Abstract
The regulation of B-cell lymphoma 2 (BCL2) protein expression in germinal center (GC) B cells has been controversial. Previous reports have indicated posttranscriptional regulation plays a dominant role. However, a number of recent studies contradicted these reports. Using real-time polymerase chain reaction (PCR) and Standardized Reverse Transcriptase-PCR (StaRT-PCR), we measured the level of mRNA expression in GC, mantle zone (MNZ), and marginal zone (MGZ) cells from laser capture microdissection. Both quantitative RT-PCR measurements of microdissected GC cells from tonsils showed that GC cells had low expression of BCL2 transcripts commensurate with the low protein expression level. These results are in agreement with microarray studies on fluorescence-activated cell sorter (FACS)-sorted cells and microdissected GC cells. We also examined BCL2 mRNA and protein expression on a series of 30 cases of diffuse large B-cell lymphoma (DLBCL) and found, in general, a good correlation. The results suggested that BCL2 protein expression is regulated at the transcriptional level in normal B cells and in the neoplastic cells in most B-cell lymphoproliferative disorders.
Introduction
BCL2 was identified in follicular lymphoma (FL) cells, the neoplastic counterpart of normal germinal center (GC) B cells, with the t(14;18)(q32;q21) chromosomal translocation.1,2 Translocation of BCL2 at 18q21 to the proximity of the immunoglobulin heavy chain (IgH) enhancer at 14q32 up-regulates BCL2 gene expression and confers resistance to apoptosis. B-cell lymphoma 2 (BCL2) protein is expressed in 89% of FLs with t(14;18), in contrast to 25% of those without t(14;18).3 Normal GC B cells have a very low level of BCL2 protein expression (Figure 1A). However, there were reports of high expression of BCL2 mRNA in GC B cells,4-6 leading to the conclusion that BCL2 protein expression is regulated at the posttranscriptional level. However, a number of recent studies contradicted the previous reports of high BCL2 mRNA expression in GC B cells, including several microarray gene expression profiling studies (Figure 1B).7-11
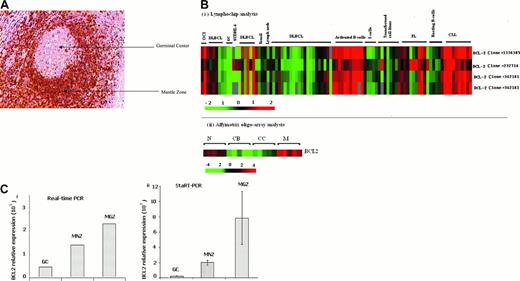
BCL2 expression in GC B cells. (A) BCL2 in paraffin-embedded reactive tonsil. The tissue section was stained with the avidin-biotin-peroxidase complex (ABC) method. GC B cells are negative for BCL2, whereas MNZ B cells show strong positive staining. Scattered BCL2-positive T cells are seen in the GC. The image was acquired using an Arcturus PixCell II microscope (Mountain View, CA); the image is 200× original magnification (20× objective) and was captured using the PixCell II Image Archiving workstation. (B) BCL2 mRNA expression on Lymphochip and oligonucleotide array. Previously published data revealed low expression of mRNA in GC B cells. (i) Lymphochip analysis.9 (ii) Affymetrix Chip analysis.10 CB indicates centroblast; CC, centrocyte; N, naive B cell; and M, memory B cell. Also note the high expression of BCL2 message in CLL and FL. Only clone 232714 measures truncated transcripts resulting from translocation of the BCL2 gene at the major breakpoint region. (C) BCL2 expression in microdissected germinal center (GC), mantle zone (MNZ), and marginal zone (MGZ) cells measured by real-time PCR (i) and StaRT-PCR (ii). Standard deviation from 3 independent experiments is shown.
BCL2 expression in GC B cells. (A) BCL2 in paraffin-embedded reactive tonsil. The tissue section was stained with the avidin-biotin-peroxidase complex (ABC) method. GC B cells are negative for BCL2, whereas MNZ B cells show strong positive staining. Scattered BCL2-positive T cells are seen in the GC. The image was acquired using an Arcturus PixCell II microscope (Mountain View, CA); the image is 200× original magnification (20× objective) and was captured using the PixCell II Image Archiving workstation. (B) BCL2 mRNA expression on Lymphochip and oligonucleotide array. Previously published data revealed low expression of mRNA in GC B cells. (i) Lymphochip analysis.9 (ii) Affymetrix Chip analysis.10 CB indicates centroblast; CC, centrocyte; N, naive B cell; and M, memory B cell. Also note the high expression of BCL2 message in CLL and FL. Only clone 232714 measures truncated transcripts resulting from translocation of the BCL2 gene at the major breakpoint region. (C) BCL2 expression in microdissected germinal center (GC), mantle zone (MNZ), and marginal zone (MGZ) cells measured by real-time PCR (i) and StaRT-PCR (ii). Standard deviation from 3 independent experiments is shown.
Microarrays do not provide a very precise measurement of mRNA level and the experiments were performed on fluorescence-activated cell sorter (FACS)-isolated cells. To obtain more precise measurements and to directly measure BCL2 mRNA expression in GC B cells, we used real-time polymerase chain reaction (PCR) and Standardized Reverse Transcriptase-PCR (StaRT-PCR) studies to determine the level of BCL2 transcripts in laser capture microdissected normal lymphoid tissues. We also compared the BCL2 protein expression in 30 cases of diffuse large B-cell lymphoma (DLBCL) with microarray-determined BCL2 mRNA expression levels. Our results clearly showed that GC B cells have low expression of BCL2 transcripts paralleling the low protein expression level. There is also a good correlation between mRNA and protein expression in DLBCL.
Study design
Laser capture microdissection (LCM)
To detect mRNA expression of BCL2 in normal GC, mantle zone (MNZ), and marginal zone (MGZ) cells, we dissected these 3 compartments from snap-frozen reactive tonsils or spleens using LCM (Arcturus PixCell II System, Mountain View, CA). DNA-free RNA was extracted using Absolutely RNA MicroRNA Isolation Kit (Stratagene, La Jolla, CA) and was reverse transcribed into cDNA for analysis. Informed consent was obtained, and the Institutional Review Board of the University of Nebraska approved this study.
Quantitative RT-PCR assay
The expression of BCL2, relative to the housekeeping gene β-actin (ACTB), was measured by real-time PCR as described previously.13 StaRT-PCR is a competitive quantitative RT-PCR assay performed by Gene Express Inc (Toledo, OH).
Immunohistochemistry for BCL2
Formalin-fixed, paraffin-embedded 5-μm tissue sections were stained with BCL2 (clone 24) antibody (Dako, Carpinteria, CA) as described previously.14 BCL2 immunostaining was scored semiquantitatively combining the intensity of staining with the proportion of cells positive for the stain (intensity of staining graded as 1+,2+, and 3+; and the % of cells positive graded as 1+ [10%-25%], 2+ [25%-75%], 3+ [> 75%]; BCL2 protein index = intensity score × numeric positive score).
Analysis of cDNA microarray data
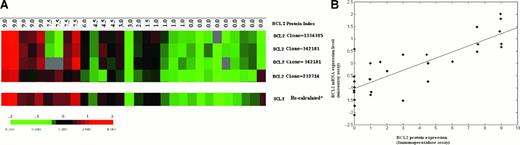
We correlated BCL2 protein expression and BCL2 mRNA level measured by cDNA microarray in a series of previously reported DLBCL cases.9 Of the 3 different BCL2 clones (232714, 342181, 1336385) on the Lymphochip microarray, only clone 232714 with a more 5′ sequence could measure message with a truncated 3′ end due to translocation at the major breakpoint region.14 However, clone 232714 is located far from the 3′ end of the transcript and may fail to accurately measure the normal BCL2 transcripts. We used the mean values measured by the BCL2 clones that are located close to the 3′ end (342181, 1336385) of the transcript to represent the gene expression level in cases where this mean value was greater than the measurement by clone 232714.
Results and discussion
BCL2 is an antiapoptosis factor important in normal B-cell development and differentiation.15 Previous reports showed that both naive and memory B cells have detectable BCL2 protein expression, whereas both centroblasts and centrocytes in normal GCs express very low levels of BCL2.7 There were a number of studies reporting that GC B cells contained abundant BCL2 mRNA and hence suggested that BCL2 protein expression may be regulated at a posttranscriptional level.4-6 We and others have observed low mRNA expression in gene expression profiling studies of purified GC B cells,9-12 which contradict previous reports. In 2 previous studies using in situ hybridization, the oligonucleotide probe used was complementary to 105 to 134 of the translated region of BCL2 containing 100% G and C sequence. A basic local alignment search tool (BLAST) search of this sequence showed 18- to 19-bp stretches that are identical to known human transcripts (eg, HRY, HSVRG1) and might lead to nonspecific binding.
We used the more accurate quantitative PCR assays on microdissected whole GC, MNZ, and MGZ cells to provide independent confirmation of the microarray data. Real-time PCR showed that naive and memory B cells expressed 4- to 10-fold more BCL2 mRNA than GC B cells, even though more T cells that expressed BCL2 mRNA were present in the GC than in the MNZ and MGZ compartments (Figure 1Ci). StaRT-PCR, a competitive quantitative PCR assay using BCL2 primers (F1 and R1) about 900 bp upstream of our real-time PCR primer pairs (F2 and R2; Table 1), confirmed the real-time PCR results (Figure 1Cii). BCL2 mRNA level in GC B cells (1.6 × 102BCL2 molecules/106 ACTB molecules) was markedly lower than in MNZ cells (1.8 × 103BCL2 molecules/106 ACTB molecules) and MGZ cells (7.8 × 103BCL2 molecules/106 ACTB molecules; Figure 1C). These results on tissue compartments were in agreement with previous reports where FACS-sorted GC B cells had much lower expression of BCL2 than in activated and resting blood B cells and naive and memory B cells9 (Figure 1B). Microarray experiments showed an increased expression of BCL2 mRNA in follicular lymphoma (FL) and chronic lymphocytic leukemia (CLL), neoplastic conditions that are known to be very frequently BCL2 protein positive. In DLBCL, BCL2 mRNA expression was more variable. However, there is good correlation between BCL2 protein expression level measured by immunohistochemistry and mRNA level measured from microarray analysis. (Figure 2B; r = 0.81).
BCL2 primers and probe sequences used in quantitative RT-PCR
Primers* . | Sequence (5′-3′) . | Length of the product . |
|---|---|---|
| F1 | TTTTAGGAGACCGAAGTCCG | 392 bp (BCL2, exon 3) StaRT-PCR |
| R1 | AGCCAACGTGCCATGTGCTA | |
| F2 | TTGCTTTACGTGGCCTGTTTC | 94 bp (BCL2, exon 3) Real-time PCR |
| R2 | GAAGACCCTGAAGGACAGCCAT | |
| BCL2 probe | FAM–CCCACCCAGAGCCCTCCTGCC–TAMRA |
Primers* . | Sequence (5′-3′) . | Length of the product . |
|---|---|---|
| F1 | TTTTAGGAGACCGAAGTCCG | 392 bp (BCL2, exon 3) StaRT-PCR |
| R1 | AGCCAACGTGCCATGTGCTA | |
| F2 | TTGCTTTACGTGGCCTGTTTC | 94 bp (BCL2, exon 3) Real-time PCR |
| R2 | GAAGACCCTGAAGGACAGCCAT | |
| BCL2 probe | FAM–CCCACCCAGAGCCCTCCTGCC–TAMRA |
F indicates forward; and R, reverse.
F1 and R1 were applied in StaRT-PCR and F2 and R2 were used in real-time PCR
The correlation between BCL2 protein and mRNA expression level. (A) Correlation of BCL2 protein index with BCL2 mRNA measured by cDNA microarray in 30 cases of DLBCL. Protein index was generated based on the scores of staining intensity and the percentage of positive cells with immunohistochemistry. RNA was measured with the Lymphochip microarray.9 Each row represents measurements by a separate cDNA clone on the microarray. Gene expression is represented by the ratio between each experimental mRNA sample to the reference mRNA standard and is depicted according to the color scale shown at the bottom. Gray indicates missing or excluded data. (B) Scatter plot between BCL2 mRNA expression on cDNA microarray and BCL2 protein index measured by immunohistochemistry. BCL2 mRNA expression was calculated as detailed in “Study design” to more accurately reflect the true expression levels (*). The least squares fit line indicates good correlation (r = 0.81) of BCL2 protein level with BCL2 mRNA level.
The correlation between BCL2 protein and mRNA expression level. (A) Correlation of BCL2 protein index with BCL2 mRNA measured by cDNA microarray in 30 cases of DLBCL. Protein index was generated based on the scores of staining intensity and the percentage of positive cells with immunohistochemistry. RNA was measured with the Lymphochip microarray.9 Each row represents measurements by a separate cDNA clone on the microarray. Gene expression is represented by the ratio between each experimental mRNA sample to the reference mRNA standard and is depicted according to the color scale shown at the bottom. Gray indicates missing or excluded data. (B) Scatter plot between BCL2 mRNA expression on cDNA microarray and BCL2 protein index measured by immunohistochemistry. BCL2 mRNA expression was calculated as detailed in “Study design” to more accurately reflect the true expression levels (*). The least squares fit line indicates good correlation (r = 0.81) of BCL2 protein level with BCL2 mRNA level.
Experiments using FACS-sorted and microdissected cell populations indicated that BCL2 protein expression paralleled BCL2 mRNA expression and BCL2 transcript is not present at a high level in GC B cells as previously reported. Hence, BCL2 protein expression in GCs is likely regulated at the transcriptional level. There is also a good correlation between BCL2 mRNA and protein expression in B-cell neoplasias, suggesting that BCL2 protein expression is similarly regulated in most B-cell malignancies (Figures 1B and 2A-B). However, our study does not exclude the presence of modified BCL2 transcripts that may fine-tune the level of protein expression in certain situations.16
Recent studies have provided some insight on the transcriptional control of the BCL2. Two promoters, P1 (5′) and P2 (3′), can regulate BCL2 transcription,17 and P1 is the active one in normal B cells. Depending on the cell type and differentiation stage, various transcription factors and regulatory elements have been demonstrated to regulate the expression of the BCL2 gene. BCL2 expression has been found to be regulated by transcription factors such as Aiolos and /or c-Myb in T cells,18,19 cyclic adenosine monophosphate (cAMP)-responsive element-binding proteins in B cells,20 and the Brn-3a pou family proteins in neuronal cells.21 Transcription factors of the nuclear factor κB (NF-kB) family, such as p50 homodimers, may contribute to the enhancement of BCL2 transcription.22 Negative regulatory elements that may interact with p53,23 the Ets family proteins in pre-B cells,24 and Rel/NF-kB in pro-B cells have also been described.25 In addition, Wilms tumor 1 (WT1) protein has been shown to repress BCL2 activity in HeLa cells and B cells.26 Further experiments on the transcriptional regulation of BCL2 in the GC will lead to a better understanding of the mechanism controlling BCL2 protein expression in B lymphocytes.
Prepublished online as Blood First Edition Paper, July 8, 2004; DOI 10.1182/blood-2004-01-0243.
Supported in part by a National Cancer Institute (NCI) grant (CA84967), Department of Health and Human Services.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Robert S Wicker for his assistance in the real-time PCR assay and James C Lynch, Li Xiao and Lynette Smith for statistical analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal