Abstract
T-cell receptor (TCR) with unique major histocompatibility complex (MHC)-unrestricted antigen-binding properties was isolated from a human T-cell clone specific for the tumor antigen MUC1. This TCR binds its epitope on the MUC1 protein without the requirement of processing and presentation. A single-chain Vα/Vβ/Cβ (scTCR) was fused to a CD3 zeta (ζ) chain to allow expression on the surface of cells of the innate (granulocytes, macrophages, natural killer [NK] cells) as well as the adaptive (T and B cells) immune system. To test the ability of the cells of the innate immune system to reject a tumor when provided with a tumor antigen-specific TCR, we reconstituted severe combined immunodeficiency (SCID) mice with bone marrow cells transduced with a retroviral vector encoding this receptor and challenged them with a MUC1-positive human tumor. These mice controlled the growth of the tumor significantly better than the control mice. We performed a similar experiment in immunocompetent mice transgenic for human MUC1. Expression of the TCR on large percentages of cells did not result in infiltration or destruction of tissues expressing MUC1. Reconstituted mice controlled the outgrowth of a MUC1-transfected but not the parental control tumor. scTCR expression appears lifelong, suggesting a successful transduction of the self-renewing stem cells. (Blood. 2005;105:4583-4589)
Introduction
Effective immunotherapy of advanced human cancer is an unrealized goal that nevertheless remains important to pursue given the failure and high toxicity of standard therapies. Immunotherapy appears more attainable in recent years due to documented successes of passively administered antibodies or T cells specific for various human tumor antigens.1-3 It has become clear from studying various approaches in animal models and in a more limited fashion in clinical trials that immunotherapy is more potent if both the innate and the adaptive cellular immune responses are efficiently engaged.4,5 Timely recognition of the tumor by the cells of the innate immune system, such as natural killer (NK) cells, granulocytes, and macrophages, appears to be a prerequisite for an efficient stimulation of tumor-specific adaptive immunity.6,7
A potentially powerful strategy for achieving simultaneous activation of the innate and the adaptive antitumor immune responses is to endow effector cells in both systems with specificity for a particular tumor antigen. Tumor-specific antibodies have been transduced into T cells (T-bodies) endowing them with tumor antigen specificity without the limitation of major histocompatibility complex (MHC) restriction.8,9 While antibodies can have exquisite specificity, one disadvantage is their high affinity of binding to antigen, which could impair infiltration of tumors,10 result in irreversible binding of an effector cell to a tumor cell, or even apoptosis of effector cells following their interaction with tumor cells. T-cell receptors (TCRs), on the other hand, have much lower binding affinity, and therefore a cell bearing a tumor-specific TCR could engage and disengage from its target multiple times and effect its function against multiple tumor cells. The limitation to using tumor-specific TCRs for cancer immunotherapy, however, is that they are MHC restricted and their use is therefore limited to a subgroup of patients with a particular HLA. Furthermore, tumor cells frequently down-regulate MHC or antigen-processing molecules11,12 and thus escape TCR-mediated recognition. Nevertheless, approaches involving transduction of cloned tumor-specific TCRs into effector T cells for adoptive immunotherapy continue to be developed.13,14 Most therapeutic attempts so far have been limited to expressing tumor-specific TCRs in T cells and have not included transduction of other cells, particularly those of the innate immune system.
We previously described a human cytotoxic T-cell line MA that specifically recognized the MUC1 tumor antigen as a whole protein expressed on the surface of epithelial adenocarcinomas.15 The epitope recognized by the MA TCR is located in each of the 20 amino acid-long tandem repeats in the extracellular domain of MUC1 (each molecule can have more than 100 such repeats). The amino acid sequence critical for recognition by the TCR is PDTRP presented at the tip of the structure we previously described as the “immunodominant knob” on the native MUC1 protein.16,17 A large number of these tandemly repeated and structurally stable PDTRP-bearing knobs on a single MUC1 molecule as well as on the neighboring MUC1 molecules on the surface of tumor cell can engage multiple TCRs and through receptor clustering signal the T cell to effect its function. Because antigen processing and presentation in MHC is not involved, this receptor can be used as a universal therapeutic reagent for targeting MUC1-positive tumors (more than 80% of all human cancers are MUC1 positive)18 in all patients regardless of their HLA type.
In this paper we describe the therapeutic potential of this MHC-unrestricted TCR engineered as a single-chain Vα/Vβ/Cβ fused to a CD3 zeta (ζ) chain and cloned into a retroviral vector. We show that lethally irradiated severe combined immunodeficiency (SCID) mice reconstituted with bone marrow (BM) cells transduced with this receptor controlled the growth of MUC1-positive human tumor xenografts. BM transduction and reconstitution experiments performed in fully immunocompetent mice showed that the presence of the ζ chain as an integral part of the single-chain TCR (scTCR) allowed cell surface expression in cells of the innate (granulocytes, monocytes, NK cells) as well as the adaptive (T and B cells) immune system. Experiments performed in human MUC1 transgenic (Tg) mice showed that expression of this TCR on high percentages of circulating blood cells did not have any negative effects on normal tissues expressing the normal form of this antigen. Expression of the scTCR appears to be lifelong, suggesting that under our transduction conditions, not only progenitor cells but also self-renewing stem cells were transduced. These results support further development of this reagent for a widely applicable gene therapy/immunotherapy of cancer.
Materials and methods
Computer modeling
The amino acid sequence of the SM3 antibody was provided by Dr J. Taylor-Papadimitriou, Imperial Cancer Research Fund. The Protein Data Bank at the Research Collaboratory for Structural Bioinformatics19 was searched for best-fit sequence alignments with the binding domain of the SM3 antibody using the basic local alignment search tool (BLAST).20 Coordinates of a crystallized human α/β TCR heterodimer were generously provided by Dr Ian Wilson, Scripps Clinic.21 Modeling studies were conducted on a Silicon Graphics Indigo workstation (Mountain View, CA). Homolog templates were mutated, and initial models were constructed using the program O.22 The programs LEaP23 and AMBER/Interface were used to import protein database (PDB) files created by O into AMBER, which was used for global energy minimizations and molecular dynamics of selected loops and modeling of ligand binding.24 The previously solved structure of the MUC1 epitope PDTRP and its 4 flanking amino acids16 was initially positioned in the antigen-binding region by rigid body docking using O. This positioning was followed by energy minimization and molecular dynamics in AMBER to allow the MUC1 epitope to position itself within the antigen-binding cleft. Figures were constructed using MOLSCRIPT and Raster3D.25,26
Construction of scTCR and expression in RBL and BWZ cells
The TCR Vα23.1Jα14.3 was cloned from the MA cytotoxic T lymphocyte (CTL) clone by reverse transcriptase-polymerase chain reaction (RT-PCR) and ligated in frame to the TCR Vβ8.3DβJβ1.2 region using a 15 amino acid (aa) flexible linker encoding 3 repeats of the sequence GGGGS. The TCR Vα23.1Jα14.3-linker-Vβ8.3DβJβ1.2 fragment was then cloned into a vector containing the human Cβ2, followed by a polypeptide linker (GDLVPRGSSRLD) and the murine CD3 ζ chain (a gift from Dr A. J. McMichael, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, United Kingdom).27 The last cysteine in the Cβ2 region was mutated to alanine using site-directed mutagenesis to prevent dimerization of the scTCR. The construct was then cloned into the pEF6 mammalian expression vector (Invitrogen, Carlsbad, CA). RBL cells were kindly provided by Dr Richard Klausner while at the National Cancer Institute (NCI), and the BWZ murine T cells were kindly provided by Dr Nilabh Shastri, University of California, Berkeley. RBL cells were grown in cDMEM-10 medium (Dulbecco modified Eagle medium [DMEM] plus 10% fetal bovine serum [FBS], 2 mM l-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, 10 μM 2-mercaptoethanol [2-ME], 1 × nonessential amino acids, and 1 × sodium pyruvate; Mediatech, Herndon, VA), while BWZ cells were grown in cRPMI-10 (RPMI and 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, 10 μM 2-ME, 1× nonessential amino acids, and 1× sodium pyruvate). RBL and BWZ cells were transfected by electroporation using Bio-Rad Gene Pulser II (Bio-Rad Laboratories, Hercules, CA) at 960 μF and 200 V settings.
Basophil degranulation assay
RBL cells or RBL-scTCR were incubated with 3H-serotonin (New England Nuclear, Boston, MA) for 24 hours. After washing, cells were transferred to plates coated with βF1 antibody or with MUC1 140-mer synthetic peptide (7 repeats of the sequence PDTRPAPGSTAPPAHGVTSA).28 Plates were centrifuged briefly and incubated for 30 minutes at 37°C. Ice-cold phosphate-buffered saline (PBS) (Sigma, St Louis, MO) was added, and the supernatant was harvested after additional centrifugation. Radioactivity was measured using a Wallac 1205 betaplate liquid scintillation counter (Wallac, Gaithersburg, MD).
IL-2 enzyme-linked immunosorbent assay (ELISA)
BWZ or BWZ-scTCR cells were plated in U-bottom 96-well plates at 1 × 105/200 μL cRPMI-10 medium, and 2 × 104 tumor cells (irradiated 60 Gy [6000 rad]) were added as stimulators. Thirty-six hours later, the amount of murine interleukin-2 (mIL-2) released in the medium was measured using a mouse IL-2 OptEIA kit (BD Pharmingen, San Diego. CA), according to the manufacturer's recommendations.
Construction of scTCR-EGFP MFG retroviral vector and production of viral supernatant
The MFG retroviral vector was kindly provided by Dr Paul Robbins (University of Pittsburgh, PA). The internal ribosome entry site-enhanced green fluorescent protein (IRES-EGFP) cassette was cloned by PCR from the pIRES2-EGFP vector (Clontech Laboratories, Palo Alto, CA) into the MFG retroviral vector downstream of the scTCR gene. The GP+E-86 ecotropic retroviral packaging cells (American Type Culture Collection, Manassas, VA) were transfected with the scTCR-EGFP MFG vector and cultured for 5 days, followed by sorting the EGFPhigh population. Sorted cells were then cultured in DMEM-15 (DMEM plus 15% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin), and 36 hours later retroviral supernatant was harvested and frozen at -80°C until use.
Retroviral transduction of BM cells
All experiments in animals were performed under an approved protocol no. 0304530A-1 of the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC). Six- to 8-week-old SCID or BALB/c mice (Jackson ImmunoResearch Laboratories, Bar Harbor, ME) were injected intraperitoneally with 150 mg/kg 5-fluorouracil (5-FU) (Invivogen, San Diego, CA), and 5 days later mice were killed and BM cells were isolated and prestimulated for 72 hours in the presence of 50 ng/mL recombinant stem cell factor (rSCF), 10 ng/mL recombinant murine IL-3 (rmIL-3), and 10 ng/mL rmIL-6 (PeproTech, Rocky Hill, NJ) in DMEM-15.29 BM cells were resuspended in retroviral supernatant supplemented with 50 ng/mL rSCF, 10 ng/mL rmIL-3, 10 ng/mL rmIL-6, and polybrene at 8 μg/mL. In all BM transduction experiments, cells were plated in 24-well plates precoated with recombinant fibronectin CH-296 fragment (Takara Bio, Madison, WI). Cells were then centrifuged for 30 minutes at room temperature at 800g in a Sorvall T6000B centrifuge (Kendro Laboratory Products, Asheville, NC) and put back in culture at 37°C. Transduction was repeated every 12 hours for a total of 4 cycles.
Reconstitution of irradiated mice with transduced BM cells and flow cytometric analyses
Transduced BM cells were resuspended in PBS at 1 × 107/mL, and 200 μL of cell suspension was injected via the tail vein into irradiated recipient mice. SCID mice received a single dose of 3.5 Gy (350 rad), while BALB/c mice were given a split dose of 9 Gy (900 rad) total (50:50) 3 hours apart. Reconstituted mice were maintained in a germ-free environment and were put on acidified water (pH 2.5) for 3 weeks after reconstitution. At 3, 6, and 11 weeks after reconstitution, 200 μL of blood was collected via tail artery and cells were stained with the appropriate anti-surface marker antibody—phycoerythrin (PE)-conjugated anti-CD3, -B220, -GR-1, -Mac-3, and -DX5; and allophycocyanin (APC)-conjugated anti-F4/80 (eBiosciences, San Diego, CA)—and analyzed on the Becton Dickinson FACSCalibur (BD Biosciences, San Jose, CA).
Tumor challenge and immunohistochemistry
Mice reconstituted with BM cells transduced with scTCR-MFG or with a control supernatant were challenged subcutaneously with various numbers of HPAF tumor cells 5 weeks after reconstitution. Tumor size was measured every 2 to 3 days using calipers. Tumors harvested from control or treated mice were fixed in 10% formalin, paraffin embedded, and sections stained with hematoxylin and eosin (H&E), antimyeloperoxidase (Labvision, Fremont, CA), anti-granzyme B (Labvision), or anti-F4/80 (eBiosciences) in the Department of Pathology core facility, University of Pittsburgh.
Fluorescent microscopic analyses of tissue sections from reconstituted mice
Six weeks after reconstitution, C57BL/6 mice or MUC1 Tg mice reconstituted with scTCR-EGFP-transduced BM cells were killed, and spleen, lung, and pancreas were harvested and fixed in 2% paraformaldehyde (PFA) in PBS. Tissues were then frozen, sectioned, and visualized for infiltration with EGFP-positive immune cells at the Center for Biologic Imaging, University of Pittsburgh.
Statistics
Statistical analysis was done using Microsoft Excel and Graphpad Prism (GraphPad Software, San Diego, CA) software.
Results
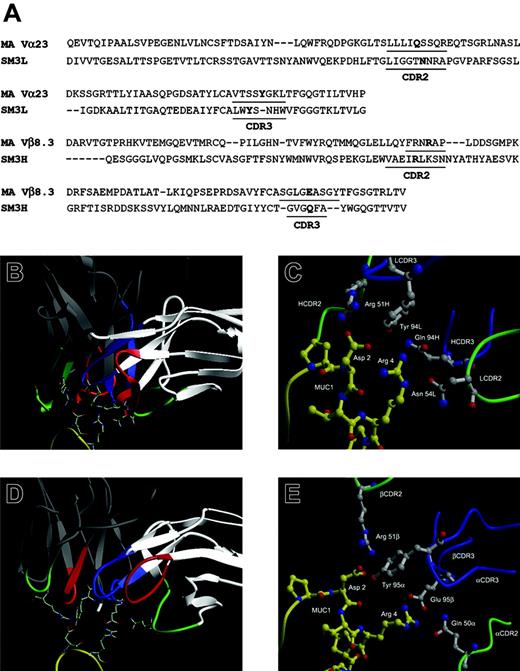
Computational modeling of MA TCR binding to the MUC1 immunodominant knob shows similarity to the binding of SM3 antibody specific for the same epitope
MUC1-specific MHC-unrestricted CTL clone MA established from a tumor-draining lymph node of a breast cancer patient was previously described.15 This clone mediated TCR-dependent killing of MUC1-positive tumor cells that was not restricted by their HLA type. It was also capable of binding synthetic, tandemly repeated MUC1 epitopes immobilized on the surface of polyactide-l-glycolide (PLGA) beads, resulting in the influx of Ca2+. Semiquantitative RT-PCR analysis and DNA sequencing revealed that the TCR responsible for this binding was composed of Vα23.1Jα14.3 and Vβ8.3DβJβ1.2. We cloned this TCR and transfected the TCR-deficient Jurkat cell line (J.RT3-T3.5)30 with a plasmid vector encoding the full-length TCR α and β chains. We confirmed its functionality and specificity by live imaging microscopy, where TCR-transfected cells, but not control cells, fluxed Ca2+ upon binding to MUC1-positive human tumor cells (Movies S1 and S2, available at the Blood website; see the Supplemental Materials link at the top of the online article).
Comparison of the CDR2 and CDR3 regions of the MUC1-specific antibody SM3 and the MA TCR and modeling their interactions with MUC1. (A) Amino acid sequence alignment of SM3 and MA TCR. Numbering corresponds to MA TCR sequence. Residues in bold are hypothesized to be involved in the binding. (B-C) SM3 and (D-E) MA TCR computer-based models of the interactions with the MUC1 epitope. The colors indicate the following: red, CDR1; green, CDR2; blue, CDR3; yellow, MUC1; white, the SM3 light chain and MA TCR α chain; gray, the SM3 heavy chain and MA TCR β chain. Contact residues are shown as stick diagrams with nitrogens in blue and oxygens in red, in close-up comparison of SM3 (C) and MA TCR (E) computer-based models of their interactions with MUC1 and labeled by chain, residue, and number in chain.
Comparison of the CDR2 and CDR3 regions of the MUC1-specific antibody SM3 and the MA TCR and modeling their interactions with MUC1. (A) Amino acid sequence alignment of SM3 and MA TCR. Numbering corresponds to MA TCR sequence. Residues in bold are hypothesized to be involved in the binding. (B-C) SM3 and (D-E) MA TCR computer-based models of the interactions with the MUC1 epitope. The colors indicate the following: red, CDR1; green, CDR2; blue, CDR3; yellow, MUC1; white, the SM3 light chain and MA TCR α chain; gray, the SM3 heavy chain and MA TCR β chain. Contact residues are shown as stick diagrams with nitrogens in blue and oxygens in red, in close-up comparison of SM3 (C) and MA TCR (E) computer-based models of their interactions with MUC1 and labeled by chain, residue, and number in chain.
We had also previously published that antibody SM3, which recognizes the same epitope,31 blocks tumor cell recognition and killing by MHC-unrestricted MUC1-specific CTLs.32,33 Considering the common epitope recognized by both SM3 and MA TCR, their common immunoglobulin (Ig)-like fold, and similarities in the complementarity-determining region-3 (CDR3) sequences (Figure 1A), we used the available sequence and structural information to model antigen binding by these 2 functionally related receptors. Both SM3 and the MA TCR were modeled by homology using appropriate antibody templates. Simulated docking of the MUC1 epitope produced minimized structures of both SM3 and MA TCR with bound ligand and predicted similar molecular determinants in the CDR2 and CDR3 regions important for binding. Our models predict that similarly positioned arginines in the corresponding Vβ and VH CDR2s interact favorably with the aspartic acid in the MUC1 epitope PDTRP (Figure 1B-E). Equivalently positioned tyrosines in the corresponding Vα and VL CDR3s stabilize the interaction with the aspartic acid of MUC1. A glutamine in the αCDR2 and a glutamic acid in the βCDR3 interact with the arginine in the epitope, as do an asparagine in the VL CDR2 and a glutamine in the VH CDR3.
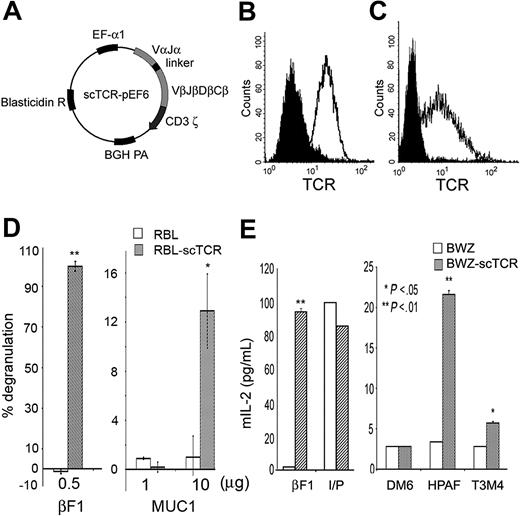
MA TCR as a single-chain (scTCR) construct fused with ζ chain is functionally expressed on the surface of T and non-T cells
To create a more practical reagent for future gene therapy/immunotherapy applications, we converted the full-length 2-chain TCR that was dependent on CD3 molecules unique to T cells for cell surface expression into a CD3-independent scTCR. The VαJα segment was fused to the VβDβJβ segment using a 15 amino acid flexible linker.34 This chimeric structure was then cloned into a vector containing the human TCR Cβ2 and the CD3 ζ transmembrane and cytoplasmic domains separated by a short linker27,35 and further subcloned into the pEF6 plasmid to create scTCR-pEF6 mammalian expression vector (Figure 2A). Transfection of the scTCR-pEF6 into a CD3- rat basophil cell line RBL35 (Figure 2B) or a CD3+ mouse T-cell lymphoma line BWZ36 (Figure 2C) resulted in high surface expression of the scTCR in both cell types.
Cells transfected with scTCR recognize MUC1-positive tumors and synthetic MUC1 antigen. (A) Mammalian expression vector encoding MA scTCR gene. The scTCR was cloned into the pEF6 vector. BGH indicates bovine growth hormone; EF-α, elongation factor α. (B) Cell surface expression of the scTCR in RBL cells or (C) BWZ cells stably transfected with the scTCR-pEF6 vector. Cells were stained with anti-TCR βF1 (open histogram) or with isotype control (filled histogram) antibody. (D) Degranulation of RBL cells or RBL-scTCR cells following stimulation with platebound βF1 antibody or MUC1 140-mer peptide. Specific degranulation is presented as percent of maximum degranulation induced by TCR cross-linking with βF1 antibody. (E) IL-2 enzyme-linked immunosorbent assay (ELISA) for BWZ or BWZ-scTCR following stimulation with platebound anti-TCR βF1 antibody, ionomycin plus PMA (I/P), DM6 (MUC1-negative tumor), HPAF, or T3M4 (MUC1-positive tumors). IL-2 in culture supernatant was measured by ELISA, and values were plotted on the y-axis as picograms per milliliter. Cells were stimulated as indicated.
Cells transfected with scTCR recognize MUC1-positive tumors and synthetic MUC1 antigen. (A) Mammalian expression vector encoding MA scTCR gene. The scTCR was cloned into the pEF6 vector. BGH indicates bovine growth hormone; EF-α, elongation factor α. (B) Cell surface expression of the scTCR in RBL cells or (C) BWZ cells stably transfected with the scTCR-pEF6 vector. Cells were stained with anti-TCR βF1 (open histogram) or with isotype control (filled histogram) antibody. (D) Degranulation of RBL cells or RBL-scTCR cells following stimulation with platebound βF1 antibody or MUC1 140-mer peptide. Specific degranulation is presented as percent of maximum degranulation induced by TCR cross-linking with βF1 antibody. (E) IL-2 enzyme-linked immunosorbent assay (ELISA) for BWZ or BWZ-scTCR following stimulation with platebound anti-TCR βF1 antibody, ionomycin plus PMA (I/P), DM6 (MUC1-negative tumor), HPAF, or T3M4 (MUC1-positive tumors). IL-2 in culture supernatant was measured by ELISA, and values were plotted on the y-axis as picograms per milliliter. Cells were stimulated as indicated.
These same 2 cell lines were used to test the function and antigen specificity of the scTCR. Transfected RBLs degranulated upon cross-linking of their scTCR with the anti-TCR antibody βF1 or upon specific recognition of the MUC1 antigen (Figure 2D). No degranulation was seen when cells were stimulated with control antigen ovalbumin (data not shown). There was no degranulation in control untransfected RBLs upon encounter with either MUC1 or βF1 antibody. Similarly, transfected BWZ cells produced a substantial amount of IL-2 when their scTCR was cross-linked with platebound βF1 antibody (Figure 2E). No significant IL-2 production was detected when BWZ-scTCR cells were incubated with the DM6 tumor cell line that did not express MUC1; however, a substantial level of IL-2 was produced upon encounter with the MUC1-positive tumor cell lines HPAF and T3M4. These 2 cell lines do not share HLA alleles, confirming that cells expressing the cloned scTCR exhibit the same MHC-unrestricted recognition of MUC1 as the original T-cell clone from which the TCR was derived. The difference in IL-2 secretion in response to these 2 tumors can be attributed to frequently observed differences in the level of expression and the extent of glycosylation of MUC1 on different tumor cell lines.
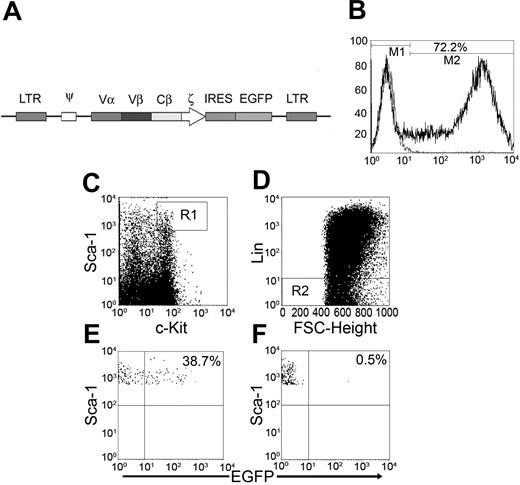
Successful transduction of BM cells that differentiate in vivo into scTCR-positive cells of multiple hematopoietic lineages
To test the antitumor activity of this MUC1-specific TCR in vivo, we cloned the scTCR into the MFG retroviral vector with an EGFP gene downstream of an IRES sequence (Figure 3A). This type of vector has been successfully used previously to track TCR-transduced cells in vivo.37 The mouse origin of the anti-human TCR monoclonal antibody bF1 that we used for the in vitro work precluded its use for detection of transduced cells in vivo due to the high background caused by the secondary anti-mouse Ig reagent. The scTCR-EGFP MFG is a bicistronic expression vector where scTCR and EGFP are expressed from the same mRNA. Transduction of NIH3T3 cells with this vector was used to confirm cytoplasmic expression of EGFP and simultaneous cell surface expression of scTCR (data not shown). We used the green fluorescence of EGFP to track scTCR-transduced cells. BM cells were isolated from 5-FU-treated BALB/c mice and transduced with the scTCR-EGFP MFG retroviral vector using fibronectin-assisted transduction protocol.38 Seventy-two percent of BM cells were successfully transduced (Figure 3B). We were especially interested in our ability to transduce hematopoietic stem cells contained within the population of cells that are Thy1.1- Lin- c-Kit+ Sca-1+.39 We found that 38% of cells of that phenotype were successfully transduced (Figure 3E).
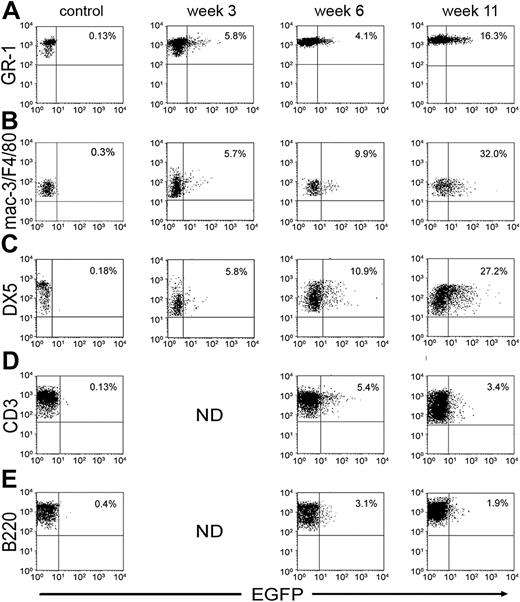
Sublethally irradiated mice received 2 × 106 BM cells via tail vein injection. At 3, 6, and 11 weeks after injection, mice were bled and the percentages of EGFP-positive cells in different cell lineages were evaluated. At week 3 after reconstitution, 5.8% of granulocytes (GR-1+) were positive for EGFP (Figure 4A). This number increased to 16.3% at week 11. Reconstitution of the monocyte/macrophage (Figure 4B) and NK cell (Figure 4C) lineage followed similar kinetics. Transduced T cells (CD3+) were seen in the periphery at week 6 after reconstitution and accounted for 5.4% (Figure 4D) of all T cells. This number dropped to 3.4% at week 11. Similarly, at 6 weeks, 3.4% of B cells (B220+) expressed EGFP (Figure 4E), and this number dropped to 1.9% at 11 weeks. This is consistent with published reports that lymphoid cells transduced with retroviral vectors have the tendency to silence expression of genes driven by the long-terminal repeat (LTR) promoter.40
Transduction of the long-term reconstituting hematopoietic stem cell population (c-Kit+ Sca-1+ Lin- Thy1.1-) with scTCR-EGFP MFG retroviral vector. (A) Schematic diagram of the scTCR-EGFP MFG retroviral vector. (B) BM cells transduced with the scTCR-EGFP MFG retroviral vector were stained on day 7 in culture for hematopoietic stem cell surface markers (c-Kit and Sca-1) and for lineage markers (Lin). Cells that expressed high levels of Sca-1 and c-Kit (C) and that lacked expression of Lin (D), R2, and R3 were gated on and were plotted against EGFP (E). FSC indicates forward scatter. (F) Mock-transduced BM cells. All cells in culture were Thy1.1- (not shown). Percentages are percentages of EGFP+ cells.
Transduction of the long-term reconstituting hematopoietic stem cell population (c-Kit+ Sca-1+ Lin- Thy1.1-) with scTCR-EGFP MFG retroviral vector. (A) Schematic diagram of the scTCR-EGFP MFG retroviral vector. (B) BM cells transduced with the scTCR-EGFP MFG retroviral vector were stained on day 7 in culture for hematopoietic stem cell surface markers (c-Kit and Sca-1) and for lineage markers (Lin). Cells that expressed high levels of Sca-1 and c-Kit (C) and that lacked expression of Lin (D), R2, and R3 were gated on and were plotted against EGFP (E). FSC indicates forward scatter. (F) Mock-transduced BM cells. All cells in culture were Thy1.1- (not shown). Percentages are percentages of EGFP+ cells.
SCID mice reconstituted with the scTCR-transduced BM controlled outgrowth of a MUC1-positive human tumor
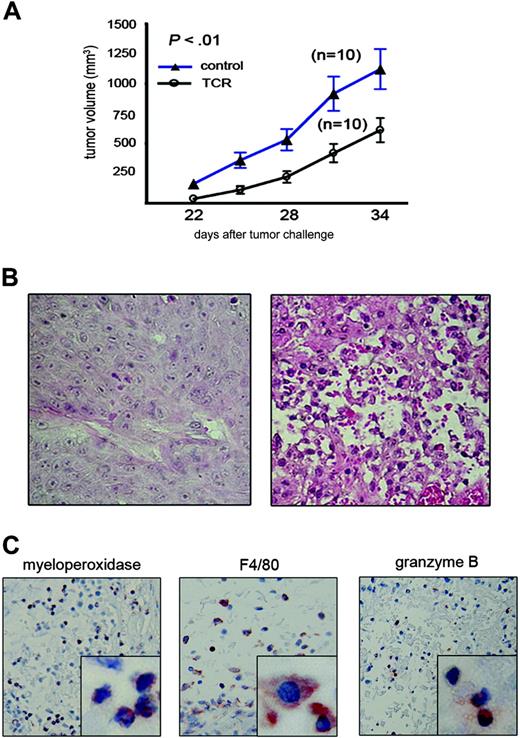
Based on our findings that the highest percentage of cells expressing the scTCR were those of the innate immune system and that this expression was most stable, we tested the potential of these cells alone to exert an antitumor effect. We reconstituted lethally irradiated SCID mice with 2 × 106 BM cells transduced with the scTCR-MFG retroviral vector or mock transduced. Expression of the scTCR was detected by RT-PCR in transduced BM cells prior to injection and in splenocytes from reconstituted mice 60 days later (Figure S1). Mice reconstituted with BM cells transduced with the scTCR retroviral vector or with control supernatant were challenged 1 month later with subcutaneous injection of the MUC1-positive human tumor cell line HPAF. Mice that received scTCR-MFG-transduced BM cells were able to inhibit growth of HPAF tumor cells compared with control mice (Figure 5A). The difference in tumor size between the 2 groups was statistically significant at each time point (P < .01). This experiment is 1 of 4 independent tumor rejection experiments that we have performed in scTCR-reconstituted SCID mice. This is the smallest antitumor effect that we have observed, but we have selected this particular example because it involved the largest number of mice per group.
Detection of scTCR-expressing cells at various times after reconstitution with transduced BM cells. Mice were bled at indicated time points, and leukocytes were stained for the appropriate cell surface markers plotted on the y-axis: (A) GR-1 for granulocytes, (B) Mac-3 or F4/80 for monocytes/macrophages, (C) DX5 for NK cells, (D) CD3 for T cells, and (E) B220 for B cells. EGFP expression is plotted on the x-axis. Percentages of EGFP-positive cells in each lineage are indicated.
Detection of scTCR-expressing cells at various times after reconstitution with transduced BM cells. Mice were bled at indicated time points, and leukocytes were stained for the appropriate cell surface markers plotted on the y-axis: (A) GR-1 for granulocytes, (B) Mac-3 or F4/80 for monocytes/macrophages, (C) DX5 for NK cells, (D) CD3 for T cells, and (E) B220 for B cells. EGFP expression is plotted on the x-axis. Percentages of EGFP-positive cells in each lineage are indicated.
SCID mice reconstituted with transduced BM cells can control the growth of the MUC1-positive tumor xenograft. (A) Control (▴) or scTCR-reconstituted (○) mice were injected subcutaneously with 2 × 106 HPAF (MUC1-positive) tumor cells. Tumor size is shown on the y-axis while days after tumor challenge is plotted on the x-axis. P values were calculated by running t test using Microsoft Excel software. Data are presented as mean ± SE. (B) H&E staining of HPAF tumor sections from control mice (left) or from scTCR-reconstituted mice (right). (C) Staining of tumor sections from scTCR-reconstituted mice for myeloperoxidase (neutrophil marker), F4/80 (monocyte/macrophage marker), or granzyme B (NK cell marker). Images were taken under × 20 magnification. Images in lower right squares were taken under × 100 magnification.
SCID mice reconstituted with transduced BM cells can control the growth of the MUC1-positive tumor xenograft. (A) Control (▴) or scTCR-reconstituted (○) mice were injected subcutaneously with 2 × 106 HPAF (MUC1-positive) tumor cells. Tumor size is shown on the y-axis while days after tumor challenge is plotted on the x-axis. P values were calculated by running t test using Microsoft Excel software. Data are presented as mean ± SE. (B) H&E staining of HPAF tumor sections from control mice (left) or from scTCR-reconstituted mice (right). (C) Staining of tumor sections from scTCR-reconstituted mice for myeloperoxidase (neutrophil marker), F4/80 (monocyte/macrophage marker), or granzyme B (NK cell marker). Images were taken under × 20 magnification. Images in lower right squares were taken under × 100 magnification.
Tumor sections from control mice were intact and homogeneous in appearance, without any infiltration by immune cells (Figure 5B, left). In contrast, tumor sections from scTCR BM-reconstituted mice were almost completely destroyed and infiltrated with various immune cells (Figure 5B, right). The predominant cells in the infiltrate were neutrophils (Figure 5C, left) followed by macrophages (Figure 5C, middle) and, to a lesser extent, NK cells (Figure 5C, right).
Lack of autoimmunity in MUC1 Tg mice reconstituted with scTCR-transduced BM cells
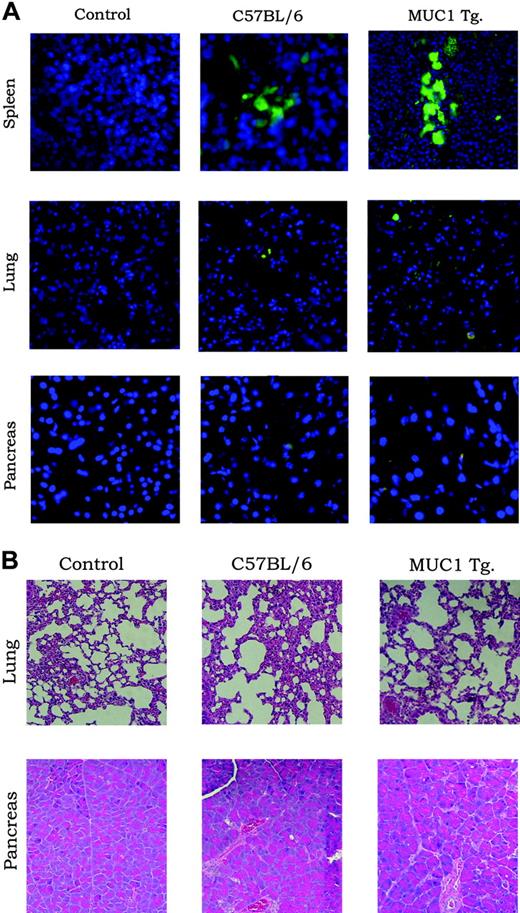
To test whether T cells expressing this TCR can develop normally in the presence of MUC1, we reconstituted C57BL/6 wild-type (wt) mice and MUC1 Tg mice with scTCR-transduced BM and 6 weeks later compared the percentages of scTCR-expressing cells. Table 1 shows that similar percentages of scTCR-positive T cells as well as other immune cells were seen in C57BL/6 and MUC1 Tg mice. Successful reconstitution of MUC1 Tg mice allowed us also to determine if the expression of this receptor could have deleterious effects on normal tissues expressing MUC1, such as the lung and the pancreas. Figure 6A shows very few EGFP-positive cells infiltrating these tissues and no difference between wt and MUC1 Tg mice. There was also no evidence of destruction of MUC1-expressing tissues in MUC1 Tg mice (Figure 6B).
scTCR-expressing cells in reconstituted C57BL/6 and MUC1 Tg mice
. | TCR reconstituted, mean % ± SD . | . | . | ||
|---|---|---|---|---|---|
| Cell type/surface marker . | Control* . | C57BL/6 . | MUC1 Tg . | ||
| T cells/CD3+ | 0.2 ± 0 | 6.4 ± 1.3 | 9.0 ± 5.7 | ||
| B cells/B220+ | 0.2 ± 0 | 9.7 ± 0.3 | 9.5 ± 1.6 | ||
| Granulocytes/GR-1+ | 0.2 ± 0.7 | 30.8 ± 21.5 | 27.4 ± 9.2 | ||
| NK cells/DX5+ | 0.1 ± 0 | 28.5 ± 4.3 | 18.5 ± 3.7 | ||
| Monocytes/F4/80+ | 0.1 ± 0.2 | 20.4 ± 5.0 | 14.6 ± 2.7 | ||
. | TCR reconstituted, mean % ± SD . | . | . | ||
|---|---|---|---|---|---|
| Cell type/surface marker . | Control* . | C57BL/6 . | MUC1 Tg . | ||
| T cells/CD3+ | 0.2 ± 0 | 6.4 ± 1.3 | 9.0 ± 5.7 | ||
| B cells/B220+ | 0.2 ± 0 | 9.7 ± 0.3 | 9.5 ± 1.6 | ||
| Granulocytes/GR-1+ | 0.2 ± 0.7 | 30.8 ± 21.5 | 27.4 ± 9.2 | ||
| NK cells/DX5+ | 0.1 ± 0 | 28.5 ± 4.3 | 18.5 ± 3.7 | ||
| Monocytes/F4/80+ | 0.1 ± 0.2 | 20.4 ± 5.0 | 14.6 ± 2.7 | ||
Analysis was performed 6 weeks after reconstitution with scTCR-transduced BM using EGFP as a marker of scTCR expression.
C57BL/6 untreated mice.
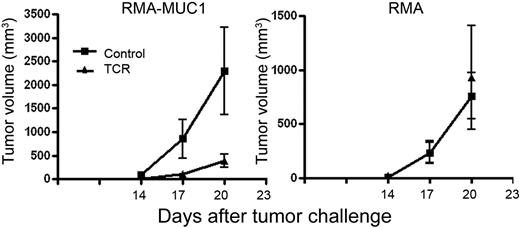
One explanation for the lack of autoimmunity in reconstituted MUC1Tg mice could be that these cells are rendered tolerant or anergic in the presence of MUC1 as a self-antigen. This was not the case, because these mice successfully controlled the growth of a mouse tumor RMA transfected with human MUC1 (RMA-MUC1) much better than the control mice. There was no inhibition of growth of the untransfected RMA control (Figure 7).
Expression of scTCR on immune cells has no deleterious effects on MUC1-positive normal tissues. (A) C57BL/6 (wild-type) and MUC1 Tg mice were reconstituted with BM cells transduced with the scTCR-EGFP MFG retroviral vector. Untreated mice served as controls. Spleen, lung, and pancreas were harvested 6 weeks after reconstitution and microscopically examined for infiltration with EGFP-positive cells (A) or stained with H&E (B) and examined for tissue destruction.
Expression of scTCR on immune cells has no deleterious effects on MUC1-positive normal tissues. (A) C57BL/6 (wild-type) and MUC1 Tg mice were reconstituted with BM cells transduced with the scTCR-EGFP MFG retroviral vector. Untreated mice served as controls. Spleen, lung, and pancreas were harvested 6 weeks after reconstitution and microscopically examined for infiltration with EGFP-positive cells (A) or stained with H&E (B) and examined for tissue destruction.
Discussion
We report here a novel reagent for immunotherapy of cancer, an MHC-unrestricted TCR specific for an epitope on the tumor antigen MUC1 engineered for expression on cells of the innate and the adaptive immune system and able to direct their effector function specifically against the tumor. The MHC-unrestricted nature of the TCR combined with the stable expression of the MUC1 tumor antigen on a large number of human tumors (more than 80%) makes therapy based on this TCR (scTCR) applicable to a large number of patients with a variety of tumors. Our experiments in SCID mice showed that the cells of the innate system alone can control tumor growth when provided with the tumor antigen-specific TCR. scTCR-expressing cells were seen as early as 3 weeks after reconstitution and were still present at high numbers more than 7 months later. Both the early presence of tumor-specific cells and their permanence would be expected to have a beneficial antitumor effect. Furthermore, the persistence of scTCR-positive cells (Figure S2) suggests that among the many different cells that were transduced in the BM there were also the long-term reconstituting hematopoietic stem cells that can continue to provide scTCR-positive progenitors and mature cells throughout the life of the animal. Expression of this TCR on large percentages of immune and other hematopoietic cells is not detrimental to the well-being of the animal. Long-term follow-up (more than 12 months) of reconstituted MUC1 Tg mice showed no signs of autoimmunity. This was to be expected because the epitope recognized by the sTCR has been shown to be expressed on the hypoglycosylated MUC1 made primarily by tumor cells.
scTCR-reconstituted MUC1 Tg mice rejected MUC1-positive tumor challenge. MUC1 Tg mice were reconstituted with BM cells transduced with scTCR (▴) or with control supernatant (▪) and challenged 6 weeks later with MUC1- tumor (RMA) or with RMA cells transfected with MUC1 (RMA-MUC1). Data are presented as mean ± SE. The number of mice at each time point ranged from 5 to 10.
scTCR-reconstituted MUC1 Tg mice rejected MUC1-positive tumor challenge. MUC1 Tg mice were reconstituted with BM cells transduced with scTCR (▴) or with control supernatant (▪) and challenged 6 weeks later with MUC1- tumor (RMA) or with RMA cells transfected with MUC1 (RMA-MUC1). Data are presented as mean ± SE. The number of mice at each time point ranged from 5 to 10.
The studies we describe here support further development of this reagent for immunotherapy of cancer. One therapeutic approach already within reach is transduction of this scTCR into BM or peripheral stem cells prior to infusion into patients who have undergone high-dose chemotherapy (HDCT). HDCT followed by autologous stem cell or BM transplant, primarily performed in breast cancer patients, has had a limited therapeutic success. The high rate of posttransplantation relapse41,42 is thought to result from the survival of some tumor cells following HDCT treatment or from infusing contaminating tumor cells with the stem cell preparation.43 Vaccination trials aimed at augmenting the immune responses to eradicate residual tumor cells following stem cell therapy showed minimal success as a result of the poor and slow reconstitution of the T-cell compartment in these patients.44,45 The recovery of the innate compartment of the immune system, particularly NK cells, occurs very rapidly after transplantation, reaching normal levels within a month after transplantation.46 We would propose that transducing BM cells with the MUC1-specific TCR prior to transplantation will result in the expression of a tumor-specific receptor on a high percentage of quickly reconstituting cells of the innate immune system, which could promptly target and destroy residual tumor cells.
Considering that MUC1 is expressed as a tumor antigen on more than 80% of all human tumors and that this TCR can theoretically direct effector cells to all such tumors in all patients, there are unlimited possibilities for its application. By varying the expression vector, one can target various effector cells for in vitro or in vivo transduction and tailor this type of gene therapy/immunotherapy to specific stages of disease and combinations with other therapies.
Prepublished online as Blood First Edition Paper, March 3, 2005; DOI 10.1182/blood-2004-10-3848.
Supported by National Institutes of Health (NIH) grant CA56103 and Department of Defense (DOD) training grant DAMD17-99-1-9352.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal