECD+ ECs. Given that this is the case, the transition from VECD+CD45- to VECD- CD45+ stages is the most important step during the commitment of hematopoietic stem cells. To our knowledge, VECDp-5′ Int1 is still the only unit that can express the gene in VECD+ hemogenic ECs but not hematopoietic cells. Indeed, GFP expression is lost during the differentiation of CD45+ hematopoietic cells from GFP+ ECs because none of the Gr1+ granulocytes, Mac1+ macrophages, or CD19+ B lymphocytes derived from GFP+ cells expressed GFP. Hence, this unit does dictate the expression of endogenous VECD during the differentiation of hemogenic ECs to hematopoietic cells and, therefore, would provide a useful tool for manipulating this step. However, we also found a small fraction of hematopoietic cells in the transgenic mouse express GFP, though its level is 5- to 10-fold lower than that of ECs. Because this population does not express endogenous VECD, this expression is ectopic. Whether this small population of cells with ectopic GFP expression represents a unique subset of hematopoietic lineage is an interesting question for the future. In conclusion, the unit described in this study is superior to previous units for a high level of EC-specific gene expression in many aspects and will be a useful tool for gene expression for vascular biology and hematology.
Prepublished online as Blood First Edition Paper, March 3, 2005; DOI 10.1182/blood-2004-09-3554.
Supported by the Leading Project Grant for Realization of Regenerative Medicine from the Ministry of Education and Science of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr A. Togawa for advice and assistance with screening the Ensemble database and Dr K. Yamaguchi for critical reading of the manuscript.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal