Abstract
Only some acute lymphoblastic leukemia (ALL) cells are thought to be capable of proliferating to maintain the leukemic clone, and these cells may be the most relevant to target with treatment regimens. We have developed a serum-free suspension culture (SC) system that supported growth of B-ALL cells from 33 patients for up to 6 weeks. ALL cells from 28 cases (85%) were expanded in this system, and growth was superior in SC than in long-term bone marrow culture. To characterize ALL progenitors, cells were sorted for expression of CD34 and CD10 or CD19 and the subfractions assayed in SC and in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice. Cells capable of long-term proliferation in vitro and NOD/SCID repopulation were derived only from the CD34+/CD10- and CD34+/CD19- subfractions, and these cells could engraft secondary recipients. The engrafted cells had the same immunophenotype and karyotype as was seen at diagnosis, suggesting they had differentiated in vivo. These results demonstrate that ALL cells capable of long-term proliferation in vitro and in vivo are CD34+/CD10-/CD19-. This suggests that cells with a more immature phenotype, rather than committed B-lymphoid cells, may be the targets for transformation in B-ALL.
Introduction
Immunologically distinct subtypes of acute lymphoblastic leukemia (ALL) are considered to reflect transformation at sequential stages of lymphocyte ontogeny.1 Philadelphia chromosome–positive (Ph+) and adult ALL appear to arise as a consequence of the transformation of multipotential progenitors.2-4 The most convincing evidence for this comes from xenotransplantation studies that showed only the CD34++/CD38- subfraction from Ph+ ALL cases could engraft nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice.4 By contrast, studies of clonality in childhood ALL suggest that the disease arises in cells already committed to the B or T lineages.5,6 Demonstration of clonality of immunoglobulin H (IgH) or T-cell receptor (TcR) rearrangements added further support to this hypothesis.7-11
However, there have been a few reports that contradict the hypothesis that the favorable/intermediate-risk leukemias arise in committed lymphoid cells.12-14 Detection of leukemia-specific karyotypes in primitive CD34+/CD19-/CD33-/CD38- cells from a B-cell precursor (BCP) ALL suggests that transformation may occur earlier in ontogeny.12 This is supported by findings of primitive CD34+/CD38- cells containing the leukemia-specific Vδ2-Dδ3 rearrangements in some cases of BCP ALL in remission.14 Nevertheless, the functional ability of these phenotypically primitive cells was not investigated in these reports.
The limited proliferative capacity of ALL cells in vitro suggests that only a small subpopulation of cells has the ability to act as progenitors in vivo to maintain the disease.2,6,15 This has important therapeutic significance, because these cells may be the most relevant targets for treatment regimens. Several reports have investigated growth of ALL blasts in short-term colony assays.16-19 However, ALL cells with colony-forming ability are low in number, and it is likely that they are not representative of the population that maintains the disease. Consequently, these assays may significantly underestimate the growth potential of ALL cells. Reports describing long-term culture of ALL cells are limited and have provided little information on the biologic characteristics of ALL progenitor cells.20-24
To increase our understanding of the biology of this disease it will be necessary to develop biologic assays that permit investigation of ALL cells with long-term proliferative ability in vitro and in vivo. We have previously used a serum-free suspension culture (SC) system to evaluate proliferation of acute myeloid leukemia (AML) cells for 8 weeks and to characterize AML progenitor cells.25-29 Here we have attempted to characterize BCP ALL progenitors by evaluating the expression of CD34, CD10, and CD19 on cells capable of long-term proliferation, in the SC assay, and NOD/SCID repopulation.
Materials and methods
Patient cells
Bone marrow (BM) cells from children (median age, 6 years; range, 1-16 years) with BCP ALL at presentation/relapse and 1 adult ALL case were obtained after informed consent and with approval of the Research Ethics Committee of the United Bristol Healthcare National Health Service Trust. Patients were selected only on the basis of availability of material for study. None of the leukemias investigated in this study had an early pre-B (CD10-) phenotype or t(9;22) or t(4;11) rearrangements. Normal bone marrow samples were obtained from donor harvests with appropriate consent. Cells were Ficoll separated to obtain a mononuclear cell (MNC) population and then frozen in Iscove modified Dulbecco medium (IMDM; Gibco, Paisley, United Kingdom) with 50% fetal calf serum (FCS; Gibco) and 10% dimethyl sulfoxide (DMSO; Manor Park Pharmaceuticals, Bristol, United Kingdom) and stored in liquid nitrogen.
ALL cell sorting
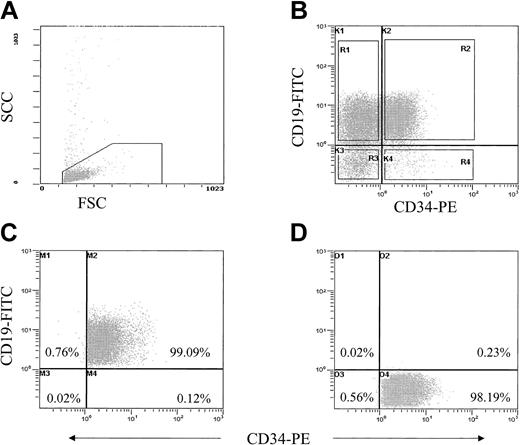
Thawed ALL cells were suspended in Hanks balanced salt solution (HBSS; Sigma-Aldrich, Poole, United Kingdom) plus 2% human albumin solution (HAS; BPL, Elstree, United Kingdom) at 107/mL. Cells were stained with monoclonal antibodies CD34-phycoerythrin (PE) and CD10- or CD19–fluorescein isothiocyanate (FITC); separate aliquots were stained with irrelevant IgG1 antibodies as isotype controls (BD Biosciences, Oxford, United Kingdom). Cells were washed twice in HBSS plus 2% HAS and maintained on ice prior to sorting. Cells were sorted using a MoFlo cell sorter (Cytomation, Hamburg, Germany) on the basis of fluorescence intensity after gating on cells with low forward and side scatter. Sort gates were set up to exclude at least 99.9% of the cells in the isotype controls, and a 10-channel gate separation was used to discriminate positive from negative fractions to ensure purity (Figure 1). The purity of all sorted subfractions, from each patient sample, was checked during and after sorting; this was always more than 98% for all populations. Cells were sorted into IMDM plus 50% FCS, maintained at 4°C, washed, resuspended at known cell concentrations, and used in SC or NOD/SCID assays.
Purification strategy for isolating cells stained with CD34/CD10 or CD19. ALL blast cells were gated on the basis of low forward and side scatter (A) and subsequently gated for expression of CD34-PE and CD10- or CD19-FITC, R1 to R4 (B). Sort gates R1 to R4 were separated by at least 10 channels, and quadrants K1, K2, and K4 in panel B were set up to exclude at least 99.9% of cells in isotype controls. (C-D) Reanalysis of CD34+/CD19+ (C) and the CD34+/CD19- subfractions (D) after sorting.
Purification strategy for isolating cells stained with CD34/CD10 or CD19. ALL blast cells were gated on the basis of low forward and side scatter (A) and subsequently gated for expression of CD34-PE and CD10- or CD19-FITC, R1 to R4 (B). Sort gates R1 to R4 were separated by at least 10 channels, and quadrants K1, K2, and K4 in panel B were set up to exclude at least 99.9% of cells in isotype controls. (C-D) Reanalysis of CD34+/CD19+ (C) and the CD34+/CD19- subfractions (D) after sorting.
In vitro assays
Suspension culture assay. Suspension cultures were initiated with unsorted ALL cells or with sorted subfractions at up to 5 × 105 cells/mL in IMDM-based serum-free medium supplemented with human insulin (10 μg/mL), transferrin (200 μg/mL; Sigma-Aldrich), and 2% HAS. The following 2 growth factor combinations were routinely used: either 20 ng/mL recombinant human Fms-like tyrosine kinase 3 (rhFlt3) ligand, 20 ng/mL rh interleukin-3 (rhIL-3), 20 ng/mL rhIL-6, 20 ng/mL rh granulocyte-macrophage colony-stimulating factor (rhGM-CSF), 20 ng/mL rhG-CSF, and 50 ng/mL stem cell factor (SCF) or 20 ng/mL rhIL-3, 10 ng/mL rhIL-7, and 50 ng/mL SCF (R&D Systems, Abingdon, United Kingdom). Cultures were incubated at 37°C in a 5% CO2 and 5% O2 humidified atmosphere and maintained for 6 weeks with weekly half-media changes. At weeks 1, 3, 5, and 6, the cells removed were analyzed by flow cytometry to determine viability and absolute cell numbers. Cells removed at weeks 2 and 4 were used for cytogenetic or morphologic analyses. Suspension cultures were maintained for 6 weeks and then harvested, total cell content was assessed, and cytogenetic/morphologic analyses performed.
Long-term marrow cultures. Stromal cultures were initiated using fresh MNC preparations from healthy donors. Cells were seeded at 5 × 104/mL in McCoy 5A medium containing 12.5% FCS, 12.5% horse serum (HS; Gibco), and 10-6 M hydrocortisone (HC; Sigma-Aldrich), incubated as described for SC, and maintained with weekly half-media changes until the stromal layer was confluent. Established layers were irradiated with 15 Gy γ-irradiation from a 137Cs source (CIS IBL-437C; Bio International, France) 24 hours prior to coculture with ALL cells. Irradiated stromas were rinsed with HC-free medium immediately after irradiation, 6 hours after irradiation, and immediately prior to subculture with ALL cells to remove HC from the system. ALL cells were seeded at 5 × 105/mL and maintained and analyzed as described for the SC assays.
Transplantation of leukemic cells into NOD/SCID mice
NOD/SCID mice were bred and maintained at the University of Bristol Animal Facility. One hour prior to transplantation, 6- to 8-week-old NOD/SCID mice were irradiated with 2 Gy acute X-rays from a linear accelerator at a rate of 7.5 cGys-1. Unsorted ALL cells and sorted subfractions were resuspended in 0.3 mL IMDM plus 5% FCS and injected into the lateral tail vein. Animals were maintained for 8 to 10 weeks or until they began to exhibit clinical symptoms of the disease. Upon killing, the gross anatomy of each mouse was inspected and femoral BM samples were removed for flow cytometric and fluorescent in situ hybridization (FISH)/histologic analyses.
In some cases, MNC preparations harvested from NOD/SCID BM were used for secondary transplantations to evaluate the self-renewal ability of sorted subfractions. For comparison with primary transplants, equal numbers of CD45+ cells were inoculated into the secondary recipients. These cells were not enriched for any particular phenotype prior to inoculation.
Evaluation of transplanted and cultured cells
Flow cytometric analysis of murine tissues. The immunophenotype of all xenografts was examined using antibodies against CD45 and appropriate lineage markers. Cell suspensions were lysed, washed, and resuspended in HBSS plus 2% HAS. In every case, cells were stained with CD45 and CD34 along with 1 or 2 of the following lineage markers—CD10, CD13, CD19, CD22, CD33 (BD Biosciences)—to compare the expression of specific markers with the original patient specimen at diagnosis/relapse. Separate aliquots were used as isotype controls. After staining, cells were washed once in buffer alone, and propidium iodide (PI; Sigma-Aldrich) at 2 μg/mL was added to the second wash. The cells were resuspended in HBSS plus 2% HAS and maintained on ice prior to analysis using a Coulter EPICS XL-MCL flow cytometer (Beckman Coulter, HighWycombe, United Kingdom). Nonviable cells were gated out based on PI uptake, and matched isotype controls were run for each tissue sample and used to define gate settings, which excluded at least 99.9% of the cells in the isotype control. Any positive value (ie, 0.1% or less) for the isotype control was then subtracted from the percentage positive in the CD45-stained samples. Cells from noninjected NOD/SCIDs were used as additional controls to verify the gate settings. We defined human cell engraftment as expression of at least 0.1% CD45+ cells in a sample. Aliquots of cells derived from murine tissue were plated onto slides for morphologic or cytogenetic analysis.
Some flow cytometric analysis was performed on cultured cells at weeks 1, 3, 5, and 6. Viability was determined by PI uptake, and absolute cell counts were established using Flow-Count Fluorospheres (Beckman Coulter) according to the manufacturer's instructions. Cells were stained with CD2, CD3, CD4, CD10, CD13, CD19, CD33, or CD34 monoclonal antibodies and prepared and analyzed as described above.
Cytogenetic and morphologic analysis. Cytogenetic analysis was performed on all patients at diagnosis, on cells harvested from cultures at weeks 2, 4, and 6, and on cells harvested from transplanted NOD/SCID marrow by the Regional Cytogenetics Unit, Southmead Hospital. At least 50 nuclei per sample were scored on preparations from cultured cells and NOD/SCID marrow. If cytogenetic analysis was not possible, cytospins of cultured cells and NOD/SCID BM were prepared. Cytospins were fixed and stained with Hema-Gurr (BDH, Poole, United Kingdom), examined, and assessed by a hematology consultant at Bristol Royal Hospital for Children.
Statistical analysis. Statistical analysis was performed using matched paired t tests or analysis of variance with the Tukey post-hoc analysis of means.
Results
Comparison of long-term culture systems
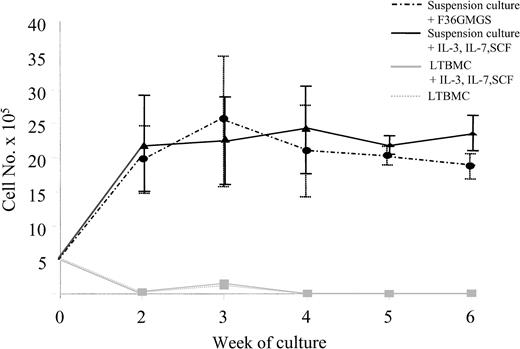
Growth of ALL cells from 9 patients (3 pre-B, 6 common ALL [c-ALL]) was compared in SC, in standard long-term BM culture (LTBMC), and in cytokine-supplemented LTBMC. SCs were supplemented with 2 different growth factor combinations: Flt3, IL-3, IL-6, GM-CSF, G-CSF, and SCF (F36GMGS) or IL-3, IL-7, and SCF. LTBMCs were supplemented with IL-3, IL-7, and SCF only. In every case, between 2-fold and more than 100-fold more viable cells were detected in SC than in LTBMC regardless of the cytokine combination used in SC. Most cells had died by day 14 in standard LTBMC (97%) and by day 21 in cytokine-supplemented LTBMC (98%). In contrast, the same cells could be maintained for 5 to 6 weeks in SC. In 8 cases, the numbers of cultured cells increased from 5 × 105 to 2.9 × 106 at week 3 with F36GMGS and to 2.4 × 106 at week 4 with IL-3, IL-7, and SCF. Cell numbers were then maintained for the remainder of the culture period. Results from these 8 cases are shown in Figure 2. Cell numbers from the ninth patient were maintained for 5 weeks and then declined.
Growth of ALL cells in different long-term culture systems. Growth of ALL cells from 9 patients were evaluated in suspension culture supplemented with F36GMGS or with IL-3, IL-7, and SCF and compared with growth in standard LTBMC and LTBMC supplemented with IL-3, IL-7, and SCF over a 6-week period. Cultures were maintained with weekly half-media changes, and the number of viable cells present at each week was determined using flow cytometry. Results from 8 patients where cell expansion was observed in culture are shown. Data are expressed as means ± standard error of measurement.
Growth of ALL cells in different long-term culture systems. Growth of ALL cells from 9 patients were evaluated in suspension culture supplemented with F36GMGS or with IL-3, IL-7, and SCF and compared with growth in standard LTBMC and LTBMC supplemented with IL-3, IL-7, and SCF over a 6-week period. Cultures were maintained with weekly half-media changes, and the number of viable cells present at each week was determined using flow cytometry. Results from 8 patients where cell expansion was observed in culture are shown. Data are expressed as means ± standard error of measurement.
Cytogenetic analysis was successfully performed on cells from 6 of the 9 cases evaluated in SC. FISH analyses at weeks 4 and 6 revealed that 45% to 100% of cells from 4 of the 6 patients had the same cytogenetic abnormality as was seen at diagnosis, while only cells with a normal karyotype were detected in 2 cases. Cells from these 2 cases were not used for any subsequent experiments detailed below. Morphologic analyses on cultured cells from the remaining 3 patients, at the same time points, revealed these to be predominantly blast cells (more than 85%). In this group of patients, there was no difference in the number of cells maintained in SC supplemented with F36GMGS or with IL-3, IL-7, and SCF (P > .13). The SC assay was subsequently used to further evaluate ALL cell proliferation from a larger sample group.
Suspension culture assay
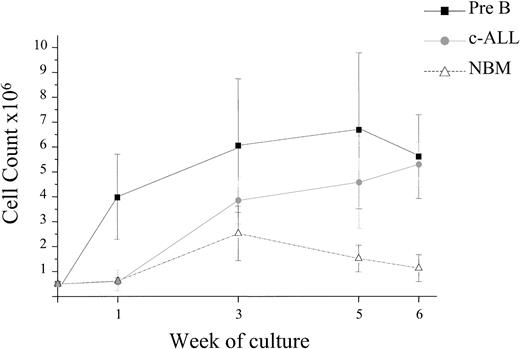
ALL blasts from 43 patients at presentation (11 pre-B, 32 c-ALL) and 4 in relapse (c-ALL) were grown in the SC assay supplemented with F36GMGS. Cells from 40 patients were maintained in this system for at least 4 weeks and up to 6 weeks in 33 of these patients. The numbers of cultured cells were expanded in 28 of these 40 patients (2- to 52-fold) from 5 × 105 up to 2.6 × 107. Proliferation of ALL cells from each subtype was observed, and there was no correlation between proliferation and subtype, prognostic indicators, or diagnostic and relapse samples (Figure 3). Growth of ALL cells was superior to that of normal BM (NBM) cells (n = 10) under these conditions (P < .03). The number of cultured BM cells increased gradually from 5 × 105 at initiation to 2.5 × 106 at week 3 and then declined to 1.5 × 106 and 1.1 × 106 at weeks 5 and 6, respectively.
ALL cell expansion in suspension culture. The proliferative capacity of ALL cells from 47 patients was evaluated in suspension culture (SC). Cells from 40 patients were maintained in the culture system for at least 4 weeks, and expansion of cell numbers was observed in 28 of these 40 cases. The graph depicts the proliferation observed in the 28 cases and that observed using cells from 10 NBM donors. Proliferation of cells from both pre-B and c-ALL subtypes was observed, and there was no correlation between proliferation, prognostic indicators, or diagnostic and relapse samples. Data are expressed as means ± standard error.
ALL cell expansion in suspension culture. The proliferative capacity of ALL cells from 47 patients was evaluated in suspension culture (SC). Cells from 40 patients were maintained in the culture system for at least 4 weeks, and expansion of cell numbers was observed in 28 of these 40 cases. The graph depicts the proliferation observed in the 28 cases and that observed using cells from 10 NBM donors. Proliferation of cells from both pre-B and c-ALL subtypes was observed, and there was no correlation between proliferation, prognostic indicators, or diagnostic and relapse samples. Data are expressed as means ± standard error.
FISH analyses were possible in 11 of the 28 patients' cells that expanded in SC and confirmed that 70% to 100% of cultured cells had the same karyotype as was seen in the patients at diagnosis. Cells from the remaining 17 patients who either had a normal karyotype (n = 10) or for whom FISH probes were not available (n = 7) were evaluated by routine morphologic analyses. In every case, the majority of the cells present were undifferentiated blasts (more than 95%).
Optimizing cytokine stimulation
We have previously shown that the cytokine combination F36GMGS at the concentrations used here was superior for supporting growth of primitive normal and AML cells in the SC system.25,29 However, growth of lymphoid cells may require alternative cytokines. Subsequent experiments were performed using cells from 12 patients that had proliferated in SC with F36GMGS and 8 cases not previously studied to compare the stimulatory effect of F36GMGS with that of IL-3, IL-7, and SCF, which is thought to stimulate lymphoid progenitor cells. Results showed that this 3-cytokine combination was as effective as the 6 cytokines for the maintenance and proliferation of ALL cells at each time point assayed over the 6-week period (P > .4, data not shown). Predictably, growth of NBM cells (n = 5) was superior in F36GMGS compared with that observed using IL-3, IL-7, and SCF. An overall expansion was observed in the 6-cytokine combination from 5 × 105 at initiation to 1.1 × 106 at week 6, while a decrease to only 3 × 105 cells at week 6 was observed in cultures stimulated with the 3 cytokines (P < .04).
Accurate phenotypic analysis is difficult on cells cultured for long periods due to autofluorescence, a phenomenon not unique to malignant cells. However, in an attempt to investigate whether the cytokine combinations, F36GMGS and IL-3, IL-7, and SCF, were supporting proliferation of particular cell types, we determined the expression of CD2, CD3, CD4, CD10, CD13, CD19, CD33, and CD34 on cultured cells from 8 c-ALL patients stimulated with each combination. At week 3, the cultured cells retained high expression of CD34 (more than 54%), CD10, and CD19 (both more than 78%) regardless of the cytokine combination used. The morphology of the cultured cells was similar under both conditions, and FISH analyses confirmed both combinations supported growth of cells with an aberrant karyotype (62% to 100%). In contrast, neither cytokine combination supported maintenance of NBM cells that expressed CD2, CD3, CD10, or CD19. CD34 expression in the NBM samples was only 1% ± 0.4% at the start of culture, and this had decreased to 0.8% ± 0.3% at week 3; instead, most cells were CD33+ (more than 83%).
Phenotypic characterization of ALL cells with long-term proliferative ability in vitro
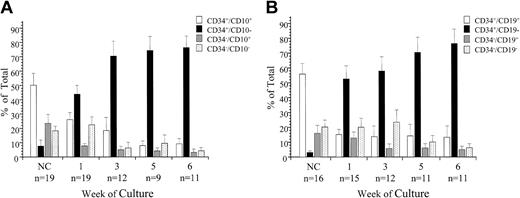
Mononuclear cells from 16 ALL patients at diagnosis (5 pre-B, 11 c-ALL) and 3 c-ALLs at relapse were sorted for expression of CD34 and CD10 and evaluated in SC (Figure 4A). Overall, most cells at sorting were CD34+/CD10+ (50% ± 8%); the CD34+/CD10- subfraction represented only 8% ± 4%. After 1 week in SC, most viable cells were derived from the CD34+/CD10- subfraction (43% ± 6%). By the third week in culture, 70% ± 10% of cells were derived from the CD34+/CD10- subfraction. This trend continued throughout the culture period, with significantly more cells derived from the CD34+/CD10- subfraction than any other (74% ± 13% and 76% ± 8% at weeks 5 and 6, respectively, F = 41**, P < .0001). In the cultures of CD34+/CD10- cells, there was a 35- to 3000-fold expansion in cell numbers, starting from an average 1.4 × 104 at initiation to an average 7.1 × 106 at week 6.
Expression of CD34 and CD10 or CD19 on ALL progenitor cells. ALL cells from 19 patients were sorted for expression of CD34 and CD10 (A), and cells from 16 patients were sorted for coexpression of CD34 and CD19 (B). The proliferative capacity of the sorted subfractions and unsorted controls was evaluated in SC. Absolute cell counts, derived from each sorted population, and unsorted controls were determined by flow cytometry at weeks 1, 3, 5, and 6. These values were then used to calculate the proportion of the total viable cells represented by each sorted population. The proportion of the total cells derived from the each sorted subfraction is presented as the mean ± SE; n indicates number of patient samples evaluated at each time point.
Expression of CD34 and CD10 or CD19 on ALL progenitor cells. ALL cells from 19 patients were sorted for expression of CD34 and CD10 (A), and cells from 16 patients were sorted for coexpression of CD34 and CD19 (B). The proliferative capacity of the sorted subfractions and unsorted controls was evaluated in SC. Absolute cell counts, derived from each sorted population, and unsorted controls were determined by flow cytometry at weeks 1, 3, 5, and 6. These values were then used to calculate the proportion of the total viable cells represented by each sorted population. The proportion of the total cells derived from the each sorted subfraction is presented as the mean ± SE; n indicates number of patient samples evaluated at each time point.
Cells from 9 of these patients (7 c-ALL and 2 pre-B) and 7 additional c-ALL patients were sorted for coexpression of CD34 and CD19 and evaluated in SC (Figure 4B). As expected, most cells at sorting were CD34+/CD19+ (55% ± 7%), and the CD34+/CD19- fraction represented only 3% ± 1%. After 7 days in culture, 52% ± 9% of cells were derived from the CD34+/CD19- subfraction, and this increased to 58% ± 9% at week 3. At weeks 5 and 6, 70% ± 10% and 77% ± 12% of cells were derived from the CD34+/CD19- subfraction (F = 11**, P < .000 01). In the cultures of CD34+/CD19- cells, there was a 2- to 6000-fold increase in cell numbers, from an average 6 × 104 at initiation to an average 1 × 107 at week 6.
Cytogenetic analyses on cells derived from the CD34+/CD10- and/or CD34+/CD19- subfractions from 15 patients at weeks 4 and 6 revealed that 58% to 100% of cultured cells had the same karyotypic abnormality as the patient at diagnosis. In addition, cytogenetic analyses performed on the CD34+/CD19- population from 3 TEL-AML1 cases (patients 56, 66, and 69) immediately after sorting revealed these cells contained the TEL-AML1 gene fusion. Morphologic analyses at week 6 on cells derived from these subfractions, from the remaining cases, showed that most were undifferentiated blasts (more than 80%) and effete cells.
In vivo NOD/SCID assay
Unsorted ALL cells from 16 patients were evaluated for their ability to engraft sublethally irradiated NOD/SCID mice (Table 1). In these cases, engraftment was achieved using 5 × 105 to 107 cells (range, 0.12%-99% CD45+). Engraftment was achieved from 4 of 4 patients evaluated using 5 × 105 unsorted cells (0.16%-59% CD45+) and from 8 of 10 patients using 106 cells (0.12%-80% CD45+). Engraftment was achieved using 5 × 106 cells from 15 patients (0.12%-92%) and in 8 of 8 patients using 107 cells (0.6%-99% CD45+). Animals injected with cells from patients 25, 31, and 53 began to show signs of morbidity 5 weeks after injection and therefore were killed and analyzed at this stage.
Engraftment characteristics of unsorted ALL cells in NOD/SCID mice
. | . | . | % CD45+ in NOD/SCID*, by no. of unsorted cells injected . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | Subtype . | Karyotype . | 5 × 105 . | 106 . | 5 × 106 . | 107 . | % FISH-positive . | |||
| 2 | c-ALL† | Complex | — | — | 1.9 | 3 | 100 | |||
| 9 | pre-B | 46,XX | — | — | 13 | — | — | |||
| 10 | c-ALL | t(12;21) | — | 0 | 0.12 | 0.6 | 80 | |||
| 11 | c-ALL | 46,XX | 0.16 | 0.3 | 8 | — | — | |||
| 18 | c-ALL | 46,XY | — | 0.12 | 0.14 | — | — | |||
| 19 | c-ALL | 58,XY | — | — | 0.15 | — | 80 | |||
| 21 | pre-B | 11q23 | — | 5 | 44 | — | 100 | |||
| 25 | pre-B | 46,XX | — | 7 | 57 | 80 | — | |||
| 28 | pre-B | 46,XY | 4 | 37 | 82 | 99 | — | |||
| 31 | c-ALL | del(9p), del(17p) | 59 | 80 | 92 | 98 | > 90 | |||
| 32 | pre-B | 46,XY | — | 0 | 0.13 | — | — | |||
| 43 | pre-B | 46,XX | — | — | 0.6 | — | — | |||
| 47 | c-ALL | del(17p) | — | — | 9.2 | 24 | 100 | |||
| 53 | pre-B | del(6p) | 2.23 | 60 | 89 | — | > 83 | |||
| 56 | c-ALL | t(12;21) | — | 3 | — | 11 | > 70 | |||
| 69 | c-ALL | t(12;21) | — | — | 2 | 29 | > 87 | |||
. | . | . | % CD45+ in NOD/SCID*, by no. of unsorted cells injected . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | Subtype . | Karyotype . | 5 × 105 . | 106 . | 5 × 106 . | 107 . | % FISH-positive . | |||
| 2 | c-ALL† | Complex | — | — | 1.9 | 3 | 100 | |||
| 9 | pre-B | 46,XX | — | — | 13 | — | — | |||
| 10 | c-ALL | t(12;21) | — | 0 | 0.12 | 0.6 | 80 | |||
| 11 | c-ALL | 46,XX | 0.16 | 0.3 | 8 | — | — | |||
| 18 | c-ALL | 46,XY | — | 0.12 | 0.14 | — | — | |||
| 19 | c-ALL | 58,XY | — | — | 0.15 | — | 80 | |||
| 21 | pre-B | 11q23 | — | 5 | 44 | — | 100 | |||
| 25 | pre-B | 46,XX | — | 7 | 57 | 80 | — | |||
| 28 | pre-B | 46,XY | 4 | 37 | 82 | 99 | — | |||
| 31 | c-ALL | del(9p), del(17p) | 59 | 80 | 92 | 98 | > 90 | |||
| 32 | pre-B | 46,XY | — | 0 | 0.13 | — | — | |||
| 43 | pre-B | 46,XX | — | — | 0.6 | — | — | |||
| 47 | c-ALL | del(17p) | — | — | 9.2 | 24 | 100 | |||
| 53 | pre-B | del(6p) | 2.23 | 60 | 89 | — | > 83 | |||
| 56 | c-ALL | t(12;21) | — | 3 | — | 11 | > 70 | |||
| 69 | c-ALL | t(12;21) | — | — | 2 | 29 | > 87 | |||
— indicates not tested.
Each value represents the mean level of CD45+ cells in the femoral marrow of at least 2 NOD/SCID mice
Relapse sample
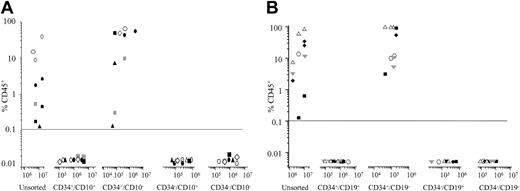
Cells from 3 pre-B (patients 9, 32, 43) and 3 c-ALL patients (patients 2, 10, 47) were sorted for expression of CD34 and CD10 and evaluated in the NOD/SCID assay (Figure 5A). Engraftment was achieved in all 6 cases with the CD34+/CD10- subfraction only (range, 0.12%-35% CD45+ using 7 × 104 to 3 × 106 cells). To achieve equivalent levels of engraftment using unsorted cells, 30- to 100-fold more cells had to be injected into the NOD/SCID recipients. In each case, there was no engraftment with the CD34+/CD10+, CD34-/CD10+, or CD34-/CD10- subfractions. A similar pattern was observed when cells from patients 9, 10, 25, and 56 and one additional c-ALL patient were sorted for coexpression of CD34 and CD19 and evaluated in the NOD/SCID assay (Figure 5B). In each case, the only cells that engrafted the NOD/SCID mice were CD34+/CD19- (range, 3%-95% using 5 × 104 to 2 × 105 cells). There was no engraftment with the other subfractions despite injecting significantly more cells. FISH analysis was possible on cells recovered from mice injected with cells from 5 patients. In every case, more than 80% of CD45+ cells had the same karyotype as that of the patient at diagnosis. Morphologic analyses on BM cells from the remaining 4 patients confirmed that more than 90% of CD45+ cells had blast morphology.
Phenotype of NOD/SCID-engrafting ALL cells. ALL cells were sorted for expression of CD34 and CD10 (n = 6) (A) or CD34 and CD19 (n = 5) (B) and evaluated in the NOD/SCID assay. Each patient is represented by a specific symbol, and each symbol depicts the engraftment obtained as measured by CD45+ cells present in the bone marrow of the NOD/SCID recipients.
Phenotype of NOD/SCID-engrafting ALL cells. ALL cells were sorted for expression of CD34 and CD10 (n = 6) (A) or CD34 and CD19 (n = 5) (B) and evaluated in the NOD/SCID assay. Each patient is represented by a specific symbol, and each symbol depicts the engraftment obtained as measured by CD45+ cells present in the bone marrow of the NOD/SCID recipients.
To evaluate self-renewal potential, BM cells from NOD/SCIDs engrafted with CD34+/CD10- cells from 4 patients and CD34+/CD19- cells from 3 patients were serially transplanted into secondary recipients. The CD34+/CD10- or CD34+/CD19- cell content of the secondary inocula was less than 15% of that in the primary transplants. In every case, similar levels of engraftment were observed in the secondary recipients as had been detected in the primary transplants (2% to 95% CD45+) using 105 to3 × 106 CD45+ cells (Table 2).
Expression of lineage-specific and differentiation markers on CD45+ cells harvested from NOD/SCID bone marrow
Patient no./recipient . | % human leukocytes CD45 . | % B lineage . | . | . | % progenitors CD34 . | % FISH-positive . | ||
|---|---|---|---|---|---|---|---|---|
| . | . | CD10 . | CD19 . | CD22 . | . | . | ||
| CD34+/CD10- | ||||||||
| 9/1° | 35 | 83 | 87 | 56 | 88 | — | ||
| 9/2° | 46 | 85 | 90 | 58 | 83 | — | ||
| 10/1° | 28 | 88 | 92 | 80 | 72 | 89 | ||
| 10/2° | 25 | 92 | 90 | 77 | 80 | 90 | ||
| 43/1° | 6 | 87 | 96 | 60 | 93 | — | ||
| 43/2° | 2 | 91 | 92 | 58 | 89 | — | ||
| 47/1° | 20 | 84 | 88 | — | 80 | 100 | ||
| 47/2° | 28 | 87 | 80 | — | 73 | 100 | ||
| CD34+/CD19- | ||||||||
| 9/1° | 12 | 86 | 91 | 63 | 85 | — | ||
| 9/2° | 15 | 80 | 83 | 55 | 83 | — | ||
| 10/1° | 79 | 92 | 95 | 84 | 90 | 91 | ||
| 10/2° | 82 | 90 | 97 | 80 | 86 | 100 | ||
| 25/1° | 95 | 78 | 80 | 75 | 79 | — | ||
| 25/2° | 95 | 82 | 85 | 80 | 75 | — | ||
Patient no./recipient . | % human leukocytes CD45 . | % B lineage . | . | . | % progenitors CD34 . | % FISH-positive . | ||
|---|---|---|---|---|---|---|---|---|
| . | . | CD10 . | CD19 . | CD22 . | . | . | ||
| CD34+/CD10- | ||||||||
| 9/1° | 35 | 83 | 87 | 56 | 88 | — | ||
| 9/2° | 46 | 85 | 90 | 58 | 83 | — | ||
| 10/1° | 28 | 88 | 92 | 80 | 72 | 89 | ||
| 10/2° | 25 | 92 | 90 | 77 | 80 | 90 | ||
| 43/1° | 6 | 87 | 96 | 60 | 93 | — | ||
| 43/2° | 2 | 91 | 92 | 58 | 89 | — | ||
| 47/1° | 20 | 84 | 88 | — | 80 | 100 | ||
| 47/2° | 28 | 87 | 80 | — | 73 | 100 | ||
| CD34+/CD19- | ||||||||
| 9/1° | 12 | 86 | 91 | 63 | 85 | — | ||
| 9/2° | 15 | 80 | 83 | 55 | 83 | — | ||
| 10/1° | 79 | 92 | 95 | 84 | 90 | 91 | ||
| 10/2° | 82 | 90 | 97 | 80 | 86 | 100 | ||
| 25/1° | 95 | 78 | 80 | 75 | 79 | — | ||
| 25/2° | 95 | 82 | 85 | 80 | 75 | — | ||
1° indicates primary NOD/SCID recipient; 2°, secondary NOD/SCID recipient; and —, not tested.
More detailed immunophenotypic analysis of the CD45+ cells removed from the NOD/SCIDs injected with CD34+/CD10- and CD34+/CD19- cells revealed that the engrafted cells had the same characteristic phenotype as the patient at diagnosis, with high levels of CD34 (more than 70%), CD10 (at least 78%), and CD19 (at least 80%). There were no significant levels of myeloid (3% or less) or T-lineage (7% or less) engraftment. Where possible, FISH analyses confirmed cells with a leukemic karyotype were transplanted from primary to secondary recipients. These findings suggest that the CD34+/CD10- or CD19- cells injected into the NOD/SCIDs have undergone some degree of differentiation in vivo, resulting in them having a similar phenotype to the bulk ALL population.
Discussion
An increased understanding of the cells that are responsible for initiation and maintenance of ALL is dependent upon the development of reproducible assay systems that allow us to study malignant cells with long-term proliferative capacity in vitro and in vivo. Previous in vitro studies on ALL have used short-term colony assays in an attempt to determine the frequency, growth characteristics, and proliferative status of ALL progenitor cells.16-19 However, because ALL progenitor cells are likely to be quiescent, the cells studied in short-term colony assays may not be the most relevant population to investigate. To study a quiescent population, long-term assays are required.
We have previously used the SC assay to characterize AML progenitor cells25-29 and shown it to be superior to LTBMC for growth of AML.25,29 Hence, we used the SC system to evaluate the growth of ALL progenitor cells and compare it with growth in standard and cytokine-supplemented LTBMC. In every case, cells grown in the LTBMC systems underwent apoptosis and very few viable cells were detected at week 3. In contrast, the same patients' cells could be expanded and maintained in SC for up to 6 weeks. FISH and morphologic analyses confirmed that most cells proliferating in SC were leukemic blasts.
Our findings contrast with previous reports that suggested a supportive stromal layer is essential for the maintenance of ALL cells in culture.20-24 However, most of these studies only evaluated growth over short times. Furthermore, defects in expression or activation of the cell adhesion molecules lymphocyte function-associated antigen-1 (LFA-1) and very late antigen-4 (VLA-4) on B-lineage ALL BM samples30 could signify a reduced dependency on the marrow microenvironment and more rapid egress into the peripheral blood. Acquisition of sequential genetic changes in this disease may precede the emergence of a dominant clone with decreased or complete absence of a requirement for stromal support. Because physical disruption of the BM architecture is more frequently seen in ALL than in AML or CML,31 it is possible that ALL cells are less stroma dependent than was previously thought; hence the enhanced survival and proliferation of ALL cells observed in the SC assay.
We used the SC assay to evaluate growth of B-ALL cells from 43 patients at diagnosis and 4 in relapse. Overall, cells from 40 patients (85%) were maintained for at least 4 weeks and for 6 weeks in 33 cases (70%); cell numbers were expanded in 28 cases. FISH and morphologic analyses confirmed the cultured cells had an abnormal karyotype or had a lymphoid blast morphology. In 3 cases the cultured cells were heterogeneous for the presence of chromosomal abnormalities (45%-60%). However, the cytogenetic analyses of these cases at diagnosis reported similar frequencies of aberrations. Therefore, our results are consistent with these findings. There were no clear correlations between growth in the SC system and prognostic indicators or particular subtypes of ALL. The growth factor combinations Flt3, IL-3, IL-6, GM-CSF, G-CSF, and SCF and IL-3, IL-7, and SCF were both effective in maintaining growth of cells that had a similar immunophenotype and karyotype to that of the patient at diagnosis. Further investigations will be necessary to determine whether proliferation of ALL cells in the SC assay can be improved by using growth factors more closely associated with lymphopoiesis, such as SCF-1. To our knowledge, this is the first report of conditions that support the long-term growth of ALL cells in such a large cohort of patients.
Subsequently we used this SC assay together with the in vivo NOD/SCID model to determine the phenotype of ALL cells with long-term proliferative capacity. Our findings indicate that the CD34+/CD10- and CD34+/CD19- subfractions represented small proportions of the total nucleated cell population, but these subfractions contained the majority of cells that were capable of proliferating for up to 6 weeks in SC. To confirm the CD34+/CD10-, CD19- phenotype of ALL progenitor cells, we evaluated the ability of the sorted subfractions to engraft NOD/SCID mice. ALL cell engraftment was only achieved with CD34+/CD10- (range, 0.12%-35% CD45+ using 7 × 104 to 3 × 106 cells) and CD34+/CD19- cells (3%-95% CD45+ using 8 × 104 to 2 × 105 cells). There was no engraftment with the CD34+/CD10+, CD34+/CD19+, or the CD34- subfractions despite injecting more than 10-fold more cells in some cases. It was possible to enrich NOD/SCID-engrafting cells by more than 2 logs using CD34+/CD10- or CD19- cells compared with using unsorted cells. Serial transplantation studies demonstrated that these cells were capable of self-renewal and expansion of the progenitor cell pool.
Our findings are in agreement with previous studies that demonstrated the presence of leukemic cells with a CD34+/CD19-12,32 or CD38-4,14 phenotype. In addition, we evaluated the functional ability of the phenotypically primitive populations and demonstrate these cells are capable of long-term proliferation in vitro and in vivo. It could be argued that the observed proliferation of CD34+/CD10- and CD19- cells was due to contaminating CD10+ or CD19+ cells in the sorted populations. However, our use of stringent sort regions meant the purity of the sorted populations was always more than 98%. Furthermore, we would have expected to observe growth in vitro and in vivo with the purified CD34+/CD10+ and CD19+ cells if our observations were due to contamination by lineage-positive cells. Because these populations had limited proliferative capacity in vitro and were not capable of engrafting NOD/SCIDs, we do not believe our findings are a result of contaminating lineage-positive cells.
Immunophenotypic results suggest that the CD34+/CD10- and CD34+/CD19- subfractions underwent some degree of differentiation in vivo, because most CD45+ cells harvested from the NOD/SCID marrow expressed CD34 and the lymphoid lineage markers CD10, CD19, and CD22, providing evidence for a hierarchic organization of ALL progenitor cells. This raises the question as to whether the cytogenetic abnormalities observed in the bulk ALL population at diagnosis are present in these phenotypically immature cells or are acquired as the cells differentiate. Due to the limited material available from the patients used in our study, it was not possible to investigate this matter thoroughly, particularly because the CD34+/CD10-, CD19- cells represented such small fractions of the blast population. However, in 3 cases, the CD34+/CD19- cells at sorting were found to contain the TEL-AML1 fusion gene at a similar incidence as the CD34+/CD19+ subfraction. In contrast, Hotfilder et al32 reported a lower incidence of the TEL-AML1 fusion gene in CD34+/CD19- cells compared with CD34+/CD19+ cells in 5 patients, and the CD34+/CD19- cells from 3 of these patients generated normal colonies in short-term assays. From this, the authors concluded that TEL-AML1 ALL originates in a CD19+ cell. These disparate observations may reflect differences in the sorting strategies used. However, patient heterogeneity cannot be disregarded, because sample size in both studies was very small. Furthermore, we evaluated the functional ability of the CD34+/CD19- cells in long-term assays routinely used for quantitation of transplantable hematopoietic stem cell content, which should not underestimate the growth potential of the ALL cells. A recent molecular study on bulk ALL cells suggests that the TEL-AML1 translocation occurs as an early leukemogenic event in an immature B cell prior to the production of a functional IgH recombination.33 Our future studies will focus on addressing these issues to determine whether these sorted cells contain the same cytogenetic abnormalities and whether they have the same genotype as the primary ALL cells.
Our results support the notion that the leukemic transformation event in this group of patients has occurred in a hematopoietic cell with a less mature phenotype than committed B cells. This is in accordance with the primitive origin of Ph+ ALL reported by Cobaleda et al,4 who also found that engrafted NOD/SCID BM cells expressed CD34, CD19, and CD38, implying they had differentiated in vivo. Together, these data suggest that regardless of the phenotype of the bulk ALL population or the prognostic indicators, the leukemogenic transformation event may take place in cells with a more primitive phenotype. In this respect, our findings are similar to published reports on the origin of AML.25-29,34,35 Our future studies will attempt to further characterize ALL progenitor cells by determining whether or not they express markers associated with normal hematopoietic progenitor cells (CD133+/CD38-) or whether these cells have undergone immunoglobulin gene rearrangements.
In summary, the data we present here suggest that the target cell for transformation in pre-B and c-ALL has a more immature phenotype than the bulk ALL cell population. Definition of leukemia progenitor cell phenotypes, their molecular characteristics, and cell cycle status are essential for improved minimal residual disease (MRD) immunosurveillance and the development of strategies that not only eliminate the bulk ALL cell population but also target the cells responsible for maintaining the disease.
Prepublished online as Blood First Edition Paper, July 8, 2004; DOI 10.1182/blood-2004-03-0901.
Supported by grants from the Leukaemia Research Fund, United Kingdom, and the National Blood Authority.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Paul Virgo, Immunology Unit, and staff of the Regional Cytogenetics Unit, Southmead Hospital, and Dr Terry Hoy and Janet Fisher, University of Wales College of Medicine cell sorting facility, for excellent technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal