Clonal expansion of mixed-lineage leukemia (MLL) rearrangements is highly indicative for acute leukemia.1-5 Here, we report the unusual finding of an MLL rearrangement in the absence of leukemia.
A 5-year-old girl was diagnosed with acute myeloid leukemia (AML) characterized by a variant t(8;21) translocation. The patient achieved remission, and is now, 4 years later, still in ongoing remission. However, a follow-up control of the bone marrow 15 months after diagnosis revealed a new clone characterized by the MLL rearrangement t(11;11)(q13;q23). To exclude the possibility of a secondary AML, numerous bone marrow control samples were subsequently investigated over a 30-month period. During this period, the amount of MLL rearranged cells was decreasing from 50% to 20% as determined by karyotype and fluorescence in situ hybridization (FISH) analysis. Morphology was unsuspicious without any sign for a secondary malignancy in all samples.
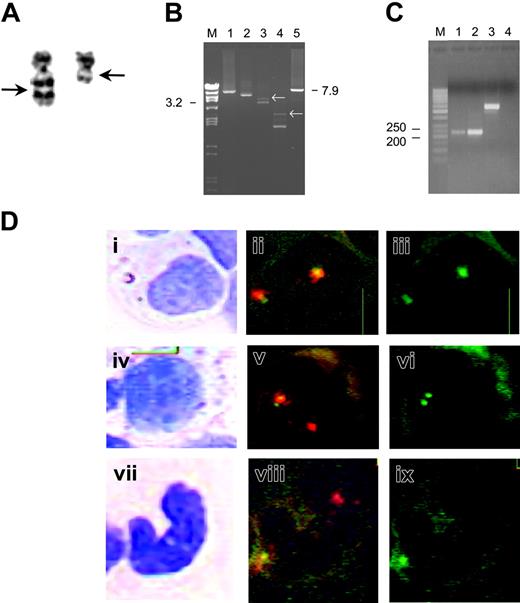
The breakpoint region of the MLL partner gene could be mapped to chromosome 11 at band q13 (Figure 1A). Using a newly developed restriction enzyme fragment length polymorphism–based cloning strategy, a new MLL partner gene was identified as ARHGEF17 (Figure 1B).6 The ARHGEF17 protein is a Rho guanine nucleotide exchange factor involved in cellular signaling.7 The fusion between the MLL and the ARHGEF17 genes occurred in introns 12 and 1, respectively. The reciprocal allele was isolated as well and fused ARHGEF17 intron 1 with MLL intron 12. The particular chimeric transcript leads to an in-frame fusion gene that expresses the corresponding fusion RNA (Figure 1C).
To discriminate between a rearrangement that already existed at diagnosis at a low level from a secondary one, molecular analysis of the chromosomal breakpoint region was performed in the diagnostic material. However, detection of the MLL-ARHGEF17 fusion gene sequence by using specific primers failed, indicating the appearance of a secondary, potentially treatment-induced rearrangement.
To further illuminate the biology of our observation, we precisely analyzed the morphology of those cells carrying the MLL-ARHGEF17 fusion gene 30 months after initial detection of the rearrangement (Figure 1D, Table 1).8 The novel MLL rearrangement could only be detected in cells within the myeloid lineage. Remarkably, all cell types of the myeloid lineage, from promyelocytes to differentiated polymorphonuclear neutrophils, were affected by the novel fusion gene, highlighting 2 conclusions: first, the MLL-ARHGEF17 rearrangement is an early clonal event in myelopoiesis, and second, the chimeric protein does not lead to a block of the myeloid differentiation.
Distribution of MLL-ARHGEF17-positive cells in the different cell types of the myeloid lineage.
. | Promyelocytes . | Myelocytes . | Metamyelocytes . | Bands . | PMNN . |
|---|---|---|---|---|---|
| t(11;11), (%) | 10 (66.6) | 178 (71.2) | 14 (63.6) | 4 (4.8) | 10 (4.2) |
| Normal (%) | 5 (34.4) | 72 (28.8) | 8 (36.4) | 80 (95.2) | 223 (95.8) |
| All | 15 | 250 | 22 | 84 | 233 |
. | Promyelocytes . | Myelocytes . | Metamyelocytes . | Bands . | PMNN . |
|---|---|---|---|---|---|
| t(11;11), (%) | 10 (66.6) | 178 (71.2) | 14 (63.6) | 4 (4.8) | 10 (4.2) |
| Normal (%) | 5 (34.4) | 72 (28.8) | 8 (36.4) | 80 (95.2) | 223 (95.8) |
| All | 15 | 250 | 22 | 84 | 233 |
Different cell types are indicated. The rows represent MLL-ARHGEF17-positive cells (marked as t(11;11)), MLL-ARHGEF17-negative cells (marked as normal), and the total number of cells of each cell type (marked as all).
Our data suggest that neither the MLL-ARHGEF17 nor the ARHGEF17-MLL gene fusion result in oncogenic dysregulation of cell growth. This is supported by the observation that the number of cells carrying the MLL rearrangement eventually started to decrease in the course of time. Nevertheless, this conclusion must be drawn very carefully, since the follow-up time of the patient is relatively short and we cannot exclude that MLL-ARHGEF17 and/or ARHGEF17-MLL contributes to leukemogenesis in combination with further additional genetic events. However, to our knowledge, this is the first report of an MLL rearrangement that results in a clonal population of cells but lacks leukemogenic behavior.
The detection of a novel MLL rearrangement and combined analysis of morphology and FISH of the bone marrow cells in the myeloid lineage. (A) Partial karyotype (G-banding) for the t(11;11)(q13;q23). Both chromosomes 11 are shown. The arrows indicate the breakpoints in the rearranged chromosomes. (B) Genomic DNA of the patient analyzed by the long-distance inverse–polymerase chain reaction (LDI-PCR) method. BamHI-digested and religated genomic DNA was tested with 5 oligonucleotides in 4 different combinations6 (lane 1: A-B, lane 2: A-C, lane 3: A-D, and lane 4: A-E; M indicates marker). The analysis included a positive control using the oligonucleotides B and F (lane 5) that amplifies a 7.9 kb DNA of the MLL breakpoint cluster region. In the analysis of the patient, non-germline amplimers can be observed (white arrows). Such non-germline amplimers were isolated. Sequence analysis of the isolated bands identified the novel MLL partner gene ARHGEF17. (C) Reverse transcriptase (RT)–PCR analysis of MLL-ARHGEF17 fusion transcript. RT-PCR analysis of the immediate breakpoint region in bone marrow cell DNA of the patient amplifying a product of 222 bp with the following primers: forward primer sequence 5′-GTCTGTTGTGAGCCCTTCC-3′, reverse primer sequence 5′-CTGCATGTAGCCCTGC ATC-3′. Lane 1: MLL-ARHGEF17, primer concentration 3.3 nmol/mL; lane 2: MLL-ARHGEF17, primer concentration 10 nmol/mL; lane 3: positive control (EWS FLI1); lane 4: negative control, M indicates marker. (D) Combined analysis of morphology and FISH. The left panels (i, iv, vii) show the cells stained with May-Grünwald-Giemsa. The middle panels (ii,v,viii) display the same cells after FISH; image acquisition was performed using a green/orange dual filter set. The right panels (iii,vi,ix) show acquisition of the same cells using a single green filter. The hybridization pattern for the nuclei without the MLL rearrangement is 2 yellow signals with the green/orange dual filter set and 2 spaced green signals with a single green filter. The hybridization pattern for the nuclei with the MLL-ARHGEF17 rearrangement is one yellow/one red signal, where the yellow signal represents the normal intact MLL gene and the green signal the rearranged MLL gene. Two closely located green signals are detected using a single green filter. The top row shows a myelocyte (i) with 2 nonrearranged MLL signals (ii,iii), the middle row shows a promyelocyte (iv) with the MLL-ARHGEF17 rearrangement (v,vi), and the bottom row shows a “band” (vii) with the MLL-ARHGEF17 rearrangement (viii,ix). Image acquisition was performed as described previously9 with the Duet system (BioView, Rehovot, Israel) at × 1000 magnification.
The detection of a novel MLL rearrangement and combined analysis of morphology and FISH of the bone marrow cells in the myeloid lineage. (A) Partial karyotype (G-banding) for the t(11;11)(q13;q23). Both chromosomes 11 are shown. The arrows indicate the breakpoints in the rearranged chromosomes. (B) Genomic DNA of the patient analyzed by the long-distance inverse–polymerase chain reaction (LDI-PCR) method. BamHI-digested and religated genomic DNA was tested with 5 oligonucleotides in 4 different combinations6 (lane 1: A-B, lane 2: A-C, lane 3: A-D, and lane 4: A-E; M indicates marker). The analysis included a positive control using the oligonucleotides B and F (lane 5) that amplifies a 7.9 kb DNA of the MLL breakpoint cluster region. In the analysis of the patient, non-germline amplimers can be observed (white arrows). Such non-germline amplimers were isolated. Sequence analysis of the isolated bands identified the novel MLL partner gene ARHGEF17. (C) Reverse transcriptase (RT)–PCR analysis of MLL-ARHGEF17 fusion transcript. RT-PCR analysis of the immediate breakpoint region in bone marrow cell DNA of the patient amplifying a product of 222 bp with the following primers: forward primer sequence 5′-GTCTGTTGTGAGCCCTTCC-3′, reverse primer sequence 5′-CTGCATGTAGCCCTGC ATC-3′. Lane 1: MLL-ARHGEF17, primer concentration 3.3 nmol/mL; lane 2: MLL-ARHGEF17, primer concentration 10 nmol/mL; lane 3: positive control (EWS FLI1); lane 4: negative control, M indicates marker. (D) Combined analysis of morphology and FISH. The left panels (i, iv, vii) show the cells stained with May-Grünwald-Giemsa. The middle panels (ii,v,viii) display the same cells after FISH; image acquisition was performed using a green/orange dual filter set. The right panels (iii,vi,ix) show acquisition of the same cells using a single green filter. The hybridization pattern for the nuclei without the MLL rearrangement is 2 yellow signals with the green/orange dual filter set and 2 spaced green signals with a single green filter. The hybridization pattern for the nuclei with the MLL-ARHGEF17 rearrangement is one yellow/one red signal, where the yellow signal represents the normal intact MLL gene and the green signal the rearranged MLL gene. Two closely located green signals are detected using a single green filter. The top row shows a myelocyte (i) with 2 nonrearranged MLL signals (ii,iii), the middle row shows a promyelocyte (iv) with the MLL-ARHGEF17 rearrangement (v,vi), and the bottom row shows a “band” (vii) with the MLL-ARHGEF17 rearrangement (viii,ix). Image acquisition was performed as described previously9 with the Duet system (BioView, Rehovot, Israel) at × 1000 magnification.
Supported by a grant from the Krebsliga of the Kanton Zurich and the Wilhelm-Sander-Stiftung.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal