Comment on Othman et al, page 4330
Again-of-function mutation of the platelet adhesion receptor GP Ibα caused by deletion of a region far removed from the ligand-binding site forces a reassessment of the mechanisms by which platelets attach to von Willebrand factor at sites of blood vessel injury.
Gain-of-function mutations affect both the platelet glycoprotein Ib/IX/V (GPIb/IX/V) complex and its primary ligand in mediating platelet adhesion, von Willebrand factor (VWF), producing either platelet-type von Willebrand disease (PT-VWD) or type 2B von Willebrand disease, respectively. These mutations paradoxically produce bleeding syndromes that can be severe, believed to be a consequence of enhanced platelet binding and rapid clearance of the largest multimers of VWF, which are the most active hemostatically. Two naturally occurring PT-VWD mutations of GPIbα involve a disulfide loop within the N-terminus of the molecule that follows a region containing 8 tandem repeats of a conserved leucine-rich motif. This region forms 1 of 2 binding patches for direct contact with VWF, with the sequence Y228 to S241 contributing 2 strands to an 8-strand intermolecular β sheet structure with VWF.1 It is postulated these interactions are stabilized by each of the 4 previously described gain-of-function mutations: 2 naturally occurring mutations, G233V and M239V, and 2 artificial mutations, D235V and K237V.2 In this issue of Blood, Othman and colleagues describe the molecular defect associated with PT-VWD in a family with 3 affected individuals. As with other cases of PT-VWD, the mutation is transmitted in an autosomal-dominant fashion. But, unlike typical PT-VWD, this mutation does not directly affect the ligand-binding region of GPIbα. Instead, the in-frame deletion of 27 nucleotides produces a molecule missing 9 amino acids from within a heavily glycosylated mucin-like spacer (the macroglycopeptide) that separates the ligand-binding domain from the plasma membrane. Because such glycosylated sequences tend to be elongated, deletion of 9 amino acids might be expected to significantly shorten the distance of the ligand-binding domain from the plasma membrane. This alone does not account for the gain of function, however, as a normal variant of GPIbα exists (the VNTR D allele) that is 4 amino acids shorter than the mutant described.3
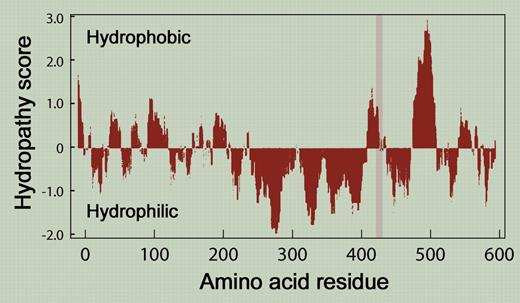
Hydropathy profile of GP Ib α by the method of Kyte and Doolittle. The gray bar denotes the deleted region.
Hydropathy profile of GP Ib α by the method of Kyte and Doolittle. The gray bar denotes the deleted region.
What else could account for the mutant's lowered threshold for VWF binding? The answer awaits clever experiments, but recent evidence indicates that the GPIb/IX/V complex has several topological requirements for optimal VWF binding, including localization to lipid rafts4 and association through its cytoplasmic domains with the adaptor protein 14-3-3ζ.5 Might it be that, rather than enhancing the affinity of individual polypeptides for VWF, the mutation changes VWF binding by altering the percentage of the GPIb/IX/V complex that associates with lipid rafts or changes the relative positioning of the cytoplasmic domains of its polypeptides? It seems unlikely that the deleted sequence positively modulates VWF binding by direct means.
One potential clue to why this particular deletion changes GPIbα's VWF-binding function while the macroglycopeptide as a whole tolerates considerable length variability is provided by examination of a hydropathy plot. Of the roughly 190 amino acids that separate the ligand-binding domain from the plasma membrane, only a short region that includes the deleted sequence is of predominantly hydrophobic character (see the figure). This may introduce a kink in the polypeptide, or produce a region that plays more than a structural role, perhaps binding either GPIbβ or GPIX as a means of regulating VWF binding. Whatever the explanation, the paper by Othman et al illustrates once again the value of studying natural mutations to gain insight into protein function. Without these patients, it is very unlikely that the deleted sequence would have been examined as a potential modulator of VWF binding. ▪

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal