Abstract
Gene therapy for a wide variety of disorders would be greatly enhanced by the development of vectors that could be targeted for gene delivery to specific populations of cells. We describe here high-efficiency targeted transduction based on a novel targeting strategy that exploits the ability of retroviruses to incorporate host cell proteins into the surface of the viral particle as they bud through the plasma membrane. Ecotropic retroviral particles produced in cells engineered to express the membrane-bound form of stem cell factor (mbSCF) transduce both human cell lines and primary cells with high efficiency in a strictly c-kit (SCF receptor)-dependent fashion. The availability of efficient targeted vectors provides a platform for the development of a new generation of therapies using in vivo gene delivery. (Blood. 2004;104: 2697-2703)
Introduction
The development of retroviral vectors that are targeted to transduce only specific cell types has been the goal of many investigators working in the field of gene therapy.1-5 The availability of such vectors would revolutionize the field, opening up new avenues for gene therapy in acquired and inherited disorders by enabling in vivo delivery of therapeutic molecules to specific populations of target cells. Novel approaches to drug delivery and for use in vaccines also become possible through display of molecules on the retroviral surface. Over a period of around a decade, however, this goal has proven highly elusive. Virtually all attempts to target specific cell types have logically focused on modification of the retroviral envelope protein, as this complex glycoprotein is established to determine viral host range and facilitate virus entry though membrane fusion.2,4,5 These modifications can be the simple substitution of the envelope protein for one from another retrovirus or other enveloped virus, such as vesicular stomatitis virus (VSV)6 , a process referred to as pseudotyping.2 More recently, attempts have been made to deliberately engineer envelope proteins to redirect their binding to new cell surface molecules.4 This has involved making N-terminal extensions, insertions, or replacements to regions of the envelope protein. Commonly used modifications have been incorporation of growth factor-binding regions7-12 or the addition of single-chain antibodies.13-17 While many of these modifications have successfully redirected viral binding, this has invariably been achieved at the expense of infectivity to the extent that targeted binding actually inhibited viral entry. This property has actually been used to advantage in an approach referred to as “inverse targeting.”10,18 A number of reports, however, have indicated that low levels of targeted transduction may be achievable by coexpression of an ecotropic envelope in conjunction with the modified envelope protein.7,9,16,19
To circumvent the problems associated with envelope modification, we have developed an alternative strategy, exploiting the natural budding mechanism of the virus to insert new binding ligands into the viral surface. We describe here successful and efficient specific targeting of transduction to human cells expressing stem cell factor (SCF) receptor (c-kit) by ecotropic retroviruses bearing surface stem cell factor.
Materials and methods
Cytokines, antibodies, and methylcellulose
All cytokines used in this study were obtained from First Link (Brierley Hill, United Kingdom), and methylcellulose used for clonogenic progenitor assays was obtained from StemCell Technologies (Vancouver, BC, Canada). Mouse anti-human c-kit antibody was obtained from R&D Systems (Abingdon, United Kingdom), mouse anti-human CD59 was from Serotec (Kidlington, United Kingdom), and anti-CD34 was from BD Bioscience (Cowley, United Kingdom). Secondary antibodies, donkey anti-goat immunoglobulin G (IgG), chicken anti-mouse IgG, and rat anti-mouse IgG were purchased from DakoCytomation Ltd (Ely, United Kingdom).
Human cell lines
The human megakaryoblastic leukemia cell line LAMA-84,20 the monomyelocytic cell line U937, and the erythroleukemia cell line TF-121 were maintained in RPMI supplemented with 10% fetal calf serum (FCS). TF-1 was supplemented with 1 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF). The human megakaryoblastic leukemia cell line MO7e22 was maintained in RPMI supplemented with 20% FCS and 10 ng/mL GM-CSF or shifted into medium without GM-CSF but containing 100 ng/mL SCF for experiments where c-kit expression was down-regulated.
Retroviral packaging and producer cell lines
Phoenix ecotropic, amphotropic (kind gifts from Dr Gary Nolan, Stanford University), and SCF-ecotropic (SCF-eco) packaging cell lines were routinely maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FCS (Invitrogen-Gibco, Paisley, United Kingdom). Retroviral producer lines were cultured in the same medium supplemented with puromycin (1 μg/mL) to select for the PINCO23 retroviral genome or G418 (1.5 mg/mL) to select for pBabe-neo (pBN) and pBabe-neo-cyclin-dependent kinase 4 (CDK4).24 The stable AM-12 retroviral producer cell line previously established in the laboratory by Chinswangwatanakul (Lewis et al24 ) was maintained in DMEM supplemented with 10% FCS and 1.5 mg/mL G418 for selection of the retroviral vector.
Construction of SCF-eco packaging cells
The membrane-bound form of stem cell factor (mbSCF) cDNA was amplified by reverse transcriptase-polymerase chain reaction (RT-PCR) from the L88.5 human stromal cell line25 and cloned into the mammalian expression vector pREP8 (Invitrogen Ltd, Paisley, United Kingdom). Phoenix ecotropic packaging cells were transfected with pREP8-mbSCF expression vector by calcium phosphate precipitation. Packaging cells were selected for the presence of the pREP8-mbSCF plasmid by culture in histidinol. Twenty clones were obtained by culture at limiting dilution and analyzed by fluorescence-activated cell sorter (FACS) for surface SCF expression. The clone with the highest level of expression was chosen for further use.
Virus binding
MO7e cells were washed twice in phosphate-buffered saline (PBS) and incubated with retroviral supernatant (100 μL) for 1 hour at 32°C. The cells were then washed twice in cold PBS and incubated with 100 μL of anti-surface subunit (SU) monoclonal antibody, 83A25, for 1 hour at 4°C. The cells were again washed twice with cold PBS and incubated with a fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse IgG antibody for 1 hour at 4°C. The cells were then washed twice in cold PBS and analyzed by flow cytometry.
CD34+ cell separation
CD34+ cells were separated from bone marrow mononuclear cells using the MiniMacs system (Miltenyi Biotec, Bisley, United Kingdom) according to manufacturer's instructions. The purity of the samples was consistently above 90%.
Estimation of retroviral titer
The pBabe vectors were titered by G418 resistance as described previously.24 Retroviral titers of the PINCO vector in the 3 Phoenix retroviral packaging cell lines were determined on NIH3T3 cells. The cells were analyzed by flow cytometry to determine the proportion of the 3T3 cells expressing enhanced green fluorescent protein (EGFP).
Supernatant transduction of hematopoietic cells
Retroviral targeting experiments on hematopoietic cell lines were transduced using a multiplicity of infection (MOI) of 5 based on the retroviral titer on NIH3T3 cells. An infection cocktail containing retroviral supernatant, 8 mg/mL polybrene, and 10% FCS in RPMI 1640 with 2.5 × 104 target cells was incubated for 24 hours at 37°C at 5% CO2. The medium was removed and replaced with fresh medium and incubated for a further 48 hours at 37°C at 5% CO2. Target cells were then washed twice with PBS, fixed, and assayed by flow cytometry.
A similar protocol was followed for CD34+ cells but the transduction cocktail was supplemented with protamine sulfate (4 μg/mL), in place of polybrene, and the recombinant human cytokines fetal liver tyrosine kinase 3 (Flt-3) ligand (100 ng/mL), interleukin 3 (IL-3; 10 ng/mL), and GM-CSF (1 ng/mL). Transduction was performed for 96 hours at 37°C in Iscove modified Dulbecco medium (IMDM) medium supplemented with 30% FCS and cells were harvested and assayed as described for cell lines.
Retroviral transduction of CD34+ primary cells by Transwell coculture
The retroviral transduction protocol used in this study was previously established by Chinswangwatanakul (Lewis et al24 ), except that Flt-3 ligand was used in place of SCF. Retroviral producers were plated in a 6-well plate at a concentration of 2 × 105 cells/well and maintained in DMEM supplemented with 10% FCS. After 24 hours, the Transwell (0.4 mm; Costar, Corning Inc, Corning, NY) was inserted into each well. Immunomagnetically isolated CD34+ cells (1 × 105) in IMDM supplemented with 30% FCS, protamine sulfate (4 μg/mL), and the cytokines IL-3 (10 ng/mL), GM-CSF (1 ng/mL), and Flt-3 ligand (100 ng/mL) were then transferred to the Transwell inserts. Transduction was carried out for 48 hours before the CD34+ cells were harvested and cultured in semisolid medium, with and without G418 selection, as described below in “Granulocyte macrophage colony-forming unit (CFU-GM) assay.”
Granulocyte macrophage colony-forming unit (CFU-GM) assay
The CD34+ transduced cells were cultured in 3 mL methylcellulose (Methocult H4230; StemCell Technologies) supplemented with “CFU-GM mix” (50 ng/mL SCF, 1 ng/mL GM-CSF, 10 ng/mL IL-3, and 100 ng/mL G-CSF) and 1.5 mg/mL G418. After mixing, the cells were plated into 35-mm diameter Petri dishes. The cultures were incubated at 37°C in humidified 5% CO2. CFU-GM colonies of more than 50 cells were scored on day 14 of incubation.
PCR
DNA from transduced MO7e cells was isolated and amplified using primers located in the EGFP sequence. Thirty-five cycles of amplification were performed. The control amplifications were performed on the same samples using primers amplifying a common segment of the α-interferon genes. The primer sequences were identified using the software package Pride (http://pride.molgen.mpg.de). Primer sequences (EGFP-forward [F], accccgaccacatgaagcagc; EGFP-reverse [R], tcgcctcgaacttcacctc; IFN-F, gaaccagtctagcacatc; and IFN-R, ggtgagctggctacgaatc) were synthesized by Sigma-Genosys (Haverhill, United Kingdom). The products were visualized in a 2% agarose gel containing 1 μg/mL ethidium bromide.
Day-14, G418-resistant CFU-GM colonies were plucked directly into a 40-mL PCR mix containing primers specific for the neo resistance gene and PCR was carried out using a nested strategy as described in Lewis et al.24 Ten microliters of the nested products were visualized, as described above.
CFU-GM replating assay
Replating assays were performed as described previously.24 On day 7 of incubation, 90 primary CFU-GM colonies growing in the presence of G418 were plucked with a Gilson P20 pipette and replated individually into 100 mL of methyl cellulose with CFU-GM mix and 1.5 mg/mL G418 in a 96-well, flat-bottom plate (60 wells per plate with the surrounding wells filled with sterile water to prevent evaporation) for secondary colony formation. The replated cultures were allowed to continue growing at 37°C, 5% CO2 in a humidified incubator, for a further 7 days at which point plates were scored for the presence and number of secondary colonies. The cumulative distribution of secondary CFU-GM colonies was analyzed in a logarithmic plot relating to log2n (the number of doublings required to produce n secondary colonies). The area under the curve (AUC), a parameter providing the kinetics of self-replication of progenitors, was calculated by the trapezium rule.
Results
Retroviruses are now known to acquire many intrinsic host cell membrane proteins as they bud through the plasma membrane.26-29 We investigated whether this property could be exploited so that molecules incorporated into the retroviral envelope could be used to redirect binding and transduction of viruses to target cells specifically expressing the appropriate receptor (Figure 1). To this end we constructed an ecotropic retroviral producer cell line expressing the membrane-bound form of human stem cell factor (mbSCF). Using Western blotting, antibody capture of virus, and proliferation assays, we established that retroviral particles derived from these modified producers efficiently incorporated mbSCF into the retroviral envelope.31
Schematic diagram of retroviral display methodology. Retroviral packaging cells were transfected with the plasmid pREP8-mbSCF. The cells were converted to producers by transfection with a retroviral genome. The SCF protein synthesized by these cells is expressed on the cell surface and incorporated into virus particles as they bud through the plasma membrane. Modified from Access Excellence at the National Health Museum30 with permission.
Schematic diagram of retroviral display methodology. Retroviral packaging cells were transfected with the plasmid pREP8-mbSCF. The cells were converted to producers by transfection with a retroviral genome. The SCF protein synthesized by these cells is expressed on the cell surface and incorporated into virus particles as they bud through the plasma membrane. Modified from Access Excellence at the National Health Museum30 with permission.
Redirection of retroviral binding
Unlike ecotropic virus, which is only permissive for infection of rodent cells, virus successfully incorporating surface SCF should be capable of binding to human cells that express the SCF receptor, c-kit. We therefore performed virus-binding assays to investigate the ability of our SCF-eco viruses to bind to the c-kit-expressing cell line, MO7e.22 As shown in the left panel of Figure 2A, anti-SU monoclonal antibody did not detect binding of normal ecotropic retrovirus to MO7e cells. Using SCF-eco virus, however, we observed a clear shift in fluorescence (Figure 2A right), indicating that the engineered virus was bound to the c-kit+ target cells. To confirm that this binding was SCF mediated, the experiment was repeated using MO7e cells that had been incubated overnight with recombinant soluble human SCF. This exposure to SCF leads to a major (approximately 10-fold) down-regulation of c-kit (Figure 2B) and also reduced SCF-eco virus binding (Figure 2C right) to a level equivalent to that of unmodified ecotropic virus (Figure 2C left).
SCF on virus particles redirects binding and facilitates transduction of MO7e cells. (A) Virus-binding assays. MO7e cells cultured in GM-CSF were incubated with retroviral supernatants, washed, and stained for the presence of bound virus by incubation with anti-SU monoclonal antibody. The lighter trace represents cells alone and the heavy trace represents that obtained in the presence of virus. The ecotropic virus is shown in the left panel and the SCF-eco virus is shown in the right panel. (B) Down-regulation of c-kit expression in response to soluble SCF. MO7e cells were cultured in the presence of GM-CSF or for 24 hours in fresh medium containing SCF and then stained for expression of surface c-kit. The light trace is cells stained with a mouse IgG1 isotype control, the dotted trace is from cells grown in GM-CSF, and the heavy trace is from cells grown for 24 hours in SCF. (C) MO7e cells cultured for 24 hours in SCF were incubated with viral supernatant and stained with anti-SU, as described in panel A. The ecotropic virus is shown in the left panel and the SCF-eco virus is shown in the right panel. (D) Transduction of MO7e cells by ecotropic retrovirus incorporating surface SCF. Cells from the c-kit+ cell line MO7e were incubated with retroviral supernatants as indicated below and transduction was evaluated by flow cytometry to detect the presence of the EGFP reporter gene. The percentage of fluorescent cells is indicated in each panel. Mock-transduced cells (i), cells transduced with amphotropic virus (ii), cells transduced with ecotropic virus (iii), and cells transduced with SCF-eco virus (iv).
SCF on virus particles redirects binding and facilitates transduction of MO7e cells. (A) Virus-binding assays. MO7e cells cultured in GM-CSF were incubated with retroviral supernatants, washed, and stained for the presence of bound virus by incubation with anti-SU monoclonal antibody. The lighter trace represents cells alone and the heavy trace represents that obtained in the presence of virus. The ecotropic virus is shown in the left panel and the SCF-eco virus is shown in the right panel. (B) Down-regulation of c-kit expression in response to soluble SCF. MO7e cells were cultured in the presence of GM-CSF or for 24 hours in fresh medium containing SCF and then stained for expression of surface c-kit. The light trace is cells stained with a mouse IgG1 isotype control, the dotted trace is from cells grown in GM-CSF, and the heavy trace is from cells grown for 24 hours in SCF. (C) MO7e cells cultured for 24 hours in SCF were incubated with viral supernatant and stained with anti-SU, as described in panel A. The ecotropic virus is shown in the left panel and the SCF-eco virus is shown in the right panel. (D) Transduction of MO7e cells by ecotropic retrovirus incorporating surface SCF. Cells from the c-kit+ cell line MO7e were incubated with retroviral supernatants as indicated below and transduction was evaluated by flow cytometry to detect the presence of the EGFP reporter gene. The percentage of fluorescent cells is indicated in each panel. Mock-transduced cells (i), cells transduced with amphotropic virus (ii), cells transduced with ecotropic virus (iii), and cells transduced with SCF-eco virus (iv).
Targeted transduction of cell lines
In contrast with many retroviruses bearing modified envelope proteins, the titer of our SCF-eco virus on murine NIH3T3 cells (2.6 × 105 infectious units [ifu]/mL) was found to be comparable to those obtained with unmanipulated eco or ampho viruses (3.9 × 105 and 2.5 × 105 ifu/mL, respectively). In all subsequent experiments, volumes of retroviral supernatant were adjusted to equalize the retroviral particle numbers accordingly.
To investigate the ability of the SCF-eco virus to specifically transduce c-kit+ cells, we first tested transduction of MO7e cells (Figure 2D). Efficient transduction of these cells using amphotropic (ampho) virus supernatant was observed (Figure 2Dii), whereas ecotropic (eco) virus, having a specific tropism for rodent cells, gave no evidence of transduction (Figure 2Diii). Conversely, the SCF-modified eco virus (SCF-eco) gave efficient transduction of these cells (Figure 2Div), achieving levels similar to those obtained with ampho virus. This suggested that the SCF-eco virus was capable of transducing c-kit+ human cells. To extend these observations, we analyzed the relative ability of the 3 virus preparations to transduce 4 cell lines with varying levels of c-kit expression (Figure 3A): MO7e and TF-1 cells, which express high levels of c-kit, LAMA-84, which has an intermediate level, and U937, essentially c-kit-. Using the high-c-kit expressers MO7e and TF-1 (Figure 3Ai-ii), the SCF-eco virus gave transduction efficiencies comparable with ampho virus (Figure 3B) and in stark contrast to the unmodified eco virus (Figure 3B), which was unable to transduce these cells. On LAMA-84 (Figure 3Aiii), the transduction by SCF-eco was significantly less efficient than before (Figure 3B), whereas on cells that do not express c-kit, the monomyelocytic cell line U937, no detectable transduction was obtained with SCF-eco or eco virus (Figure 3B). These experiments clearly demonstrated a relationship between transduction efficiency of the SCF-eco virus and the level of c-kit expression, suggesting that the SCF-c-kit interaction was mediating transduction.
Transduction of human hematopoietic cell lines by SCF-eco retrovirus is c-kit dependent. (A) Staining of 4 cell lines—MO7e, TF-1 cells, LAMA-84, and U937 cells—for surface expression of c-kit. The open traces show the mouse IgG1 isotype control used for setting the fluorescence gate. (B) The 4 lines expressing different levels of c-kit were transduced (mean ± SEM) with ampho, eco, and SCF-eco retroviral supernatants as indicated. In each case, the data were obtained from 6 independent transductions and were corrected for the background level of fluorescence obtained with mock-transduced cells.
Transduction of human hematopoietic cell lines by SCF-eco retrovirus is c-kit dependent. (A) Staining of 4 cell lines—MO7e, TF-1 cells, LAMA-84, and U937 cells—for surface expression of c-kit. The open traces show the mouse IgG1 isotype control used for setting the fluorescence gate. (B) The 4 lines expressing different levels of c-kit were transduced (mean ± SEM) with ampho, eco, and SCF-eco retroviral supernatants as indicated. In each case, the data were obtained from 6 independent transductions and were corrected for the background level of fluorescence obtained with mock-transduced cells.
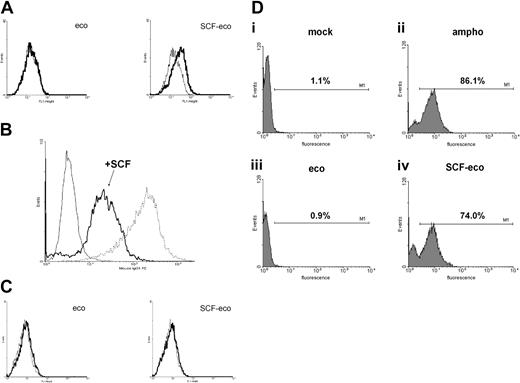
To confirm the role of the virus-associated SCF in the transduction process, we repeated the transduction of MO7e cells but this time in the presence of saturating amounts of competing soluble recombinant SCF. The presence of soluble competitor virtually eradicated SCF-eco transduction of MO7e cells (Figure 4A). DNA was extracted from the transduced cell populations used in this experiment and subjected to analysis by PCR with virus-specific primers to check for the presence of the retroviral genome (Figure 4B). Consistent with the transduction data, a positive PCR signal was obtained from cells transduced with ampho or SCF-eco virus but not mock-transduced cells. Although a weaker signal resulted from cells transduced with SCF-eco virus in the presence of growth factor, the conditions used for the PCR make it unlikely that this was due to the reduced efficiency of transduction.
Transduction of a c-kit+ cell line is inhibited by soluble SCF. (A) Transduction of MO7e cells, as shown in Figure 3, but performed in the presence [ampho (S) and SCF-eco (S)] or absence (SCF-eco) of 100 ng/mL human recombinant soluble SCF. Data are mean values from 2 independent transductions. (B) PCR analysis of DNA isolated from transduced cells shown in panel A. Lanes 2 to 4 show transductions performed in the presence of soluble SCF. (Top) PCR amplification using EGFP primers. (Bottom) PCR amplification of the same DNA samples using IFN-α primers. Lane 1, the 100-base-pair ladder; lane 2/a, amphotropic virus; lane 3/m, mock-transduced cells; lane 4/S-e, SCF-eco virus; and lane 5/S-e, SCF-eco virus.
Transduction of a c-kit+ cell line is inhibited by soluble SCF. (A) Transduction of MO7e cells, as shown in Figure 3, but performed in the presence [ampho (S) and SCF-eco (S)] or absence (SCF-eco) of 100 ng/mL human recombinant soluble SCF. Data are mean values from 2 independent transductions. (B) PCR analysis of DNA isolated from transduced cells shown in panel A. Lanes 2 to 4 show transductions performed in the presence of soluble SCF. (Top) PCR amplification using EGFP primers. (Bottom) PCR amplification of the same DNA samples using IFN-α primers. Lane 1, the 100-base-pair ladder; lane 2/a, amphotropic virus; lane 3/m, mock-transduced cells; lane 4/S-e, SCF-eco virus; and lane 5/S-e, SCF-eco virus.
Targeted transduction of primary hematopoietic progenitors
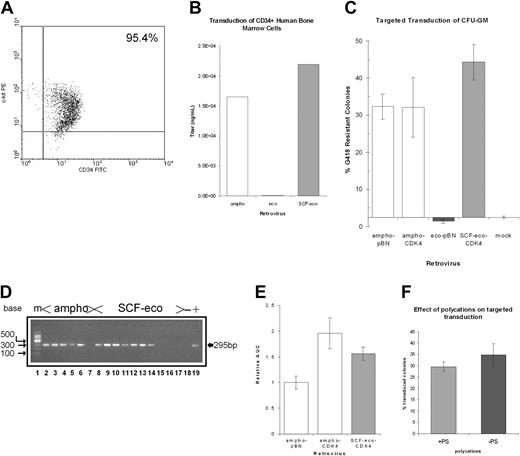
We then investigated the ability of these retroviruses to transduce primary human cells. Hematopoietic progenitors were obtained from aspirates of normal bone marrow and CD34+ cells isolated by magnetic microbead selection (MiniMacs). The CD34+ populations were tested for representation of c-kit and found to be greater than 90% positive (Figure 5A). Transduction of these cells with viral supernatants followed a very similar pattern to that seen previously with the cell lines. Both ampho and SCF-eco virus displayed similar transduction efficiencies, whereas the unmodified ecotropic virus gave virtually no transduction (Figure 5B).
Efficient transduction of c-kit+ primary human hematopoietic cells by SCF-eco retrovirus. (A) The c-kit staining of CD34+ normal bone marrow. Normal human bone marrow-derived CD34+ cells were isolated by immunomagnetic separation and stained for CD34 with FITC-conjugated monoclonal antibody and for c-kit with a phycoerythrin (PE) conjugate. The proportion of doubly stained cells is indicated in the top right quadrant. (B) Targeted transduction of CD34+ bone marrow cells. CD34+ cells were transduced with retroviral supernatants and assayed for expression of EGFP by flow cytometry. Transduction efficiency is shown relative to the results obtained with ampho virus and data are derived as for Figure 3 but from 2 independent marrows transduced in duplicate. (C) Targeted transduction of CFU-GM. CD34+ cells were cocultured with retroviral producers using Transwells. The transduced cells were plated in semisolid media containing G418 to select for colonies expressing the retrovirally encoded neo gene, and resistant colonies were counted. The data (mean ± SEM) are derived from 3 independent marrows transduced in duplicate. (D) Nested PCR analysis of colonies surviving in semisolid media supplemented with G418 using primers that detect the neo gene. The figure shows data from a representative sample of the colonies analyzed. Lane 1, 100-bp ladder size marker; lanes 2-7, cells transduced with ampho virus; lanes 8-17, cells transduced with SCF-eco virus; lane18/-, semisolid media only; lane 19/+, plasmid DNA containing retroviral genome. (E) Replating assay on G418-resistant colonies. Colonies surviving in semisolid media containing G418 were replated to assay expression of the CDK4 gene encoded by the retroviral genome. Replating activity, expressed as area under the curve (AUC), was compared with that obtained from an identical retroviral vector transducing only the neo resistance marker. (F) Effect of polycations on targeted transduction. CD34+ progenitors were transduced as in panel C but in the presence (+PS) or absence (-PS) of 4 μg/mL protamine sulfate; transduction was determined, as before, from the percentage of colonies resistant to G418. The data shown are a representative example of the experiment that was performed 3 times.
Efficient transduction of c-kit+ primary human hematopoietic cells by SCF-eco retrovirus. (A) The c-kit staining of CD34+ normal bone marrow. Normal human bone marrow-derived CD34+ cells were isolated by immunomagnetic separation and stained for CD34 with FITC-conjugated monoclonal antibody and for c-kit with a phycoerythrin (PE) conjugate. The proportion of doubly stained cells is indicated in the top right quadrant. (B) Targeted transduction of CD34+ bone marrow cells. CD34+ cells were transduced with retroviral supernatants and assayed for expression of EGFP by flow cytometry. Transduction efficiency is shown relative to the results obtained with ampho virus and data are derived as for Figure 3 but from 2 independent marrows transduced in duplicate. (C) Targeted transduction of CFU-GM. CD34+ cells were cocultured with retroviral producers using Transwells. The transduced cells were plated in semisolid media containing G418 to select for colonies expressing the retrovirally encoded neo gene, and resistant colonies were counted. The data (mean ± SEM) are derived from 3 independent marrows transduced in duplicate. (D) Nested PCR analysis of colonies surviving in semisolid media supplemented with G418 using primers that detect the neo gene. The figure shows data from a representative sample of the colonies analyzed. Lane 1, 100-bp ladder size marker; lanes 2-7, cells transduced with ampho virus; lanes 8-17, cells transduced with SCF-eco virus; lane18/-, semisolid media only; lane 19/+, plasmid DNA containing retroviral genome. (E) Replating assay on G418-resistant colonies. Colonies surviving in semisolid media containing G418 were replated to assay expression of the CDK4 gene encoded by the retroviral genome. Replating activity, expressed as area under the curve (AUC), was compared with that obtained from an identical retroviral vector transducing only the neo resistance marker. (F) Effect of polycations on targeted transduction. CD34+ progenitors were transduced as in panel C but in the presence (+PS) or absence (-PS) of 4 μg/mL protamine sulfate; transduction was determined, as before, from the percentage of colonies resistant to G418. The data shown are a representative example of the experiment that was performed 3 times.
We also tested the ability of the different viruses to transduce granulocyte macrophage colony-forming cells (GM-CFCs). For this we packaged a different vector, one that encoded a neo gene conferring resistance to the drug G418. Transductions were performed by Transwell coculture24 ; this allows prolonged exposure to target cells but prevents cell-cell contact. A typical transduction result using bone marrow-derived CD34+ cells is shown in Table 1 and the overall results are summarized in Figure 5C. As observed previously with this transduction system, a high proportion of G418-resistant colonies were obtained with ampho virus.24 Nevertheless, an equivalent level of transduction was obtained using the SCF-eco virus, whereas virtually no G418-resistant colonies resulted from transduction with eco virus. To confirm that the G418-resistant colonies were truly arising from SCF-eco virus transduction of CFU-GM, some of the resistant colonies were analyzed by PCR (Figure 5D). In 15 of 20 colonies amplified using primers specific for the neo gene, a positive signal was obtained, whereas for the ampho virus, 12 of 14 colonies tested proved positive. These values are consistent (χ2 test; P = .34 and P = .72, respectively) with the frequencies of positive-scoring colonies (95%) previously observed in validating of this technique.24 This indicated that retroviral vector genome carried in the SCF-eco retrovirus particles was present in the G418-resistant colonies.
Transduction of CFU-GM from human CD34+ cells
. | Transduction 1 . | . | . | Transduction 2 . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Retrovirus . | −G418 . | +G418 . | +G418 . | −G418 . | +G418 . | +G418 . | %resistant . | ||||
| Mock transduced | 240 | 6 | 2 | 294 | 15 | 5 | 2.5 | ||||
| Ampho-pBN | 77 | 22 | 21 | — | — | — | 27.9 | ||||
| Ampho-CDK4 | 286 | 28 | 33 | 170 | 94 | — | 25.4 | ||||
| Eco-pBN | 250 | 2 | 6 | — | — | — | 1.6 | ||||
| SCF-eco-CDK4 | 259 | 93 | 119 | 227 | 91 | 94 | 40.8 | ||||
. | Transduction 1 . | . | . | Transduction 2 . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Retrovirus . | −G418 . | +G418 . | +G418 . | −G418 . | +G418 . | +G418 . | %resistant . | ||||
| Mock transduced | 240 | 6 | 2 | 294 | 15 | 5 | 2.5 | ||||
| Ampho-pBN | 77 | 22 | 21 | — | — | — | 27.9 | ||||
| Ampho-CDK4 | 286 | 28 | 33 | 170 | 94 | — | 25.4 | ||||
| Eco-pBN | 250 | 2 | 6 | — | — | — | 1.6 | ||||
| SCF-eco-CDK4 | 259 | 93 | 119 | 227 | 91 | 94 | 40.8 | ||||
—indicates____.
To provide a functional test of our targeting strategy, we took advantage of the fact that the retroviral vector used in these studies also included the cDNA for the cyclin-dependent kinase, CDK4. Previous work has shown that expression of this gene in CFU-GM progenitors can increase their frequency of self-renewal in a replating assay.24 To evaluate whether the activity of the CDK4 gene could be detected in SCF-eco-transduced colonies, some of these were replated and the replating efficiency was compared with colonies transduced with either the equivalent ampho virus or an ampho vector containing only the neo resistance marker. The colony-forming cells transduced by viruses encoding CDK4 did not differ significantly from each other in replating activity (P = .27), whereas cells transduced with both ampho CDK4 and SCF-eco-CDK4 showed an increased frequency of replating (P < .05) in comparison with those transduced with the control pBN vector, indicating activity of the CDK4 gene (Figure 5E).
In some targeting strategies, retroviral gene delivery in vitro has been found to be particularly dependent on the presence of polycations such as polybrene or lipofectamine during transduction.32 Although it is not possible to extrapolate this to transduction in vivo,33 reliance on such agents could compromise the ability of targeted retroviruses to deliver genes in vivo. We therefore tested the ability of our SCF-targeted viruses to deliver the neo gene to CD34+ progenitors in the absence of such agents. Our standard protocols for transduction of CD34+ cells include protamine sulfate. Omission of protamine sulfate from these transductions was found to have no effect on the percentage of GM colonies resistant to G418 (Figure 5F), indicating that efficient targeted transduction in our system was not dependent on the presence of polycations.
Discussion
We have described here a novel approach to retroviral targeting that exploits the natural ability of retroviruses to incorporate host cell surface proteins into the lipid envelope of the virus. By producing ecotropic virus from packaging cells that were engineered to express human membrane-bound SCF on their surface we have successfully created viruses that infect human cells with high efficiency in a strictly SCF-receptor-dependent fashion. Most significantly, we have shown this to be true not just in cell lines but also in primary cells that are potential targets for therapeutic interventions; over half the GM colony-forming cells in the CD34+ population were transduced by our SCF-eco virus. Moreover, through transduction with vectors harboring the CDK4 gene, we have demonstrated that this targeting system can be used to functionally alter the properties of the targeted cells.
Many attempts have been made previously to retarget retroviral vectors to alternative cell surface receptors, including the SCF receptor,10,34 with a view to achieving cell type specificity in target cell transduction. In essentially all cases this has been through engineering of the retroviral envelope protein itself. Unfortunately, attempts to alter the binding specificity of retroviral envelopes have nearly always compromised their ability to mediate fusion with the host cell membrane, leading to unsuccessful retroviral transduction.5 Retroviral envelope proteins are thought to undergo major conformational changes during fusion4 and these are most likely inhibited or hindered by addition to or altering of the envelope sequences. Targeting strategies relying on coexpression of a modified envelope and an ecotropic envelope7,9,16,19 have provided some very limited success but, importantly, may still be severely compromised by difficulties in forming envelope trimers and in clustering of envelope proteins, steps crucial for retroviral entry.4 This interpretation is supported by the observation that when ecotropic virus was redirected to asialoglycoprotein receptors by chemical modification with lactose, a small molecule that should not cause steric hindrance, efficient targeted transduction of a human liver cell line was demonstrated.35
By leaving the retroviral envelope protein intact and separating virus binding from that of fusion, our approach is able to circumvent all these problems. A similar premise has been the basis for studies in which retroviral envelope was substituted by influenza hemagglutinin (HA) to provide a fusion function.36,37
Regarding the precise mechanism of targeted entry in our system, the role played by the ecotropic envelope protein is of interest. It seems likely that this is providing a fusion function for viral entry, as envelope-free HIV virus-like particles (VLPs) can be made in abundance but have no infectivity, even though they are able to bind to target cells.38 It is unclear though whether the ecotropic receptor is unusual in its apparent ability to fuse independently of binding. One clue may come from the fact that, unlike many other onco-retroviruses, ecotropic virus entry has been shown in some circumstances to be pH dependent, indicating that rather than interaction at the plasma membrane, an endosomal entry route is possible.39 Late endosomes have a low internal pH, which could be instrumental in triggering the conformational changes required for fusion.
Endocytosis would be an advantageous route of entry, as late endosomes are primarily perinuclear40 ; this may differ from amphotropic viruses that are pH-independent for entry. This difference in entry route of targeted and amphotropic viruses might explain the somewhat reduced level of reporter gene expression we observed with targeted transduction. This may result in lower copy number transductions or, perhaps, access to different sites of nuclear integration leading to lower levels of expression.
Targeted vectors in particular would be released from the necessity to perform transductions ex vivo and could herald a new phase of development of gene therapy protocols using in vivo gene delivery. This would be particularly felt in the arena of acquired disease but also has major ramifications for treating inherited disorders.
Prepublished online as Blood First Edition Paper, July 15, 2004; DOI 10.1182/blood-2003-10-3717.
Supported by a Chevening Award of the British Council and a Malaysian National Cancer Council Fellowship (A.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr Simon Wagner for his comments on the manuscript and Dr Yasuhiro Takeuchi for his kind gift of 83A25 anti-SU monoclonal antibody.



![Figure 4. Transduction of a c-kit+ cell line is inhibited by soluble SCF. (A) Transduction of MO7e cells, as shown in Figure 3, but performed in the presence [ampho (S) and SCF-eco (S)] or absence (SCF-eco) of 100 ng/mL human recombinant soluble SCF. Data are mean values from 2 independent transductions. (B) PCR analysis of DNA isolated from transduced cells shown in panel A. Lanes 2 to 4 show transductions performed in the presence of soluble SCF. (Top) PCR amplification using EGFP primers. (Bottom) PCR amplification of the same DNA samples using IFN-α primers. Lane 1, the 100-base-pair ladder; lane 2/a, amphotropic virus; lane 3/m, mock-transduced cells; lane 4/S-e, SCF-eco virus; and lane 5/S-e, SCF-eco virus.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/9/10.1182_blood-2003-10-3717/6/m_zh80210468400004.jpeg?Expires=1767762857&Signature=Jje6KyLqaHJNdQW5wixTNl-l8DQ7tPTDGsYiyL0XrsDwbq4NYxHLx5Zc1DHZRdQzQPrVo6d8sMargLsdiKwWXBS5FzyIuBwd0ZrMLWCOw4bW2AIuAvhBtQ9NszN8FppsSN7sfryZSu0Bx7OjLNrdA7j8bipWUMJGll4IknTLvjmNdhNKYsP7OM3~orPCQ3JE1lnYThXf4kE32QWSLOO1Hu855ycG9JNzI6m6FKRxnex8UgGgc42uZg~Cr7-bI6FKDM3tYBjsSpvYL7Fc1wfwItlBgi16~5Crqaz8IBcT8Z2CSerFnd8T4g~qjS05Q6WgGBU0SqmnaD1uP31g3D99lA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal