Abstract
Inhibitor of DNA binding (Id) proteins function as inhibitors of members of the basic helix-loop-helix family of transcription factors and have been demonstrated to play an important role in regulating lymphopoiesis. However, the role of these proteins in regulation of myelopoiesis is currently unclear. In this study, we have investigated the role of Id1 and Id2 in the regulation of granulopoiesis. Id1 expression was initially up-regulated during early granulopoiesis, which was then followed by a decrease in expression during final maturation. In contrast, Id2 expression was up-regulated in terminally differentiated granulocytes. In order to determine whether Id expression plays a critical role in regulating granulopoiesis, Id1 and Id2 were ectopically expressed in CD34+ cells by retroviral transduction. Our experiments demonstrate that constitutive expression of Id1 inhibits eosinophil development, whereas in contrast neutrophil differentiation was modestly enhanced. Constitutive Id2 expression accelerates final maturation of both eosinophils and neutrophils, whereas inhibition of Id2 expression blocks differentiation of both lineages. Transplantation of β2-microglobulin-/- nonobese diabetic severe combined immunodeficient (NOD/SCID) mice with CD34+ cells ectopically expressing Id1 resulted in enhanced neutrophil development, whereas ectopic expression of Id2 induced both eosinophil and neutrophil development. These data demonstrate that both Id1 and Id2 play a critical, although differential role in granulopoiesis.
Introduction
Basic helix-loop-helix (bHLH) transcription factors have been demonstrated to play a critical role in a wide variety of developmental processes, including neurogenesis,1 myogenesis,2 and hematopoiesis.3 The family of bHLH proteins consists of 3 distinct groups. The first encompasses the E-proteins, which consist of 4 different members: E2-2, HeLa E-box-binding factor (HEB), and the E2a-encoded proteins, E21 and E47. They are ubiquitously expressed, and can both homodimerize and heterodimerize with the second class of bHLH transcription factors, which are cell type– or tissue-specific.4 This group of tissue-specific transcription factors, also referred to as Class B proteins, consists of several factors that are involved in development of distinct cell types. For example, human achaete-scute homolog-1 (HASH-1),5 neurogenin,6 and NeuroD7 are all involved in neurogenic development, whereas MyoD8 and myogenin9,10 are required for normal muscle differentiation. Class B bHLH transcription factors have also been shown to play an important role in hematopoiesis. For example, both stem-cell leukemia/T-cell acute lymphoblastic leukemia-1 (SCL/Tal-1)3 and lymphoblastic leukemia derived sequence 1 (Lyl-1)11 are required for normal hematopoiesis and play a role in development of T-cell acute lymphoblastic leukemia.
Heterodimerization of bHLH transcription factors results in the formation of a parallel 4-helix bundle that allows both DNA binding domains to associate with the E-box recognition site.12 Transcriptional activity of these transcription factors is inhibited by a third group of bHLH transcription factors, the Id proteins, which lack the basic DNA binding domain. To date, 4 known Id proteins have been identified13-18 that are closely related in their HLH regions but differ in their expression patterns.19,20 It has been demonstrated that Id1 mRNA levels are often high in proliferating cells, but is down-regulated in quiescent or differentiating cells.21 In contrast, Id2 mRNA levels have been shown to be up-regulated during differentiation of the leukemic HL60 cell line toward either granulocytes or macrophages.22 Id proteins have recently been demonstrated to play a critical role in lymphopoiesis, for example, by inhibiting B-lymphocyte development at an early stage of differentiation, as has been demonstrated for Id1, Id2, and Id3.23,24 Furthermore, T-lymphocyte differentiation is inhibited by Id3.25,26 In contrast, natural killer (NK) cell development is positively regulated by both Id227,28 and Id3,25 suggesting a role for Id proteins in regulating specific lineage choices during lymphopoiesis.
In this study we have investigated for the first time the role of Id1 and Id2 in regulation of granulocyte differentiation utilizing a human ex vivo granulocyte differentiation model. Our data demonstrate that both Id1 and Id2 play an important role in regulating proliferation during granulocyte differentiation. Furthermore, constitutive expression of Id1 blocked eosinophil differentiation and induced neutrophil development, whereas conversely, Id2 expression enhanced both eosinophil and neutrophil differentiation. In addition, these results were confirmed using an in vivo transplantation model. Human neutrophil development was induced in the bone marrow of β2-microglobulin-/- nonobese diabetic severe combined immunodeficient (NOD/SCID) mice engrafted with cells ectopically expressing either Id1 or Id2, whereas eosinophil development was only enhanced in mice that underwent transplantation with cells ectopically expressing Id2. These data demonstrate that both Id1 and Id2 have distinct functions during myelopoiesis.
Materials and methods
Isolation and culture of human CD34+ cells
CD34+ cells were isolated as previously described.29 In brief, mononuclear cells were isolated from umbilical cord blood by density centrifugation over a ficoll-paque solution (density 1.077 g/mL). Magnetic activated cell sorting (MACS; Miltenyi Biotech, Auburn, CA) using a hapten-conjugated antibody against CD34, which was coupled to beads, was used to isolate CD34+ cells. CD34+ cells were cultured in Iscoves modified Dulbecco medium (IMDM; Gibco, Paisley, United Kingdom) supplemented with 10% fetal calf serum (FCS), 50 μM β-mercaptoethanol, 10 U/mL penicillin, 10 μg/mL streptomycin, and 2 mM glutamine at a density of 0.3 × 106 cells/mL. Cells were differentiated toward eosinophils upon addition of stem cell factor (SCF; 50 ng/mL), FMS-related tyrosine kinase 3 (FLT-3) ligand (50 ng/mL), granulocyte macrophage–colony-stimulating factor (GM-CSF; 0.1 nM), interleukin 3 (IL-3; 0.1 nM), and IL-5 (0.1 nM). Every 3 days, cells were counted and fresh medium was added to a density of 0.5 × 106 cells/mL. After 3 days of differentiation, only IL-3 and IL-5 were added to the cells. Neutrophil differentiation was induced upon addition of SCF (50 ng/mL), FLT-3 ligand (50 ng/mL), GM-CSF (0.1 nM), IL-3 (0.1 nM), and G-CSF (30 ng/mL). After 6 days of culture only G-CSF was added to the cells. CD34+ cells used in transplantation studies were cultured for 3 days in IMDM containing the cytokines SCF (50 ng/mL), FLT-3 ligand (50 ng/mL) and thrombopoietin (TPO; 10 ng/mL). Cord blood samples were collected from healthy donors after informed consent was provided according to the Declaration of Helsinki. Protocols were approved by the local ethics committee of the University Medical Center in Utrecht.
Viral transduction of CD34+ cells
A bicistronic retroviral DNA construct was used, expressing either Id1 (kindly provided by Dr H. Spits, AMC, Amsterdam, The Netherlands) or Id2, and an internal ribosomal entry site (IRES) followed by the gene encoding for enhanced green fluorescent protein (LZRS-eGFP). In addition, for knock-down experiments, a bicistronic retroviral DNA construct (pRetrosuper; kindly provided by Dr H. Spits, Amsterdam, the Netherlands) consisting of an RNAi probe against Id2 and a gene encoding for GFP was used.30 LZRS-eGFP retrovirus was produced by transient transfection of the retroviral packaging cell line, Phoenix-ampho,31 by calcium phosphate coprecipitation. Cells were plated in 6-cm dishes, 24 hours before transfection. At 5 minutes prior to transfection, 25 μM chloroquine diphosphate was added to the cells. A total of 10 μg DNA was used per transfection. Medium was refreshed 16 hours after transfection. After an additional 24 hours, viral supernatants were collected and filtered through a 0.45-μm filter. CD34+ cells were transduced in 24-well dishes precoated with 20 μg/cm2 recombinant human fibronectin fragment CH-296 (RetroNectin; Takara, Otsu, Japan) for 2 hours, and 2% bovine serum albumin (BSA) for 30 minutes. Transduction was performed by addition of 0.5 mL viral supernatant to 0.5 mL medium containing 0.5 × 106 cells. At 24 hours after transduction, 0.7 mL medium was removed from the cells and 0.5 mL fresh virus supernatant was added together with 0.5 mL fresh medium.
Histochemical staining of eosinophil and neutrophil precursors
May-Grünwald-Giemsa staining was used to analyze both differentiating eosinophils and neutrophils. Cytospins were prepared from 5 × 104 differentiating granulocytes and were fixed in methanol for 3 minutes. After fixation, cytospins were stained in a 50% eosin methylene blue solution according to May-Grn̋wald (Sigma-Aldrich GmbH, Seelze, Germany) for 20 minutes, rinsed in water for 5 seconds, and the nuclei were counterstained with 10% Giemsa solution (Merck kGaA, Darmstadt, Germany) for 15 minutes. During eosinophil differentiation, cells could be characterized as differentiating from blast cells toward promyelocyte type I, promyelocyte type II, myelocyte, metamyelocyte, and finally mature eosinophils with segmented nuclei. These stages can be distinguished by the size of the cells, ratio of cytoplasm versus nucleus present, presence of azurophilic granules, appearance of eosinophilic granules, and the shape of the nuclei. Differentiated eosinophils were characterized as cells belonging to the stages of myelocyte, metamyelocyte, and mature eosinophils. Neutrophil differentiation can also be characterized by distinct stages from myeloblast, promyelocyte I, promyelocyte II, myelocyte, and metamyelocytes toward neutrophils with banded or segmented nuclei. Differentiated neutrophils were characterized as cells containing either banded or segmented nuclei. Micrographs were made using an Axiostar Plus upright microscope (Zeiss, Göttingen, Germany) and a Canon Powershot G5 digital camera (Canon, Moofdorp, the Netherlands). Magnification of the micrographs is ×1000. Cells embedded in Entellan (Merck, Darmstadt, Germany) were analyzed using an Axiostar Plus microscope with a Plan-neofluor 100×/1.30 oil (a = 0.20 mm) objective (Zeiss, Göttingen, Germany) and immersion oil 518C (Zeiss, Darmstadt, Germany). Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) was used to prepare the micrographs for composition.
Immunohistochemical staining of eosinophil and neutrophil precursors
Eosinophil and neutrophil differentiation was also analyzed by intracellular staining of eosinophil peroxidase and lactoferrin, respectively. Cells were first washed in phosphate-buffered saline (PBS) and resuspended in 100 μL 0.5% formaldehyde. After a 15-minute incubation at 37°C, 900 μL ice-cold methanol was added to the cells. Cells were washed with PBS after 30 minutes of incubation on ice and resuspended in 0.5% BSA. After a 10-minute incubation at room temperature, cells were washed and neutrophil progenitors were resuspended in phycoerythrin (PE)–conjugated anti-lactoferrin (Immunotech, Marseille, France) and incubated for another 25 minutes. Eosinophil progenitors were incubated with anti–eosinophil peroxidase (Oncogene Research Products, Calbiochem, Cambridge, MA) and incubated for 25 minutes. Eosinophil progenitors were subsequently washed and incubated for 25 minutes with a PE-conjugated goat antimouse antibody (Pharmingen, San Diego, CA). Cells were again washed and lactoferrin- and eosinophil peroxidase–positive cells were detected by fluorescence-activated cell sorting (FACS) analysis (FACS Vantage, Becton Dickinson, Alphen a/d Rijn, The Netherlands).
Western blot analysis
Western blot analysis was performed using standard techniques. In brief, for detection of Id1 and Id2 expression, differentiating granulocytes were lysed in Laemmli buffer (0.12 M Tris HCl pH 6.8, 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.05 μg/μL bromophenol blue, and 35 mM β-mercaptoethanol), and boiled for 5 minutes. Equal amounts of total lysate were analyzed by 15% SDS–polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to Immobilon-P (Millipore, Bedford, MA) and incubated with blocking buffer (Tris buffered saline/Tween20) containing 5% lowfat milk for 16 hours at 4°C before incubating with either an Id1 antibody (Santa Cruz, Santa Cruz, CA), an Id2 antibody (Santa Cruz), or an antibody against β-actin (Santa Cruz) for 2 hours in the same buffer. Subsequently, blots were incubated with peroxidase-conjugated secondary antibodies for 1 hour. Enhanced chemical luminescence (ECL) was used as a detection method according to the manufacturer's protocol (Amersham Pharmacia, Amersham, United Kingdom).
Reverse transcriptase–polymerase chain reaction (RT-PCR) assays
RNA was isolated from granulocyte progenitors as previously described,32 and reverse transcribed using Iscript cDNA synthesis kit (Biorad, Hercules, CA) after DNase treatment with DNAfree (Ambion, Cambridgeshire, United Kingdom) according to the manufacturer's protocol. Real-time PCR reactions were performed in a 20 μL mixture containing 5 μL cDNA, 10 μL SYBR Green mastermix (Biorad) and 0.8 μM of each primer. Amplification and detection of SYBR Green was performed with a MyIQ Icycler (Biorad) under the following conditions: 1 minute at 95°C, followed by 45 cycles of 10 seconds at 95°C, 20 seconds at 61°C, and 25 seconds at 72°C. Results were normalized for the housekeeping gene HPRT and were expressed as fold regulation compared with CD34+ cells.
3H-thymidine incorporation assays
CD34+ cells were transduced 2 days after isolation from umbical cord blood. At 24 hours after transduction, eGFP-positive cells were separated from the nontransduced cells. A quantity of 40 000 cells, resuspended in normal culture medium (see “Isolation and culture of human CD34+ cells”) were incubated with 5 μCi/mL (185 kBq/mL) 3H-thymidine for 72 hours at days 8, 11, and 14. The total amount of 3H-thymidine incorporated over a 9-day period was calculated, and data were depicted as the fold regulation of the total amount of incorporated 3H-thymidine compared with control cells.
Transplantation of human CD34+ cells into β2-microglobulin-/- NOD/SCID mice
The β2-microglobulin-/- NOD/SCID33 mice were bred and maintained under sterile conditions in microisolator cages and provided with autoclaved food and acidified water containing 111 mg/L L-cyclopropyl-6-fluoro-1,4-dihydro-4-oxo-7-(1-piperazinyl)-3-quinolinecarboxylic acid. All animal procedures were performed with consent from the local ethics committee at Lund University.
Mice, 8 to 10 weeks old, were sublethally irradiated with 350 cGy administered from a 137Cs source, and underwent transplantation via tail vein injections with approximately 500 000 unsorted retrovirally transduced cord blood–derived hematopoietic progenitors along with 1.106 irradiated (1500 cGy) CD34-depleted cord blood–derived accessory cells.34 After 6 weeks, the mice were killed and both tibiae and femora were flushed. Bone marrow cells were stained with anti–huCD45–allophycocyanin (APC) and a mixture of antibodies against myeloid antigens including anti–huCD33-PE, anti–huCD15-PE, anti–huCDw123-PE, and anti–huCD235-PE (all from Becton Dickinson). The samples were analyzed, and from bone marrow samples positive for human cells, eGFP-positive CD45+ myeloid cells were sorted on a FACS Vantage (Becton Dickinson, San Jose, CA) and cytospins prepared. May-Grünwald-Giemsa staining was used to analyze both mature eosinophils and neutrophils.
Statistics
A paired t test was performed to compare the difference in the percentage eGFP-positive cells between day 7 and day 17 and the fold induction of proliferation, and a Student t test was performed to compare the difference in differentiation and annexin-positive cells between the controls and cells transduced with either Id1 or Id2 or Id2 RNAi. A P value of .05 or less was considered significant.
Results
Id1 and Id2 protein expression is regulated during granulocyte differentiation
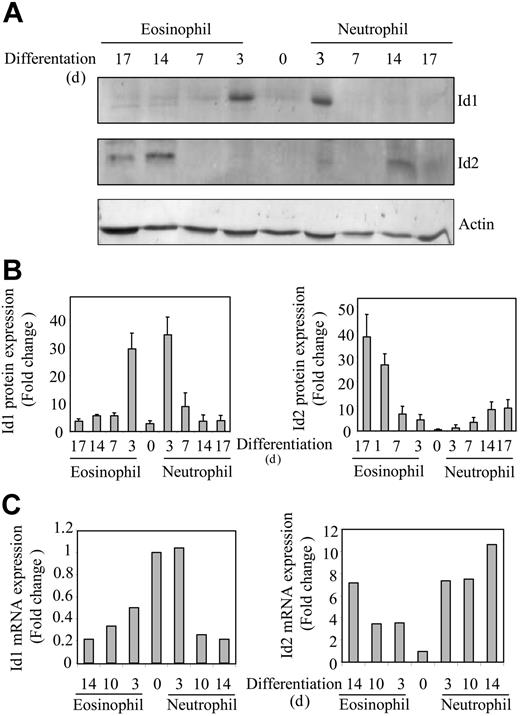
CD34+ progenitor cells, isolated from umbilical cord blood, were differentiated toward eosinophils or neutrophils in presence of the cytokines IL-3 and IL-5, or G-CSF, respectively, as described in “Materials and methods.” In order to investigate whether protein expression of Id1 and Id2 was also regulated during granulopoiesis, protein lysates were made from differentiating eosinophils and neutrophils. Proteins were separated by SDS-PAGE, Western blotting was performed using an antibody against either Id1 or Id2 (Figure 1A), and protein levels were quantified (Figure 1B). Id1 expression could not be clearly detected in freshly isolated CD34+ progenitors; however, after 3 days of either eosinophil or neutrophil differentiation, Id1 protein levels were transiently up-regulated. During extended culture the expression levels were again down-regulated. In contrast, during the early stages of eosinophil differentiation, Id2 could not be detected, whereas levels were up-regulated during final maturation. During neutrophil differentiation on the other hand, Id2 protein expression was very low although modestly up-regulated during terminal differentiation. To confirm if this was reflected at the level of mRNA, real-time quantitive PCR analysis was performed (Figure 1C). Id1 mRNA was expressed in CD34+ cells and down-regulated during terminal eosinophil and neutrophil differentiation, whereas Id2 mRNA was up-regulated during terminal differentiation. However, in contrast to the protein levels, mRNA expression of Id1 at day 0 is higher, suggesting that although mRNA is expressed, translation is apparently not initiated in these CD34+ progenitors. These experiments suggest a possible difference in the regulation of eosinophil and neutrophil differentiation by Id proteins.
Expression of Id1 and Id2 is regulated during granulocyte differentiation. Umbilical cord blood–derived CD34+ cells were differentiated toward eosinophils or neutrophils in presence of the cytokines FLT-3 ligand, SCF, GM-CSF, IL-3 and IL-5, or G-CSF. After 3 days of culture, eosinophil progenitors were cultured in the presence of IL-3 and IL-5, whereas neutrophil progenitors were cultured in the presence of G-CSF alone after 6 days of culture. (A) Protein lysates were made immediately after isolation from cord blood and after 3, 7, 14, and 17 days of differentiation. Western blot analysis was performed with an antibody against Id1 (top blot), an antibody against Id2 (middle blot), and, as a control for equal loading, an antibody against β-actin (bottom blot). The experiment shown is representative of 3 independent experiments. (B) Protein levels were quantified using ImageQuant software (Amersham Biosciences, Freiburg, Germany), and data are depicted as an average of 3 independent experiments plus or minus the standard error of the mean (SEM). (C) RNA was isolated from cells differentiating toward eosinophils and neutrophils at the time points indicated. Quantitative real-time PCR reactions were performed using gene-specific primers for Id1 (left) and Id2 (right). Values were corrected for the expression of HPRT in all samples. The experiment shown is representative of 3 independent experiments.
Expression of Id1 and Id2 is regulated during granulocyte differentiation. Umbilical cord blood–derived CD34+ cells were differentiated toward eosinophils or neutrophils in presence of the cytokines FLT-3 ligand, SCF, GM-CSF, IL-3 and IL-5, or G-CSF. After 3 days of culture, eosinophil progenitors were cultured in the presence of IL-3 and IL-5, whereas neutrophil progenitors were cultured in the presence of G-CSF alone after 6 days of culture. (A) Protein lysates were made immediately after isolation from cord blood and after 3, 7, 14, and 17 days of differentiation. Western blot analysis was performed with an antibody against Id1 (top blot), an antibody against Id2 (middle blot), and, as a control for equal loading, an antibody against β-actin (bottom blot). The experiment shown is representative of 3 independent experiments. (B) Protein levels were quantified using ImageQuant software (Amersham Biosciences, Freiburg, Germany), and data are depicted as an average of 3 independent experiments plus or minus the standard error of the mean (SEM). (C) RNA was isolated from cells differentiating toward eosinophils and neutrophils at the time points indicated. Quantitative real-time PCR reactions were performed using gene-specific primers for Id1 (left) and Id2 (right). Values were corrected for the expression of HPRT in all samples. The experiment shown is representative of 3 independent experiments.
Id1 inhibits eosinophil differentiation but enhances neutrophil development
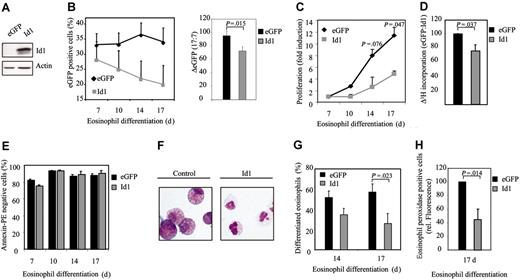
To determine whether expression of Id1 plays a critical role in regulating eosinophil differentiation, a bicistronic retroviral DNA construct coexpressing eGFP and Id1 was used to generate retrovirus and subsequently to infect CD34+ progenitors (Figure 2A). Cells were subsequently differentiated toward eosinophils for 17 days. The percentage of eGFP-positive cells corresponds to the percentage of transduced cells and was determined by FACS analysis (Figure 2B). During differentiation, a decrease in the percentage of eGFP-positive cells after transduction with Id1 was observed, relative to control (Figure 2B, right panel). This suggests that Id1 might play a role in inhibiting proliferation during eosinophil differentiation. In order to determine whether this is indeed the case, proliferation was determined both by counting the total amount of eGFP-positive cells (Figure 2C) as wells as by performing 3H-thymidine incorporation assays (Figure 2D). Transduction of cells with Id1 indeed resulted in decreased proliferation compared with cells transduced with eGFP alone. In order to investigate whether the inhibition of proliferation in cells ectopically expressing Id1 was due to enhanced apoptosis, annexin V staining was performed in transduced cells. No significant difference could be observed in the percentage of annexin V–positive cells between cells transduced with empty vector alone or cells transduced with Id1 (Figure 2E). In order to investigate whether Id1 also plays a role in eosinophil differentiation per se, cells were again transduced with either eGFP or Id1. After 14 and 17 days of differentiation, eGFP-positive cells were sorted by FACS from the nontransduced cells and cytospins were prepared (Figure 2F). The morphology of the differentiating cells was analyzed by May-Grünwald-Giemsa staining as described in “Materials and methods.” The percentage of differentiated eosinophils after transduction with Id1 was dramatically inhibited compared with cells transduced with eGFP alone (Figure 2G). Interestingly, after 17 days of differentiation, the majority of the cells ectopically expressing Id1, cultured in presence of IL-5, were neutrophil progenitors as defined by May-Grünwald-Giemsa staining (myelocytes, metamyelocytes) and the remainder of the cells had differentiated toward eosinophils but appeared blocked at the promyelocytic type I and II stages. In contrast, after 17 days of normal eosinophil development, the majority of the cells were mature myelocytes and metamyeloyctes (Table 1). Similar results were obtained using the granule protein eosinophil peroxidase as a marker for eosinophil differentiation (Figure 2H).
Stages of eosinophil differentiation in CD34+ cells transduced with eGFP or Id1
. | eGFP . | . | Id1 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I | 17.6 ± 4.8 | 22.8 ± 4.6 | 40.9 ± 4.9 | 53.4 ± 10.3 | ||
| Promyelocyte II | 31.7 ± 6.2 | 21.9 ± 5.7 | 25.4 ± 4.6 | 21.4 ± 6.7 | ||
| Myelocyte | 47.4 ± 5.6 | 51.5 ± 9.9 | 30.2 ± 3.9 | 25.9 ± 10.1 | ||
| Metamyelocyte/mature | 2.9 ± 2.1 | 1.9 ± 1.4 | 2.3 ± 2.2 | 0.1 ± 0.1 | ||
. | eGFP . | . | Id1 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I | 17.6 ± 4.8 | 22.8 ± 4.6 | 40.9 ± 4.9 | 53.4 ± 10.3 | ||
| Promyelocyte II | 31.7 ± 6.2 | 21.9 ± 5.7 | 25.4 ± 4.6 | 21.4 ± 6.7 | ||
| Myelocyte | 47.4 ± 5.6 | 51.5 ± 9.9 | 30.2 ± 3.9 | 25.9 ± 10.1 | ||
| Metamyelocyte/mature | 2.9 ± 2.1 | 1.9 ± 1.4 | 2.3 ± 2.2 | 0.1 ± 0.1 | ||
After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. Eosinophil differentiation was expressed as percentage of myeloblast, promyelocyte I, promyelocyte II, myelocyte, metamyelocyte, and mature eosinophils within the eosinophil lineage. Data are expressed as an average of 7 independent experiments plus or minus the standard error of the mean (SEM).
Eosinophil differentiation is inhibited by Id1. (A) After a 17-day period of differentiation, protein lysates were prepared from eosinophil progenitors transduced with eGFP or Id1 and a Western blot was performed with an N-terminal antibody against Id1 (top panel) and, as a control for equal loading, an antibody against β-actin (bottom panel). CD34+ cells were transduced with eGFP or Id1 and cultured in the presence of IL-5 for 17 days. (B, left) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. ♦ indicates eGFP; ▦, Id1. Results are expressed as an average of 3 independent experiments plus or minus SEM. (Right) The difference between the percentage of eGFP-positive cells after 7 and 17 days of differentiation is depicted. ▪ indicates eGFP; ▦, Id1. (C) Proliferation was determined by counting the eGFP-positive cells. ♦ indicates eGFP; ▦, Id1. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (D) Proliferation was determined by performing 3H-thymidine incorporation assays. Data were depicted as a ratio between cells transduced with empty vector alone and cells transduced with Id1. ▪ indicates eGFP; ▦, Id1. Data were expressed as an average of 3 independent experiments plus or minus SEM. (E) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Bar shading is as in panel D. Data were expressed as an average of 3 independent experiments plus or minus SEM. (F) CD34+ cells were transduced with eGFP or Id1. After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (G) Eosinophil differentiation was expressed as a percentage of differentiated eosinophils within the eosinophil lineage. Data were expressed as an average of 7 independent experiments plus or minus SEM. (H) Eosinophil differentiation was also analyzed by staining the granule protein eosinophil peroxidase after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of eosinophil peroxidase between cells transduced with empty vector alone or cells transduced with Id1. Data were expressed as an average of 3 independent experiments plus or minus SEM. (G-H) Bar shading is as in panel D.
Eosinophil differentiation is inhibited by Id1. (A) After a 17-day period of differentiation, protein lysates were prepared from eosinophil progenitors transduced with eGFP or Id1 and a Western blot was performed with an N-terminal antibody against Id1 (top panel) and, as a control for equal loading, an antibody against β-actin (bottom panel). CD34+ cells were transduced with eGFP or Id1 and cultured in the presence of IL-5 for 17 days. (B, left) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. ♦ indicates eGFP; ▦, Id1. Results are expressed as an average of 3 independent experiments plus or minus SEM. (Right) The difference between the percentage of eGFP-positive cells after 7 and 17 days of differentiation is depicted. ▪ indicates eGFP; ▦, Id1. (C) Proliferation was determined by counting the eGFP-positive cells. ♦ indicates eGFP; ▦, Id1. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (D) Proliferation was determined by performing 3H-thymidine incorporation assays. Data were depicted as a ratio between cells transduced with empty vector alone and cells transduced with Id1. ▪ indicates eGFP; ▦, Id1. Data were expressed as an average of 3 independent experiments plus or minus SEM. (E) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Bar shading is as in panel D. Data were expressed as an average of 3 independent experiments plus or minus SEM. (F) CD34+ cells were transduced with eGFP or Id1. After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (G) Eosinophil differentiation was expressed as a percentage of differentiated eosinophils within the eosinophil lineage. Data were expressed as an average of 7 independent experiments plus or minus SEM. (H) Eosinophil differentiation was also analyzed by staining the granule protein eosinophil peroxidase after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of eosinophil peroxidase between cells transduced with empty vector alone or cells transduced with Id1. Data were expressed as an average of 3 independent experiments plus or minus SEM. (G-H) Bar shading is as in panel D.
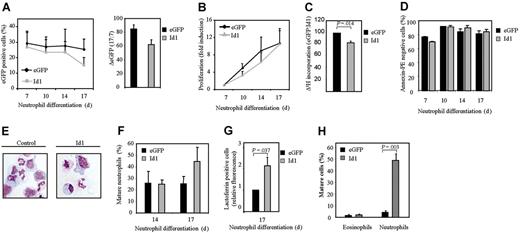
The data described in Figure 2 suggest that Id1 plays an important role in normal eosinophil differentiation. To determine whether this protein plays an important role in granulopoiesis in general, we analyzed the effect of constitutive Id1 expression during neutrophil maturation (Figure 3A). Interestingly, in contrast to eosinophilopoiesis, differentiation during neutrophil development (Figure 3E-F, Table 2) was modestly enhanced in cells ectopically expressing Id1. The level of the neutrophil-specific granule protein lactoferrin was also significantly enhanced during differentiation in cells ectopically expressing Id1 (Figure 3G). In order to investigate whether Id1 also plays a significant role in regulation of lineage development in vivo, CD34+ cells transduced with either eGFP or Id1 were transplanted into β2-microglobulin-/- NOD/SCID mice (Figure 3H). Six weeks after injection, eGFP-positive human myeloid cells were sorted and cytospins prepared (see “Materials and methods”). The morphology of the cells was analyzed by May-Grünwald-Giemsa staining. No difference could be observed in eosinophil development between bone marrow cells ectopically expressing Id1 or empty vector alone. However, this is probably explained by the fact that the percentage of mature eosinophils in the bone marrow of mice that underwent transplantation with CD34+ cells was very low (1.5%), making it impossible to observe any significant inhibition in differentiation. In contrast, transplantation of cells ectopically expressing Id1 resulted in a striking increase in the percentage of mature neutrophils in vivo (Figure 3H). These data strongly suggest that Id1 plays a distinct role in regulation of the 2 lineages: down-regulation of Id1 appears a requirement for eosinophil differentiation, but apparently is not required for neutrophil maturation.
Stages of neutrophil differentiation in CD34+ cells transduced with eGFP or Id1
. | eGFP . | . | Id1 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I and II | 40.4 ± 19.8 | 34.8 ± 9.5 | 37.3 ± 12.9 | 32.2 ± 14.9 | ||
| Myelocyte | 16.2 ± 3.8 | 27.8 ± 7.8 | 13.7 ± 3.7 | 18.9 ± 4.4 | ||
| Metamyelocyte/banded | 35.1 ± 12.4 | 28.8 ± 0.5 | 40.5 ± 8.1 | 38.8 ± 13.5 | ||
| Segmented | 10.9 ± 3.9 | 10.5 ± 5.0 | 10.0 ± 2.1 | 10.5 ± 1.6 | ||
. | eGFP . | . | Id1 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I and II | 40.4 ± 19.8 | 34.8 ± 9.5 | 37.3 ± 12.9 | 32.2 ± 14.9 | ||
| Myelocyte | 16.2 ± 3.8 | 27.8 ± 7.8 | 13.7 ± 3.7 | 18.9 ± 4.4 | ||
| Metamyelocyte/banded | 35.1 ± 12.4 | 28.8 ± 0.5 | 40.5 ± 8.1 | 38.8 ± 13.5 | ||
| Segmented | 10.9 ± 3.9 | 10.5 ± 5.0 | 10.0 ± 2.1 | 10.5 ± 1.6 | ||
After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. Neutrophil differentiation was expressed as percentage of myeloblast, promyelocyte I, promyelocyte II, myelocyte, and metamyelocyte toward neutrophils with banded and segmented nuclei within the neutrophil lineage. Data are expressed as an average of 3 independent experiments plus or minus SEM.
Constitutive Id1 expression enhances neutrophil differentiation. CD34+ cells were transduced with eGFP or Id1 and cultured in the presence of G-CSF for 17 days. (A, left) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. ♦ indicates eGFP; ▦, Id1. Results are expressed as an average of 3 independent experiments plus or minus SEM. (Right) The difference between the percentage of eGFP-positive cells after 7 and 17 days of differentiation is depicted. ▪ indicates eGFP; ▦, Id1. (B) Proliferation was determined by counting the eGFP-positive cells. ♦ indicates eGFP; ▴, Id1. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (C) Proliferation was determined by performing 3H-thymidine incorporation assays. Data are depicted as a ratio between cells transduced with empty vector alone or cells transduced with Id1. ▪ indicates eGFP; ▦, Id1. Data are expressed as an average of 3 independent experiments plus or minus SEM. (D) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Bar shading is as in panel C. Data are expressed as an average of 3 independent experiments plus or minus SEM. (E) CD34+ cells were transduced with eGFP or Id1. After 14 and 17 days of neutrophil differentiation, transduced cells were separated from the nontransduced cells, and cytospins were prepared and stained with May-Grünwald-Giemsa solution. (F) Neutrophil differentiation was expressed as a percentage of differentiated neutrophils within the neutrophil lineage. Data were expressed as an average of 3 independent experiments plus or minus SEM. (G) Neutrophil differentiation was also analyzed by staining the granule protein lactoferrin after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of lactoferrin between cells transduced with empty vector alone or cells transduced with Id1. Data were expressed as an average of 3 independent experiments plus or minus SEM. (H) CD34+ cells, cultured in the presence of the cytokines SCF, FLT-3 ligand, and TPO, were transduced with empty vector alone or with Id1. After 3 days of culture, cells were injected into β2-microglobulin-/- NOD/SCID mice. Six weeks after injection, mice were killed, eGFP-positive human myeloid cells were sorted, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (F-H) Bar shading is as in panel C.
Constitutive Id1 expression enhances neutrophil differentiation. CD34+ cells were transduced with eGFP or Id1 and cultured in the presence of G-CSF for 17 days. (A, left) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. ♦ indicates eGFP; ▦, Id1. Results are expressed as an average of 3 independent experiments plus or minus SEM. (Right) The difference between the percentage of eGFP-positive cells after 7 and 17 days of differentiation is depicted. ▪ indicates eGFP; ▦, Id1. (B) Proliferation was determined by counting the eGFP-positive cells. ♦ indicates eGFP; ▴, Id1. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (C) Proliferation was determined by performing 3H-thymidine incorporation assays. Data are depicted as a ratio between cells transduced with empty vector alone or cells transduced with Id1. ▪ indicates eGFP; ▦, Id1. Data are expressed as an average of 3 independent experiments plus or minus SEM. (D) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Bar shading is as in panel C. Data are expressed as an average of 3 independent experiments plus or minus SEM. (E) CD34+ cells were transduced with eGFP or Id1. After 14 and 17 days of neutrophil differentiation, transduced cells were separated from the nontransduced cells, and cytospins were prepared and stained with May-Grünwald-Giemsa solution. (F) Neutrophil differentiation was expressed as a percentage of differentiated neutrophils within the neutrophil lineage. Data were expressed as an average of 3 independent experiments plus or minus SEM. (G) Neutrophil differentiation was also analyzed by staining the granule protein lactoferrin after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of lactoferrin between cells transduced with empty vector alone or cells transduced with Id1. Data were expressed as an average of 3 independent experiments plus or minus SEM. (H) CD34+ cells, cultured in the presence of the cytokines SCF, FLT-3 ligand, and TPO, were transduced with empty vector alone or with Id1. After 3 days of culture, cells were injected into β2-microglobulin-/- NOD/SCID mice. Six weeks after injection, mice were killed, eGFP-positive human myeloid cells were sorted, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (F-H) Bar shading is as in panel C.
Constitutive Id2 expression induces granulocyte differentiation
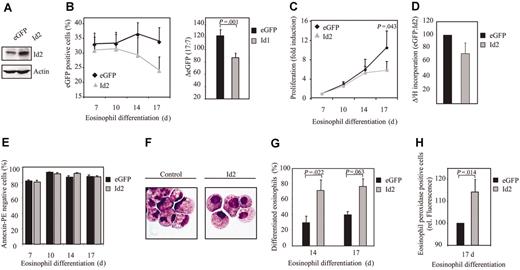
In contrast to Id1, Id2 expression was up-regulated during eosinophil differentiation. In order to investigate the relevance of this observation, umbilical cord blood–derived CD34+ cells were transduced with either eGFP or Id2 (Figure 4A). We observed a decrease in the percentage of eGFP-positive cells after transduction with Id2, compared with controls (Figure 4B). This suggests that Id2, similar to Id1, might play a role in regulating proliferation during eosinophil differentiation. To test this hypothesis, proliferation was determined by counting the total amount of eGFP-positive cells and by performing 3H-thymidine incorporation assays. Indeed, transduction of cells with Id2 resulted in a decreased proliferation compared with cells transduced with eGFP alone (Figure 4C-D), without inducing enhanced levels of apoptosis (Figure 4E). Cells were subsequently transduced with either eGFP or Id2 to investigate whether Id2 plays a role in eosinophil differentiation per se. After 14 and 17 days of differentiation, eGFP-positive cells were sorted by FACS from the nontransduced cells and cytospins were prepared (Figure 4F). Differentiation was analyzed by May-Grünwald-Giemsa staining as described in “Materials and methods.” The percentage of differentiated eosinophils within the eosinophil lineage was enhanced in cells ectopically expressing Id2 compared with the control cells (Figure 4G, Table 3). The percentage of juvenile eosinophils was similar in cells constitutively expressing Id2 compared with control cells, suggesting that it is only final maturation that is regulated by Id2 (Table 3). Similar results were obtained using the granule protein eosinophil peroxidase as a marker for eosinophil differentiation (Figure 4H). Thus, early constitutive expression of Id2 results in an increased percentage of differentiated eosinophils.
Stages of eosinophil differentiation in CD34+ cells transduced with eGFP or Id2
. | eGFP . | . | Id2 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I | 26.2 ± 14.0 | 16.9 ± 12.0 | 7.9 ± 3.2 | 15.2 ± 2.2 | ||
| Promyelocyte II | 24.7 ± 7.9 | 42.8 ± 7.9 | 20.0 ± 7.3 | 9.1 ± 3.8 | ||
| Myelocyte | 26.6 ± 7.3 | 35.3 ± 11.0 | 61.5 ± 4.2 | 61.9 ± 4.2 | ||
| Metamyelocyte/mature | 3.1 ± 1.7 | 4.4 ± 2.0 | 5.1 ± 1.1 | 9.6 ± 4.8 | ||
. | eGFP . | . | Id2 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I | 26.2 ± 14.0 | 16.9 ± 12.0 | 7.9 ± 3.2 | 15.2 ± 2.2 | ||
| Promyelocyte II | 24.7 ± 7.9 | 42.8 ± 7.9 | 20.0 ± 7.3 | 9.1 ± 3.8 | ||
| Myelocyte | 26.6 ± 7.3 | 35.3 ± 11.0 | 61.5 ± 4.2 | 61.9 ± 4.2 | ||
| Metamyelocyte/mature | 3.1 ± 1.7 | 4.4 ± 2.0 | 5.1 ± 1.1 | 9.6 ± 4.8 | ||
After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. Eosinophil differentiation was expressed as a percentage of myeloblast, promyelocyte I, promyelocyte II, myelocyte, metamyelocyte, and mature eosinophils within the eosinophil lineage. Data are expressed as an average of 3 independent experiments plus or minus SEM.
Constitutive Id2 expression enhances eosinophil differentiation. (A) After a 7-day period of culture, protein lysates were prepared from eosinophil progenitors transduced with eGFP or Id2 and a Western blot was performed with an N-terminal antibody against Id2 (top panel) and, as a control for equal loading, an antibody against β-actin (bottom panel). CD34+ cells were transduced with eGFP or Id2 and cultured in the presence of IL-5 for 17 days. (B) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. Results are expressed as an average of 3 independent experiments plus or minus SEM. The difference between the percentage of eGFP-positive cells after 17 and 7 days of differentiation was depicted in the right panel. (C) Proliferation was determined by counting the eGFP-positive cells. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (D) Proliferation was determined by performing 3H-thymidine incorporation assays. Data are depicted as a ratio between cells transduced with empty vector alone and cells transduced with Id1. Data are expressed as an average of 3 independent experiments plus or minus SEM. (E) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Data are expressed as an average of 3 independent experiments plus or minus SEM. (F) CD34+ cells were transduced with eGFP or Id2. After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (G) Eosinophil differentiation was expressed as a percentage of differentiated eosinophils within the eosinophil lineage. Data are expressed as an average of 3 independent experiments plus or minus SEM. (H) Eosinophil differentiation was also analyzed by staining the granule protein eosinophil peroxidase after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of eosinophil peroxidase between cells transduced with empty vector alone or cells transduced with Id2. Data are expressed as an average of 3 independent experiments plus or minus SEM.
Constitutive Id2 expression enhances eosinophil differentiation. (A) After a 7-day period of culture, protein lysates were prepared from eosinophil progenitors transduced with eGFP or Id2 and a Western blot was performed with an N-terminal antibody against Id2 (top panel) and, as a control for equal loading, an antibody against β-actin (bottom panel). CD34+ cells were transduced with eGFP or Id2 and cultured in the presence of IL-5 for 17 days. (B) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. Results are expressed as an average of 3 independent experiments plus or minus SEM. The difference between the percentage of eGFP-positive cells after 17 and 7 days of differentiation was depicted in the right panel. (C) Proliferation was determined by counting the eGFP-positive cells. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (D) Proliferation was determined by performing 3H-thymidine incorporation assays. Data are depicted as a ratio between cells transduced with empty vector alone and cells transduced with Id1. Data are expressed as an average of 3 independent experiments plus or minus SEM. (E) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Data are expressed as an average of 3 independent experiments plus or minus SEM. (F) CD34+ cells were transduced with eGFP or Id2. After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (G) Eosinophil differentiation was expressed as a percentage of differentiated eosinophils within the eosinophil lineage. Data are expressed as an average of 3 independent experiments plus or minus SEM. (H) Eosinophil differentiation was also analyzed by staining the granule protein eosinophil peroxidase after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of eosinophil peroxidase between cells transduced with empty vector alone or cells transduced with Id2. Data are expressed as an average of 3 independent experiments plus or minus SEM.
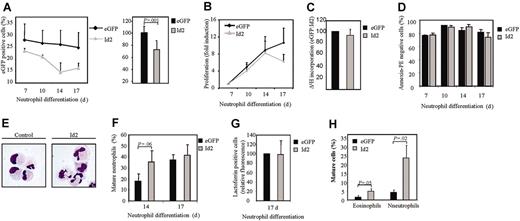
In order to investigate whether Id2 is also capable of regulating neutrophil differentiation, cells were transduced with either eGFP or Id2 and were subsequently differentiated in the presence of G-CSF toward neutrophils for 17 days (Figure 5A). Transduction of cells with Id2 resulted in a slight decrease in proliferation (Figure 5B-C) without enhanced apoptosis (Figure 5D), whereas neutrophil maturation was significantly accelerated (Figure 5E-G, Table 4). To investigate the role of Id2 in regulation of lineage development in vivo, CD34+ cells transduced with either eGFP or Id2 were transplanted into β2-microglobulin-/- NOD/SCID mice (Figure 5H). Ectopic expression of Id2 resulted in significantly increased levels of both mature eosinophils and neutrophils in the myeloid compartment of the bone marrow. These data demonstrate that Id2 expression regulates both eosinophil and neutrophil development.
Stages of neutrophil differentiation in CD34+ cells transduced with eGFP or Id2
. | eGFP . | . | Id2 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I and II | 43.9 ± 11.5 | 26.5 ± 6.7 | 32.8 ± 6.7 | 19.7 ± 5.1 | ||
| Myelocyte | 24.7 ± 7.9 | 15.9 ± 7.7 | 16.8 ± 6.4 | 17.5 ± 6.4 | ||
| Metamyelocyte/banded | 25.7 ± 9.7 | 46.4 ± 6.2 | 35.7 ± 8.4 | 47.9 ± 4.2 | ||
| Segmented | 2.7 ± 1.9 | 8.6 ± 2.1 | 8.5 ± 5.4 | 13.4 ± 4.8 | ||
. | eGFP . | . | Id2 . | . | ||
|---|---|---|---|---|---|---|
. | Day 14 . | Day 17 . | Day 14 . | Day 17 . | ||
| Myeloblast/promyelocyte I and II | 43.9 ± 11.5 | 26.5 ± 6.7 | 32.8 ± 6.7 | 19.7 ± 5.1 | ||
| Myelocyte | 24.7 ± 7.9 | 15.9 ± 7.7 | 16.8 ± 6.4 | 17.5 ± 6.4 | ||
| Metamyelocyte/banded | 25.7 ± 9.7 | 46.4 ± 6.2 | 35.7 ± 8.4 | 47.9 ± 4.2 | ||
| Segmented | 2.7 ± 1.9 | 8.6 ± 2.1 | 8.5 ± 5.4 | 13.4 ± 4.8 | ||
After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. Neutrophil differentiation was expressed as a percentage of myeloblast, promyelocyte I, promyelocyte II, myelocyte, and metamyelocyte toward neutrophils with banded and segmented nuclei within the neutrophil lineage. Data are expressed as an average of 5 independent experiments plus or minus SEM.
Constitutive Id2 expression induces neutrophil differentiation. CD34+ cells were transduced with eGFP or Id2 and cultured in the presence of G-CSF for 17 days. (A) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. Results are expressed as an average of 3 independent experiments plus or minus SEM. The difference between the percentage of eGFP-positive cells after 17 and 7 days of differentiation is depicted in the right panel. (B) Proliferation was determined by counting the eGFP-positive cells. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (C) Proliferation was determined by performing 3H-thymidine incorporation assays. Data were depicted as a ratio between cells transduced with empty vector alone and cells transduced with Id1. Data are expressed as an average of 3 independent experiments plus or minus SEM. (D) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Data are expressed as an average of 3 independent experiments plus or minus SEM. (E) CD34+ cells were transduced with eGFP or Id2. After 14 and 17 days of neutrophil differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (F) Neutrophil differentiation was expressed as a percentage of maturing neutrophils within the neutrophil lineage. Data are expressed as an average of 5 independent experiments plus or minus SEM. (G) Neutrophil differentiation was also analyzed by staining the granule protein lactoferrin after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of lactoferrin between cells transduced with empty vector alone or cells transduced with Id2. Data are expressed as an average of 3 independent experiments plus or minus SEM. (H) CD34+ cells, cultured in the presence of the cytokines SCF, FLT-3 ligand, and TPO, were transduced with empty vector alone or with Id2. Afer 3 days of culture, cells were injected into β2-microglobulin(–/–) NOD/SCID mice. Six weeks after injection, mice were killed, eGFP-positive human myeloid cells were sorted, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution.
Constitutive Id2 expression induces neutrophil differentiation. CD34+ cells were transduced with eGFP or Id2 and cultured in the presence of G-CSF for 17 days. (A) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. Results are expressed as an average of 3 independent experiments plus or minus SEM. The difference between the percentage of eGFP-positive cells after 17 and 7 days of differentiation is depicted in the right panel. (B) Proliferation was determined by counting the eGFP-positive cells. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. (C) Proliferation was determined by performing 3H-thymidine incorporation assays. Data were depicted as a ratio between cells transduced with empty vector alone and cells transduced with Id1. Data are expressed as an average of 3 independent experiments plus or minus SEM. (D) The percentage of apoptotic, transduced cells was determined with annexin V–PE by FACS analysis after 7, 10, 14, and 17 days of differentiation. Data are expressed as an average of 3 independent experiments plus or minus SEM. (E) CD34+ cells were transduced with eGFP or Id2. After 14 and 17 days of neutrophil differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. (F) Neutrophil differentiation was expressed as a percentage of maturing neutrophils within the neutrophil lineage. Data are expressed as an average of 5 independent experiments plus or minus SEM. (G) Neutrophil differentiation was also analyzed by staining the granule protein lactoferrin after 17 days of culture. Data were depicted as the ratio of the levels (arithmetric mean) of lactoferrin between cells transduced with empty vector alone or cells transduced with Id2. Data are expressed as an average of 3 independent experiments plus or minus SEM. (H) CD34+ cells, cultured in the presence of the cytokines SCF, FLT-3 ligand, and TPO, were transduced with empty vector alone or with Id2. Afer 3 days of culture, cells were injected into β2-microglobulin(–/–) NOD/SCID mice. Six weeks after injection, mice were killed, eGFP-positive human myeloid cells were sorted, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution.
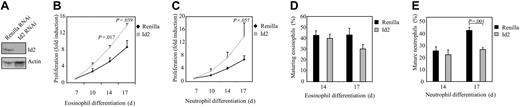
Since Id2 expression is up-regulated during granulocyte differentiation, and ectopic expression of this protein induces granulopoiesis, it was of interest to examine the effect of inhibition of Id2 expression on eosinophil and neutrophil development. In order to investigate this, a bicistronic retroviral DNA construct coexpressing eGFP and either Id2 RNAi or control Renilla RNAi was used to generate retrovirus and subsequently to infect CD34+ cells. To demonstrate that the construct was functional and able to knock down Id2 expression, cells were cotransfected with an Id2 expression construct and either pRetrosuper Renilla RNAi or pRetrosuper Id2 RNAi (Figure 6A). Id2 expression was dramatically down-regulated in cells transfected with Id2 RNAi compared with cells transfected with Renilla RNAi as a control.
CD34+ cells were subsequently transduced and differentiated toward eosinophils and neutrophils. The effect of Id2 knock-down in granulocyte progenitors on the proliferation of these cells was examined by counting the number of eGFP-positive cells (Figure 6B-C). Inhibition of Id2 expression induced proliferation during both eosinophil and neutrophil development.
To investigate whether Id2 knock-down could affect granulocyte differentiation per se, transduced cells were separated from the nontransduced cells and cytospins prepared after 14 and 17 days of differentiation. Terminal maturation of both eosinophils and neutrophils was inhibited when Id2 expression was blocked (Figure 6D-E). Granulopoiesis was primarily blocked at the promyelocyte stage of differentiation (data not shown). These data demonstrate that up-regulation of Id2 protein levels during granulopoiesis is required for maturation.
Discussion
In this study, we have investigated the role of Id proteins during granulopoiesis. Our data demonstrate that expression of Id proteins is differentially regulated during eosinophil and neutrophil differentiation. Id1 protein levels are initially up-regulated during early granulopoiesis and down-regulated during final maturation. In contrast, Id2 expression is highly up-regulated during eosinophil differentiation, and is moderately enhanced in terminally differentiated neutrophils. Furthermore, we also demonstrate that ectopic expression of Id1 inhibits both proliferation and differentiation during eosinophil development, whereas neutrophil differentiation was modestly enhanced by Id1. In contrast, ectopic expression of Id2 inhibits proliferation and stimulates final maturation of granulocytes, whereas inhibition of Id2 expression induces proliferation and inhibits maturation of both eosinophil and neutrophil progenitors. In addition, our results also demonstrate that both Id1 and Id2 play an important role in regulation of lineage development in vivo. Eosinophil development was enhanced in cells ectopically expressing Id2, whereas neutrophil development was enhanced in cells ectopically expressing either Id1 or Id2. Although we observed little effect of Id1 expression in eosinophil differentiation in vivo, these experiments were hampered by the very low levels of mature eosinophils generated in this NOD/SCID model system.
Id2 knock-down in granulocyte progenitors inhibits eosinophil and neutrophil differentiation. (A) Protein lysates were prepared from COS cells cotransfected with pSG5-Id2 and pRetrosuper Renilla RNAi or pRetrosuper Id2 RNAi. A Western blot was performed with an N-terminal antibody against Id2 (top panel) and, as a control for equal loading, an antibody against β-actin (bottom panel). CD34+ cells were transduced with Renilla RNAi or Id2 RNAi and cultured in the presence of IL-5 or G-CSF for 17 days. Proliferation during eosinophil (B) and neutrophil (C) differentiation was determined by counting the eGFP-positive cells. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. CD34+ cells were transduced with eGFP or Id2. After 14 and 17 days of eosinophil (D) and neutrophil (E) differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. Data are expressed as an average of 6 independent experiments plus or minus SEM.
Id2 knock-down in granulocyte progenitors inhibits eosinophil and neutrophil differentiation. (A) Protein lysates were prepared from COS cells cotransfected with pSG5-Id2 and pRetrosuper Renilla RNAi or pRetrosuper Id2 RNAi. A Western blot was performed with an N-terminal antibody against Id2 (top panel) and, as a control for equal loading, an antibody against β-actin (bottom panel). CD34+ cells were transduced with Renilla RNAi or Id2 RNAi and cultured in the presence of IL-5 or G-CSF for 17 days. Proliferation during eosinophil (B) and neutrophil (C) differentiation was determined by counting the eGFP-positive cells. Results are expressed as fold increase in cell numbers compared with day 7 as an average of 3 independent experiments plus or minus SEM. CD34+ cells were transduced with eGFP or Id2. After 14 and 17 days of eosinophil (D) and neutrophil (E) differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. Cytospins were stained with May-Grünwald-Giemsa solution. Data are expressed as an average of 6 independent experiments plus or minus SEM.
Id proteins are considered as positive regulators of proliferation in a variety of cell types.35-39 Interestingly, however, our data demonstrate that Id proteins may also inhibit proliferation during granulopoiesis. Id proteins primarily function by heterodimerization with bHLH transcription factors, thereby inhibiting their transcriptional activity.40 It has been demonstrated that ectopic expression of Id1 in mouse erythroleukemia (MEL) cells abrogates the formation of a complex between the bHLH transcription factors SCL/Tal1 and E2A in those cells.41 SCL/Tal1 is primarily expressed in hematopoietic cells,42,43 and plays an important role in both primitive and definitive hematopoiesis during embryogenesis and is embryonic lethal, as observed in SCL-deficient mice.44,45 In the adult hematopoietic system, SCL has been demonstrated to play an important role in the generation of erythrocytes and megakaryocytes.46 In addition, enforced expression of SCL in human CD34+ hematopoietic stem cells resulted in enhanced colony growth and size during megakaryocyte and erythrocyte development, which was attributed to a shortened cell cycle time.47 SCL is highly expressed in hematopoietic stem cells and is rapidly down-regulated during early stages of monocyte and granulocyte development.48 Ectopic expression of SCL in a granulocytic cell line (HL60 cells) results in enhanced levels of proliferation and inhibits differentiation. We observed an inhibition in proliferation during eosinophil differentiation in CD34+ hematopoietic progenitors ectopically expressing Id1 and Id2. It is tempting to speculate that Id proteins in these hematopoietic precursors associates with SCL and, thereby, inhibit proliferation.
It has recently been suggested that enhanced differentiation induced by Id proteins is regulated by an alternative, bHLH-independent mechanism. Recent studies have revealed that Id proteins are also capable of association with several other classes of proteins besides bHLH transcription factors. These include mouse Id-associated protein 1 (MIDA1),49,50 Paired-box protein 2 (Pax2), Pax5, Pax8,51 c-ets-1,52,53 and members of the family of retinoblastoma proteins.54-56 Knock-down of MIDA1 (mouse Id-associated 1) by treatment with antisense RNA resulted in decreased levels of proliferation in erythroleukemic cells.57 It has been demonstrated that Id1 association with MIDA1 results in disruption of its binding to DNA,50 suggesting that association of Id1 with MIDA1 can inhibit proliferation. Whereas MIDA1 expression has been observed only in a small number of cell types, including erythroid leukemic cells, it would be of interest to investigate whether MIDA1 is expressed in human hematopoietic precursors in general.
Id proteins have been demonstrated to play an important role in negatively regulating differentiation of several hematopoietic lineages, including lymphocyte development. Id2-deficient mice exhibit enhanced levels of mature B lymphocytes in the spleen, indicating that Id2 expression may inhibit B-cell development.37 Furthermore, both Id158 and Id324 have been shown to inhibit B-lymphocyte development at an early stage of differentiation. In addition to its role in regulating B-cell development, Id3 has also been demonstrated to down-regulate T-lymphocyte differentiation.25,26 Similarly, our data clearly demonstrate that ectopic expression of Id1 inhibits eosinophil differentiation, and suggests that Id1 needs to be down-regulated to enable eosinophilopoiesis.
Although Id proteins, in general, are considered to be inhibitors of differentiation, recent studies have suggested that they are also capable of positively regulating differentiation in specific cell types, such as during NK cell development. In Id2-deficient mice, for example, NK cell development was completely abolished.27,28 In addition, studies with Id2-deficient mice revealed that Id2 expression is required for the development of a specific subset of dendritic cells.59 Our data demonstrate that Id2 expression positively regulates granulopoiesis, and supports the hypothesis that Id proteins can have different cell type–specific functions.
While different members of the family of Id proteins have been demonstrated to inhibit similar bHLH proteins, the question is raised as to why ectopic expression of Id1 and Id2, in both our ex vivo differentiation model as well as in vivo, have distinct effects. In contrast to Id1, Id2 can associate with retinoblastoma proteins and thereby abrogate retinoblastoma-mediated growth arrest.55,56 In addition, Id proteins possess several potential phosphorylation sites, and it has been demonstrated that Id2, Id3, and Id4, but not Id1, can be phosphorlyated by the cyclin D–CDK2 complex.60 It is thought that phosphorylation of Id2 is required for cell cycle progression, and ectopic expression of a nonphosphorylatable Id2 mutant was found to inhibit cell growth.61 It is therefore tempting to speculate that phosphorylation of Id proteins might differentially regulate their functions during terminal differentiation. Furthermore, it has been demonstrated that there are differences in efficiency in association between different Id proteins and bHLH transcription factors. Id2, for example, is less efficient in associating with bHLH transcription factors compared with Id1.61 In addition, Id1 associates more efficiently with PAX5, required for B-cell development, compared with Id2. The differences in ability to bind to other proteins might play an important role in differentially regulating differentiation of certain cell types.
Ectopic expression of Id1 in cells cultured in the presence of IL-5 blocked eosinophil differentiation at an early stage. Interestingly, an increase in the percentage of cells belonging to the neutrophil lineage could be observed in some experiments (Figure 2E). Neutrophil differentiation in the presence of G-CSF was also modestly enhanced by ectopic expression of Id1. In contrast, Id2 expression enhances granulocyte differentiation in general in both our ex vivo and in vivo differentiation models. Although the regulatory mechanisms by which Id proteins regulate differentiation of specific cell types remains to be investigated, these data clearly demonstrate that correct expression of both Id1 and Id2 plays a critical, though differential, role in granulocyte differentiation of primary human hematopoietic cells.
Prepublished online as Blood First Edition Paper, February 8, 2005; DOI 10.1182/blood-2004-12-4883.
Supported by research grant no. UU 2001-2491 from the Dutch Cancer Society (M.B., H.W.M.v.D., L.P.V.), and a Center of Excellence grant from the Swedish Foundation for Strategic Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr G. Nolan (Stanford University, Stanford, CA) for kindly providing us with the LZRS construct and the phoenix-ampho packaging cell line. We would also like to thank Prof H. Spits (AMC, Amsterdam, The Netherlands) for providing us with the Id1 retroviral construct and the pRetrosuper construct.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal