Abstract
The deregulated, oncogenic tyrosine kinase Bcr-Abl causes chronic myeloid leukemia (CML). Imatinib mesylate (Gleevec, STI571), a Bcr-Abl kinase inhibitor, selectively inhibits proliferation and promotes apoptosis of CML cells. Despite the success of imatinib mesylate in the treatment of CML, resistance is observed, particularly in advanced disease. The most common imatinib mesylate resistance mechanism involves Bcr-Abl kinase domain mutations that impart varying degrees of drug insensitivity. AP23464, a potent adenosine 5′-triphosphate (ATP)–based inhibitor of Src and Abl kinases, displays antiproliferative activity against a human CML cell line and Bcr-Abl–transduced Ba/F3 cells (IC50 = 14 nM; imatinib mesylate IC50 = 350 nM). AP23464 ablates Bcr-Abl tyrosine phosphorylation, blocks cell cycle progression, and promotes apoptosis of Bcr-Abl–expressing cells. Biochemical assays with purified glutathione S transferase (GST)–Abl kinase domain confirmed that AP23464 directly inhibits Abl activity. Importantly, the low nanomolar cellular and biochemical inhibitory properties of AP23464 extend to frequently observed imatinib mesylate–resistant Bcr-Abl mutants, including nucleotide binding P-loop mutants Q252H, Y253F, E255K, C-terminal loop mutant M351T, and activation loop mutant H396P. AP23464 was ineffective against mutant T315I, an imatinib mesylate contact residue. The potency of AP23464 against imatinib mesylate–refractory Bcr-Abl and its distinct binding mode relative to imatinib mesylate warrant further investigation of AP23464 for the treatment of CML.
Introduction
The initial, chronic stage of chronic myeloid leukemia (CML) is strictly dependent on signals emanating from the deregulated protein tyrosine kinase Bcr-Abl.1 Although the Bcr-Abl signaling cascade is incompletely understood, the premise that CML can be treated by selective inhibition of Bcr-Abl kinase activity has been validated in the clinic.2-4 Imatinib mesylate (Gleevec, STI571), a protein tyrosine kinase inhibitor with a narrow specificity profile (Abl, ARG, Kit, and platelet-derived growth factor receptor [PDGFR]), has remarkable single agent activity in patients with CML and is now the first-line treatment for the disease.5,6 In addition, this molecular-targeted therapy is effective against other cancers that depend on signaling through imatinib mesylate–sensitive protein tyrosine kinases.7-12
An important clinical concern pertaining to imatinib mesylate therapy is relapse after an initial response, particularly in patients with advanced phase CML. For example, among patients with accelerated phase enrolled in a phase 2 clinical trial, the incidence of disease progression at 24 months was 50%.13 Between 60% and 90% of patients who acquire imatinib mesylate resistance harbor one or more specific mutations in the kinase domain of Bcr-Abl that impair the ability of imatinib mesylate to inhibit Bcr-Abl kinase activity. These mutations presumably affect drug binding without eliminating adenosine 5′-triphosphate (ATP) binding or kinase activity.14-19
Clinically observed mutations have been identified within several regions of the Bcr-Abl kinase domain. In this study, we examined 6 common kinase domain variants that collectively account for at least 60% of reported Bcr-Abl mutations in relapsed patients: Q252H, Y253F, E255K, T315I, M351T, and H396P. The panel spans a range of residual imatinib mesylate sensitivities (IC50 = 900-4400 nM)20 and encompasses several functionally distinct kinase domain regions, including the nucleotide binding P-loop (Q252H, Y253F, E255K), 2 imatinib mesylate contact residues (Y253F and T315I), the base supporting the activation loop (M351T), and the activation loop (H396P).21,22
Currently, there is considerable interest in developing alternative Abl kinase inhibitors capable of inhibiting the Bcr-Abl kinase domain mutants observed in relapsed patients. Using structure-based drug design and focused synthetic libraries of trisubstituted purine analogs, we identified AP23464 as a potent inhibitor of Abl and Src-family kinases. AP23464 displayed potent inhibitory activity against Src-family kinases and Abl kinase (IC50 ≤ 1 nM), as well as lower activity on a small subset of additional kinases, with specificity for these kinases versus more than 40 others tested (manuscript in preparation).
On the basis of its impressive potency as an inhibitor of Src family and Abl kinases, we tested AP23464 against human leukemic cell lines and found it to selectively inhibit proliferation, block cell cycle progression, and promote apoptosis in Bcr-Abl–expressing K562 cells. These effects were accompanied by the elimination of cellular tyrosine phosphorylation of Bcr-Abl and its downstream signaling effectors signal transducer and activator of transcription 5 (STAT-5) and CrkL. AP23464 was also tested against Abl kinase domains carrying single mutations that cause imatinib mesylate resistance. Biochemical assays with purified wild-type (WT) and mutant glutathione S transferase (GST)–Abl kinase fusion proteins demonstrated potent inhibition of Abl autophosphorylation and Abl-catalyzed peptide substrate phosphorylation. The one exception was the Abl mutant T315I. In cell-based assays, the inhibitor effectively blocked proliferation of Ba/F3 cells expressing either WT Bcr-Abl or 5 of the most common imatinib mesylate–resistant Bcr-Abl variants observed in the clinical setting, but not mutant T315I. Consistent with cell proliferation results, annexin V binding assays confirmed that AP23464 induced apoptosis of Ba/F3 cells expressing either WT Bcr-Abl or AP23464-sensitive Bcr-Abl variants. Similarly, immunoblot analysis showed that AP23464 reduced cellular tyrosine phosphorylation of WT Bcr-Abl and imatinib mesylate–resistant Bcr-Abl mutants except T315I. AP23464 is a promising lead compound for the treatment of patients with CML with Bcr-Abl kinase domain mutations that confer imatinib mesylate resistance.
Materials and methods
Reagents
Stock solutions (10 mM in dimethyl sulfoxide) of AP23464 (C26H30N5O2P; MW = 475.5) were stored at –20° C. Experiments were performed with serial dilutions of the 10 mM stock. AP23464 was identified by using the combined tools of computational modeling studies, small focused library synthesis, and biologic testing to optimize the purine substituents at the 2, 6, and 9 positions for maximum Src specificity and potency. Full synthetic details for AP23464 will be presented elsewhere.
Cell lines
K562 (Bcr-Abl–positive human erythroleukemia, ATCC no. CCL-243) or HL60 (Bcr-Abl–negative human acute myeloid leukemia, ATCC no. CCL-240) cells were obtained from the American Type Culture Collection (Manassas, VA), and maintained in Iscoves modified Dulbecco medium containing 4 mM L-glutamine, 1.5 g/L sodium bicarbonate, and 10% (K562) or 20% (HL60) fetal calf serum (FCS). Stable Ba/F3 cell lines expressing full-length wild-type Bcr-Abl or Bcr-Abl with kinase domain point mutations were previously generated in our laboratory.20 Ba/F3 cells expressing Bcr-Abl or Bcr-Abl mutants were maintained in RPMI 1640 growth media supplemented with 200 μM L-glutamine, 10% FCS, penicillin (200 U/mL), and streptomycin (200 μg/mL). Parental Ba/F3 cells were cultured in the same media supplemented with WEHI-conditioned media as a source of interleukin 3 (IL-3).
Cell proliferation assays
Exponentially growing K562 or HL60 cells were plated, in triplicate, at 1 × 104 cells per 96-well plate and incubated in the presence of AP23464 or imatinib mesylate (0-10 μM) for 3 days. Cell proliferation was measured with an methanethiosulfonate (MTS)–based viability assay (CellTiter 96 Aqueous One Solution Reagent; Promega, Madison, WI), as previously described.20 Parental Ba/F3 cells (supplemented with IL-3) or Ba/F3 cells expressing WT or mutant Bcr-Abl protein were plated in quadruplicate at 5 × 103 cells/well in 96-well plates with AP23464 included in the media. The MTS assay was performed daily to confirm exponential growth of the untreated cells (0, 24, 48 hours). Results from day 2 were used to construct best-fit curves and calculate the cellular IC50 in Microsoft Excel (Redmond, WA). The mean based on 4 replicates was calculated in the absence of inhibitor and for each concentration of AP23464. Means ± SE were generated from 3 independent experiments and reported as the percentage absorbance of control. Additional experiments (0-10 μM AP23464) were carried out to determine IC50 values for parental Ba/F3 cells and Ba/F3 cells expressing Bcr-Abl mutant T315I.
Apoptosis assays
K562 cells or HL60 cells (5 × 105 cells/well) were incubated in 6-well plates for 48 hours in 4 mL media containing AP23464, harvested, stained with Alexa Fluor 488 annexin V (Molecular Probes, Eugene, OR) according to manufacturer's instructions, and analyzed on a FACSort flow cytometer. FL-1 histograms were generated by using CellQuest software (BD Biosciences, San Jose, CA).
Each Ba/F3 cell line (1.0 × 105 cells/well; > 95% viability) was distributed into a 24-well plate and cultured in the presence of AP23464 for 48 hours. Harvested cells were stained with annexin V–phycoerythrin (PE) and 7-amino-actinomycin D by using the Guava Nexin apoptosis kit (Guava Technologies, Hayward, CA) according to the manufacturer's instructions and examined by flow cytometry. Data collection and analysis were done by using a Guava Technologies PCA instrument equipped with Cytosoft software (Guava Technologies). Results based on 3 independent experiments are reported as averages ± SE.
Cell cycle analysis
K562 cells or HL60 cells (5 × 105 cells/well) were incubated in duplicate in 6-well plates for 24 hours in 2 mL medium containing AP23464. Harvested cells were fixed in 70% ethanol, stained with propidium iodide, and analyzed on a FACSort flow cytometer to determine DNA content. The relative percentages of cells in G1, S, or G2/M phase were calculated from FL-2 histograms using ModFit LT software (Verity Software House, Topsham, ME).
Immunoblotting
K562 cells (1 × 106) were incubated for 4 hours in 20 mL media in the presence of AP23464. Cells were collected by centrifugation and lysed in phosphate-buffered saline containing 1% NP40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 10 mM NaF, 1 mM Na3VO4, and Complete Mini Protease Inhibitor Cocktail tablets (Roche Applied Science, Indianapolis, IN). Lysates were separated by SDS–polyacrylamide gel electrophoresis (PAGE) on 10% gels (50 μg protein/lane). Immunoblot analysis for phospho-Bcr-Abl (Tyr245), phospho-STAT5 (Tyr694), phospho-CrkL (Tyr207), and eIF4E (protein loading control) was performed by using a Pathscan Bcr/Abl Multiplex Western Detection Kit (Cell Signaling Technology, Beverly, MA).
Parental Ba/F3 cells or Ba/F3 cells expressing WT or mutant Bcr-Abl (4 × 106 cells/well) were plated in 2.5 mL RPMI media containing escalating concentrations of AP23464, then incubated for 3 hours at 37° C. Cells were collected by centrifugation, washed, and disrupted in lysis buffer (Cell Signaling Technology) containing 1 mM Na3VO4, 20 μM phenylarsine oxide (Calbiochem, San Diego, CA), and Complete tablets. For immunoblot analysis, tyrosine-phosphorylated Bcr-Abl was detected with mouse monoclonal antiphosphotryrosine antibody 4G10 (Upstate Biotechnology, Waltham, MA). Bcr-Abl expression was detected with mouse monoclonal anti-Abl antibody 8E9 (BD Biosciences Pharmingen, San Diego, CA) or Ab-2 (Oncogene Science, Cambridge, MA). Tyrosinephosphorylated Src family kinase members were detected with rabbit polyclonal antiphospho-Src family antibody CST2101 (Cell Signaling Technology) according to the manufacturer's instructions. As a loading control, total Lyn expression levels were determined with rabbit polyclonal anti-Lyn antibody sc-15 (Santa Cruz Biotechnology, Santa Cruz, CA).
Kinase autophosphorylation assays with GST-Abl kinase domains
Kinase assays using GST-Abl fusion proteins (c-Abl amino acids 220-498) were performed as described23 with minor modifications. GST-Abl kinase fusion proteins were released from glutathione-Sepharose beads prior to use in the kinase assay, and the ATP concentration in the kinase assay was 5 μM. Identical procedures were used for all experiments. Abl immunoblots to demonstrate equal protein loading were performed with α-Abl Ab-2 (Oncogene Science) as previously described.23
In vitro peptide substrate phosphorylation assays with GST-Abl kinase domains
The effect of AP23464 (0-320 nM) on GST-Abl kinase activity was assessed by using a synthetic peptide substrate (Abltide: EAIYAAPFAKKK; Upstate Biotechnology). Assays were carried out in duplicate at 30° C for 15 minutes in 25 μL reaction mixture: 8 mM MOPS (3(N-morpholino) propanesulfonic acid), pH 7, 0.2 mM EDTA (ethylenediaminetetraacetic acid), 50 μM Abltide, 30 mM MgCl2, 10 mM β-glycerol phosphate, 1 mM EGTA (ethylene glycol tetraacetic acid), 0.002% Brij-35, 0.4 mM DTT (dithiothreitol), 0.2 mg/mL BSA (bovine serum albumin), 0.4 mM sodium orthovanadate, 10 nM WT or mutant GST-Abl kinase, and 100 μM ATP/γ-32[P]ATP (5000 cpm/pmol). Reactions were terminated by transferring a portion of the reaction mixture onto a p81 phosphocellulose filter and immersing in 0.75% phosphoric acid. Filters were washed 3 times in 0.75% phosphoric acid, rinsed in acetone, and air dried; phosphate incorporation was determined by scintillation counting. All results were corrected for background binding to the filters, as determined by omitting peptide substrate from the kinase reaction. Time course experiments to establish the linear range of enzymatic activity preceded kinase assays. Recombinant human Abl (residues 27-end) and Abl T315I (residues 27-end) used in preliminary experiments were purchased from Upstate Biotechnology (Waltham, MA).
Results
AP23464 inhibits proliferation, blocks cell cycle progression, and induces apoptosis of Bcr-Abl–positive K562 but not Bcr-Abl–negative HL60 leukemic cells
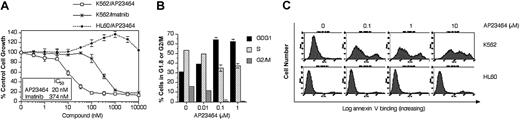
To assess the potency and specificity of AP23464 for Bcr-Abl, its effects on the Bcr-Abl–positive K562 cell line were compared with those on the Bcr-Abl–negative HL60 leukemic cell line in proliferation, cell cycle, and apoptosis assays. Cell proliferation assays showed that AP23464 inhibited the growth of Bcr-Abl–expressing K562 cells in vitro with a potency that was significantly greater than that of imatinib mesylate (Figure 1A). In contrast, the proliferation of non-Bcr-Abl–expressing HL60 cells was not inhibited in the presence of AP23464 (Figure 1A).
AP23464 inhibits proliferation, blocks cell cycle progression, and induces apoptosis of Bcr-Abl–positive K562 but not Bcr-Abl–negative HL60 leukemic cells. (A) Proliferation of K562 (Bcr-Abl–positive) or HL60 (Bcr-Abl–negative) cells in the presence of escalating concentrations of AP23464 or imatinib mesylate (0-10 000 nM). Cell growth was assessed by MTS-based viability assay. (B) Cell cycle analysis of K562 cells in the presence of AP23464. Cells (5 × 105) were incubated in duplicate for 24 hours in 4 mL medium containing AP23464 (0, 0.01, 0.1, 1 μM). Fixed cells were stained with propidium iodide and analyzed on a FACSort flow cytometer to determine DNA content. The relative percentages of cells in G1, S, or G2/M phase were calculated from FL-2 histograms by using ModFit LT software. (C) Effect of AP23464 (0, 0.1, 1, or 10 μM) on induction of apoptosis in K562 (Bcr-Abl–positive) cells or HL60 (Bcr-Abl–negative) cells. Alexa Fluor 488 annexin V–stained cells were analyzed on a FACSort flow cytometer and histograms were generated by using CellQuest software.
AP23464 inhibits proliferation, blocks cell cycle progression, and induces apoptosis of Bcr-Abl–positive K562 but not Bcr-Abl–negative HL60 leukemic cells. (A) Proliferation of K562 (Bcr-Abl–positive) or HL60 (Bcr-Abl–negative) cells in the presence of escalating concentrations of AP23464 or imatinib mesylate (0-10 000 nM). Cell growth was assessed by MTS-based viability assay. (B) Cell cycle analysis of K562 cells in the presence of AP23464. Cells (5 × 105) were incubated in duplicate for 24 hours in 4 mL medium containing AP23464 (0, 0.01, 0.1, 1 μM). Fixed cells were stained with propidium iodide and analyzed on a FACSort flow cytometer to determine DNA content. The relative percentages of cells in G1, S, or G2/M phase were calculated from FL-2 histograms by using ModFit LT software. (C) Effect of AP23464 (0, 0.1, 1, or 10 μM) on induction of apoptosis in K562 (Bcr-Abl–positive) cells or HL60 (Bcr-Abl–negative) cells. Alexa Fluor 488 annexin V–stained cells were analyzed on a FACSort flow cytometer and histograms were generated by using CellQuest software.
Activation of downstream effectors that promote cell cycle progression is a primary function of Bcr-Abl. The effect of AP23464 on this process was examined by flow cytometry. Incubation of K562 cells for 24 hours in the presence of AP23464 caused a strong accumulation in the G0/G1 cell cycle phase, with increases first apparent at 10 nM and maximal by 100 nM AP23464 (Figure 1B). These effects were not observed in HL60 cells cultured under identical conditions (data not shown).
Bcr-Abl signaling is central to the survival of CML cells.24 In transformed cells, the Bcr-Abl oncoprotein participates in maintenance of an antiapoptotic environment through regulation of Bcl-2 family members25-27 and tumor necrosis factor–related apoptosis-inducing ligand.24,28,29 In our study, incubation of K562 cells for 48 hours in the presence of AP23464 (100 nM) induced apoptosis as measured by annexin V staining followed by flow cytometry analysis (Figure 1C). AP23464 did not induce apoptosis of HL60 cells even at a concentration of 10 μM. These results were confirmed with 2 further apoptosis assays: agarose gel–based detection of apoptotic DNA ladder formation and enzyme-linked immunosorbent assay (ELISA) quantification of cytoplasmic mononucleosomes and oligonucleosomes (data not shown).
Exposure of K562 cells to AP23464 inhibits tyrosine phosphorylation of Bcr-Abl and its downstream targets STAT5 and CrkL
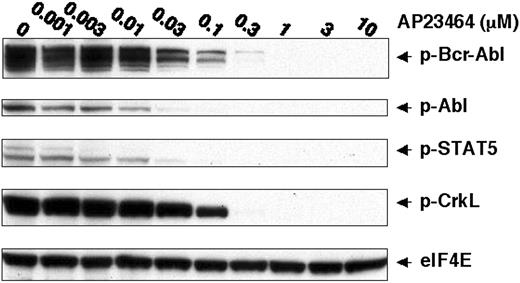
To further establish a direct link between the pronounced cellular effects of AP23464 and inhibition of Bcr-Abl tyrosine kinase activity, we examined the tyrosine phosphorylation status of Bcr-Abl and downstream substrates STAT5 and CrkL.1,30-34 Specifically, immunoblot analysis of lysates from AP23464-treated cells with antiphospho-STAT5 (Tyr694) and antiphosphoCrkL (Tyr207) monoclonal antibodies revealed a concentration-dependent reduction in phosphorylation of these Bcr-Abl targets (Figure 2). Tyrosine phosphorylation of STAT5 was almost completely blocked by 30 nM AP23464, in close agreement with the concentration range leading to antiproliferative effects in K562 cells. A higher AP23464 concentration (300 nM) was required to ablate tyrosine phosphorylation of CrkL. Bcr-Abl tyrosine phosphorylation levels were simultaneously monitored and found to be drastically reduced at a concentration of 30 nM AP23464 (Figure 2). Together, these data establish that AP23464-mediated effects on proliferation, cell cycle progression, and apoptosis of K562 cells correlate with a dramatic reduction in Bcr-Abl kinase activity.
AP23464 inhibits tyrosine phosphorylation of Bcr-Abl and downstream targets STAT-5 and CrkL. K562 cells (1 × 106) were incubated for 4 hours in 20 mL media in the presence of AP23464 (0, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, or 10 μM). Total cell lysates were prepared and separated by SDS-PAGE on 10% gels (50 μg protein/lane). Western blot analysis for phospho–Bcr-Abl, phospho-STAT5, phospho-CrkL, and eIF4E (protein loading control) was performed by using a Pathscan Bcr/Abl Multiplex Western Detection Kit (Cell Signaling Technology). Immunoblotting with nonphosphospecific antibodies confirmed that total levels of Bcr-Abl, STAT5, and CrkL were unchanged (data not shown).
AP23464 inhibits tyrosine phosphorylation of Bcr-Abl and downstream targets STAT-5 and CrkL. K562 cells (1 × 106) were incubated for 4 hours in 20 mL media in the presence of AP23464 (0, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, or 10 μM). Total cell lysates were prepared and separated by SDS-PAGE on 10% gels (50 μg protein/lane). Western blot analysis for phospho–Bcr-Abl, phospho-STAT5, phospho-CrkL, and eIF4E (protein loading control) was performed by using a Pathscan Bcr/Abl Multiplex Western Detection Kit (Cell Signaling Technology). Immunoblotting with nonphosphospecific antibodies confirmed that total levels of Bcr-Abl, STAT5, and CrkL were unchanged (data not shown).
AP23464 inhibits tyrosine phosphorylation of isolated WT Abl kinase domain and isolated kinase domain mutants Y253F, E255K, M351T, and H396P but not T315I
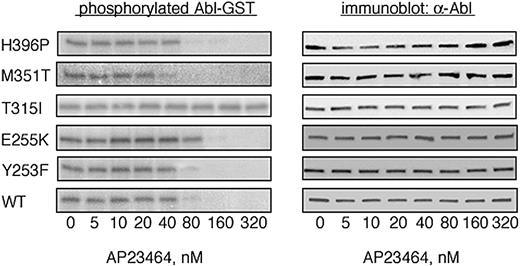
To determine whether AP23464 is capable of directly inhibiting the kinase activity of purified Abl kinase in a cell-free system, we carried out biochemical experiments with the use of GST-Abl kinase fusion proteins. Expressed proteins corresponding to WT Bcr-Abl as well as imatinib mesylate–resistant mutants Y253F, E255K, T315I, M351T, and H396P were purified by glutathione-Sepharose chromatography and used in biochemical autophosphorylation assays as previously described.23 All of the Abl kinase fusion proteins except GST-T315I exhibited concentration-dependent inhibition in response to AP23464 (Figure 3). Treatment of GST-T315I with higher AP23464 concentrations had minimal inhibitory effect on autophosphorylation activity (IC50 > 5 μM). Among the AP23464-sensitive enzymes, nearly complete inhibition of autophosphorylation was observed at concentrations at or above 80 nM (IC50 range, 31-61 nM) in every case except GST-E255K, in which autophosphorylation was completely blocked at a concentration of 160 nM (Figure 3; IC50 = 110 nM; Table 1).
AP23464 inhibits tyrosine phosphorylation of isolated wild-type and mutated Abl kinase domains except T315I. Autophosphorylation of the Abl kinase domain was analyzed for WT, Y253F, E255K, T315I, M351T, and H396P in the presence of escalating concentrations of AP23464 (0-320 nM) as previously described.23 Representative kinase assay gels from 1 of 2 independent experiments are shown. Abl blots demonstrate equal protein loading.
AP23464 inhibits tyrosine phosphorylation of isolated wild-type and mutated Abl kinase domains except T315I. Autophosphorylation of the Abl kinase domain was analyzed for WT, Y253F, E255K, T315I, M351T, and H396P in the presence of escalating concentrations of AP23464 (0-320 nM) as previously described.23 Representative kinase assay gels from 1 of 2 independent experiments are shown. Abl blots demonstrate equal protein loading.
AP23464 IC50 (nM) values for cellular proliferation, cellular Bcr-Abl tyrosine phosphorylation, GST-Abl kinase autophosphorylation, and GST-Abl kinase peptide substrate phosphorylation
. | AP23464 IC50, nM . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | Ba/F3 cellular assays . | . | Purified GST-Abl kinase assays . | . | |||
. | Cellular proliferation . | Bcr-Abl tyrosine phosphorylation . | Autophosphorylation . | Peptide substrate . | |||
| Vector only | 8275 | — | — | — | |||
| Wild type | 14 | 13 | 61 | 6 | |||
| Q252H | 18 | 12 | ND | ND | |||
| Y253F | 26 | 28 | 54 | 13 | |||
| E255K | 94 | 57 | 110 | 14 | |||
| T3151 | 9050 | > 5000 | > 5000 | > 5000 | |||
| M351T | 24 | 30 | 31 | 8 | |||
| H396P | 8 | 9 | 54 | 11 | |||
. | AP23464 IC50, nM . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | Ba/F3 cellular assays . | . | Purified GST-Abl kinase assays . | . | |||
. | Cellular proliferation . | Bcr-Abl tyrosine phosphorylation . | Autophosphorylation . | Peptide substrate . | |||
| Vector only | 8275 | — | — | — | |||
| Wild type | 14 | 13 | 61 | 6 | |||
| Q252H | 18 | 12 | ND | ND | |||
| Y253F | 26 | 28 | 54 | 13 | |||
| E255K | 94 | 57 | 110 | 14 | |||
| T3151 | 9050 | > 5000 | > 5000 | > 5000 | |||
| M351T | 24 | 30 | 31 | 8 | |||
| H396P | 8 | 9 | 54 | 11 | |||
ND indicates not done; —, not applicable.
AP23464 inhibits peptide substrate tyrosine phosphorylation by GST-WT and GST-Abl kinase domain mutants except GST-T315I
GST-Abl kinase domain fusion proteins were also used for in vitro peptide substrate phosphorylation assays. All GST-Abl kinase fusion proteins except GST-T315I were efficiently inhibited within a narrow IC50 range of 6 to 14 nM (Table 1). The IC50 for GST-T315I was not reached at the highest tested concentration of AP23464 (5 μM). In this assay, unlike the corresponding autophosphorylation assay, the catalytic activity of GST-E255K was inhibited in the same concentration range as the other GST-Abl mutants and GST-WT. Results from preliminary experiments with commercially available recombinant, full-length human Abl (IC50 = 7 nM) and Abl mutant T315I (IC50 > 5 μM) were in line with IC50 values obtained with corresponding GST-Abl fusion proteins.
AP23464 inhibits proliferation of cells expressing WT Bcr-Abl or mutated, imatinib mesylate–resistant Bcr-Abl
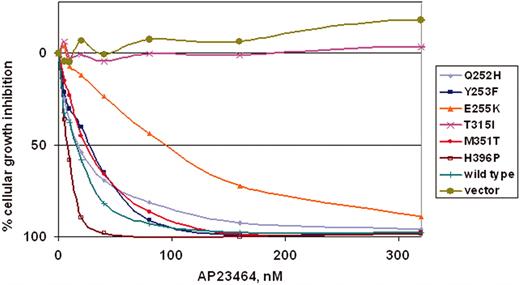
To determine whether the kinase inhibition observed in biochemical assays would translate into antiproliferative effects, a panel of Ba/F3 murine pro–B-cell lines expressing either WT Bcr-Abl p210 or mutated Bcr-Abl p210 was tested. The 6 mutants (Q252H, Y253F, E255K, T315I, M351T, and H396P) represent commonly observed clinical mutations, display a range of residual imatinib mesylate sensitivity (IC50 range, 930-4400 nM)20 and reside in several distinct functional regions within the kinase domain. Cells were cultured for 48 hours in the presence of escalating concentrations of AP23464 (0-320 nM). AP23464 exhibited a concentration-dependent antiproliferative effect on cells expressing WT Bcr-Abl and 5 kinase domain mutants: Q252H, Y253F, E255K, M351T, and H396P (Figure 4). As shown in Table 1, IC50 values for these mutants clustered in the range of 8 to 28 nM except in the case of mutant E255K (94 nM), which was consistently less sensitive to AP23464. In summary, 4 mutants exhibited IC50 values within a 2-fold range of WT Bcr-Abl, whereas E255K required approximately 7-fold higher AP23464 dosage relative to WT. AP23464 had no effect on parental Ba/F3 cells or cells expressing mutant T315I within this concentration range; further experiments with AP23464 (0-10 μM) established IC50 values of approximately 8 and 9 μM for parental cells and T315I cells, respectively (Table 1).
AP23464 inhibits proliferation of Ba/F3 cells expressing WT Bcr-Abl or Bcr-Abl mutants Q252H, Y253F, E255K, M351T, and H396P, but not mutant T315I or parental Ba/F3 cells. Parental Ba/F3 cells supplemented with IL-3 or Ba/F3 cells expressing WT or mutant Bcr-Abl protein were plated in quadruplicate at 5 × 103 cells/well in 96-well plates with AP23464 included in the media (0, 5, 10, 20, 40, 80, 160, or 320 nM). Results from day 2 MTS assays were used to construct best-fit curves and calculate the cellular IC50. The mean based on 4 replicates was calculated in the absence of inhibitor and for each concentration of AP23464. Means ± SE were generated from 3 independent experiments and reported as the percentage absorbance of control. Error bars are omitted for clarity.
AP23464 inhibits proliferation of Ba/F3 cells expressing WT Bcr-Abl or Bcr-Abl mutants Q252H, Y253F, E255K, M351T, and H396P, but not mutant T315I or parental Ba/F3 cells. Parental Ba/F3 cells supplemented with IL-3 or Ba/F3 cells expressing WT or mutant Bcr-Abl protein were plated in quadruplicate at 5 × 103 cells/well in 96-well plates with AP23464 included in the media (0, 5, 10, 20, 40, 80, 160, or 320 nM). Results from day 2 MTS assays were used to construct best-fit curves and calculate the cellular IC50. The mean based on 4 replicates was calculated in the absence of inhibitor and for each concentration of AP23464. Means ± SE were generated from 3 independent experiments and reported as the percentage absorbance of control. Error bars are omitted for clarity.
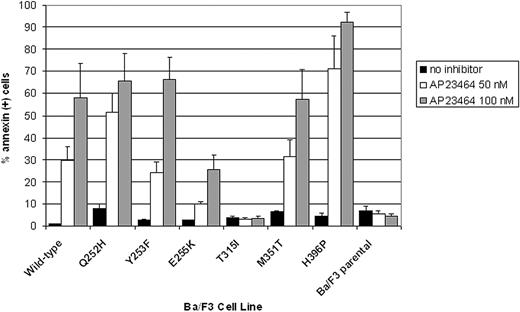
AP23464 induces apoptosis of Ba/F3 cells expressing WT Bcr-Abl or kinase domain mutants except T315I
Ba/F3 cells expressing WT or mutated Bcr-Abl as well as parental Ba/F3 cells were cultured for 48 hours in the presence of AP23464 (0, 50, 100 nM). The percentages of apoptotic cells were then assessed by annexin V staining and flow cytometry. In agreement with cellular proliferation assay results, apoptosis was induced in a concentration-dependent manner in cells expressing WT Bcr-Abl or any of the kinase domain mutants except T315I (Figure 5). At the lower AP23464 concentration (50 nM), cells expressing Bcr-Abl E255K were approximately 3-fold less responsive to the inhibitor than cells expressing WT Bcr-Abl. In contrast, the percentage of annexin-positive Bcr-Abl H396P cells following treatment with AP23464 (50 nM) was consistently at least 2-fold higher than for WT Bcr-Abl cells treated in the same manner. Parental Ba/F3 cells and Ba/F3 cells expressing Bcr-Abl mutant T315I did not undergo apoptosis above control levels in response to AP23464 (highest concentration tested, 800 nM).
Treatment with AP23464 induces apoptosis of Ba/F3 cells expressing WT Bcr-Abl, Q252H, Y253F, E255K, M351T, and H396P, but not T315I or parental Ba/F3 cells. Each Ba/F3 cell line was cultured in the presence of AP23464 (0, 50, or 100 nM) for 48 hours, harvested, stained with annexin V–PE and 7-aminoactinomycin D according to the manufacturer's instructions (Guava Nexin apoptosis kit), and examined by flow cytometry. Data collection and analysis of results were performed with a Guava Technologies PCA instrument equipped with Cytosoft software. Results based on 3 independent experiments are reported as averages ± SE.
Treatment with AP23464 induces apoptosis of Ba/F3 cells expressing WT Bcr-Abl, Q252H, Y253F, E255K, M351T, and H396P, but not T315I or parental Ba/F3 cells. Each Ba/F3 cell line was cultured in the presence of AP23464 (0, 50, or 100 nM) for 48 hours, harvested, stained with annexin V–PE and 7-aminoactinomycin D according to the manufacturer's instructions (Guava Nexin apoptosis kit), and examined by flow cytometry. Data collection and analysis of results were performed with a Guava Technologies PCA instrument equipped with Cytosoft software. Results based on 3 independent experiments are reported as averages ± SE.
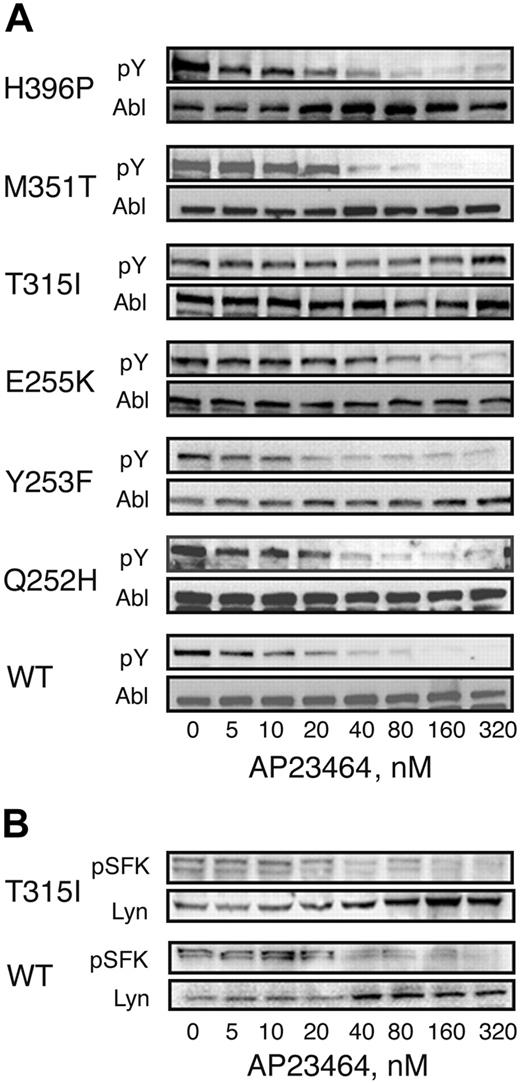
AP23464 inhibits Bcr-Abl tyrosine phosphorylation in Ba/F3 cells expressing WT Bcr-Abl or kinase domain mutants except T315I
Ba/F3 cells expressing WT or mutated Bcr-Abl as well as parental Ba/F3 cells were cultured for 3 hours in the presence of AP23464 (0-320 nM). Whole-cell lysates were separated by SDS-PAGE and subjected to immunoblot analysis by using the antiphosphotyrosine monoclonal antibody 4G10. Total Bcr-Abl levels were assessed by immunoblot analysis using Abl monoclonal antibody 8E9. Treatment with AP23464 significantly reduced Bcr-Abl tyrosine phosphorylation levels for all mutants except T315I (Figure 6A). The IC50 was below 30 nM for Bcr-Abl and all mutants except E255K (IC50, 57 nM) and T315I (IC50 > 5000 nM; Table 1). As expected, no Bcr-Abl signal was detected in lysates from untransfected, parental Ba/F3 cells (data not shown). To examine the effects of AP23464 (0-320 nM) on Src family kinase signaling, lysates were subjected to immunoblot analysis by using the polyclonal antiphospho-Src family antibody CST2101 (Cell Signaling Technology). Total Lyn levels were assessed by immunoblot analysis anti-Lyn antibody sc-15 (Santa Cruz Biotechnology). Treatment with AP23464 significantly reduced Src family kinase tyrosine phosphorylation levels in Ba/F3 cells expressing either wild-type Bcr-Abl or Bcr-Abl mutant T315I (Figure 6B).
Cellular Bcr-Abl tyrosine phosphorylation is reduced in the presence of AP23464 except in the case of T315I. Parental Ba/F3 cells or Ba/F3 cells expressing WT or mutant Bcr-Abl (4 × 106 cells/well) were incubated in escalating concentrations of AP23464 (0, 5, 10, 20, 40, 80, 160, or 320 nM) for 3 hours at 37° C. Clarified whole-cell lysates were subjected to SDS-PAGE followed by Western blot analysis. (A) Mouse monoclonal antiphosphotryrosine antibody 4G10 (Upstate Biotechnology) was used to determine levels of tyrosine-phosphorylated Bcr-Abl. Total Bcr-Abl expression levels were determined with mouse monoclonal anti-Abl antibody 8E9 (BD Biosciences Pharmingen) or Ab-2 (Oncogene Science). (B) Rabbit polyclonal antiphospho-Src family antibody (Cell Signaling Technology) was used to determine levels of tyrosine-phosphorylated Src family kinase members. As a loading control, total Lyn expression levels were determined with rabbit polyclonal anti-Lyn antibody sc-15 (Santa Cruz Biotechnology).
Cellular Bcr-Abl tyrosine phosphorylation is reduced in the presence of AP23464 except in the case of T315I. Parental Ba/F3 cells or Ba/F3 cells expressing WT or mutant Bcr-Abl (4 × 106 cells/well) were incubated in escalating concentrations of AP23464 (0, 5, 10, 20, 40, 80, 160, or 320 nM) for 3 hours at 37° C. Clarified whole-cell lysates were subjected to SDS-PAGE followed by Western blot analysis. (A) Mouse monoclonal antiphosphotryrosine antibody 4G10 (Upstate Biotechnology) was used to determine levels of tyrosine-phosphorylated Bcr-Abl. Total Bcr-Abl expression levels were determined with mouse monoclonal anti-Abl antibody 8E9 (BD Biosciences Pharmingen) or Ab-2 (Oncogene Science). (B) Rabbit polyclonal antiphospho-Src family antibody (Cell Signaling Technology) was used to determine levels of tyrosine-phosphorylated Src family kinase members. As a loading control, total Lyn expression levels were determined with rabbit polyclonal anti-Lyn antibody sc-15 (Santa Cruz Biotechnology).
Discussion
Imatinib mesylate, a specific inhibitor of the Abl tyrosine kinase, is an effective drug for the treatment of CML; however, resistance has been observed. Because the prominent mechanism of imatinib mesylate resistance involves reactivation of the Bcr-Abl kinase, a strategy for re-establishing response in these patients is the use of alternative Abl inhibitors.
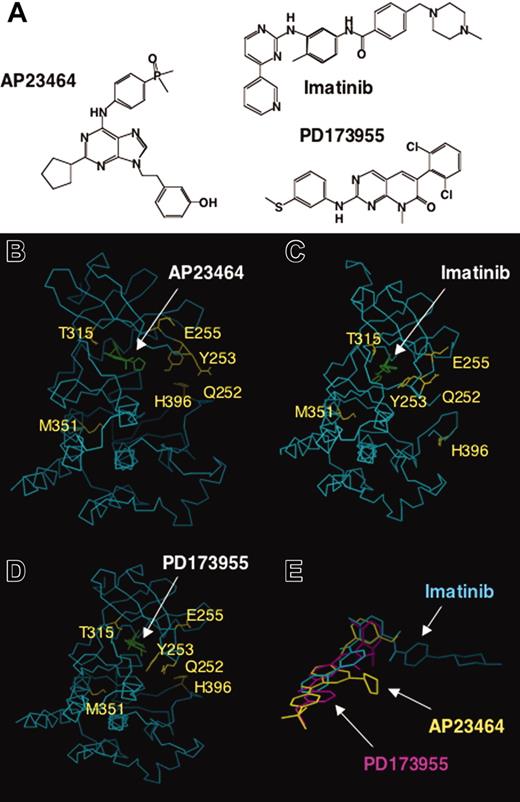
We report that AP23464, a novel 2,6,9-trisubstituted purine analog (Figure 7A), potently blocks proliferation and cell cycle progression and induces apoptosis of Bcr-Abl–positive K562 cells but not Bcr-Abl–negative HL60 cells. Tyrosine phosphorylation of Bcr-Abl as well as downstream Bcr-Abl targets, STAT5 and CrkL, was reduced on treatment of K562 cells with AP23464. To examine whether the effects observed with K562 cells were directly attributable to inhibition of Abl kinase activity, these studies were followed by enzymatic assays with isolated Abl kinase domains. We found that AP23464 blocked both autophosphorylation and peptide substrate phosphorylation by isolated WT Abl kinase and all imatinib mesylate–resistant Abl kinase domains tested with the exception of T315I. Preliminary studies indicate that both c-Kit and PDGFR are also inhibited by AP23464, but with lower potency than Src and Abl.
Comparative chemical structures and 3-dimensional (3-D) molecular models of ATP-based inhibitor AP23464 and the previously described inhibitors imatinib mesylate and PD173955 complexed with Abl kinase. (A) Chemical structures of AP23464, imatinib mesylate, and PD173955. (B) Three-dimensional molecular model of AP23464 complexed with Abl kinase in its ATP-binding active conformation. Residues that result in significantly decreased imatinib mesylate inhibitory potency if mutated are highlighted. (C) X-ray structure of imatinib mesylate complexed with Abl kinase in an induced fit conformation as previously described.21 (D) X-ray structure of PD173955 complexed with Abl kinase in an induced fit conformation as previously described.21 (E) Three-dimensional molecular model overlay of AP23464, imatinib mesylate, and PD173955 in their Abl kinase bound conformations, illustrating differences in their use of 3-D chemical space arising from varying structures and modes of binding. See “Discussion” for further description of the inhibitors, molecular modeling, and correlation with Abl kinase mutant structure-activity studies.
Comparative chemical structures and 3-dimensional (3-D) molecular models of ATP-based inhibitor AP23464 and the previously described inhibitors imatinib mesylate and PD173955 complexed with Abl kinase. (A) Chemical structures of AP23464, imatinib mesylate, and PD173955. (B) Three-dimensional molecular model of AP23464 complexed with Abl kinase in its ATP-binding active conformation. Residues that result in significantly decreased imatinib mesylate inhibitory potency if mutated are highlighted. (C) X-ray structure of imatinib mesylate complexed with Abl kinase in an induced fit conformation as previously described.21 (D) X-ray structure of PD173955 complexed with Abl kinase in an induced fit conformation as previously described.21 (E) Three-dimensional molecular model overlay of AP23464, imatinib mesylate, and PD173955 in their Abl kinase bound conformations, illustrating differences in their use of 3-D chemical space arising from varying structures and modes of binding. See “Discussion” for further description of the inhibitors, molecular modeling, and correlation with Abl kinase mutant structure-activity studies.
To address the effectiveness of AP23464 against imatinib mesylate–resistant Bcr-Abl variants in a cellular setting, Ba/F3 cells expressing WT Bcr-Abl or 1 of 6 clinically relevant Bcr-Abl kinase domain mutants were tested. The panel included mutants from distinct functional locations within the Bcr-Abl kinase that confer a range of residual imatinib mesylate sensitivity. AP23464 inhibited proliferation of Ba/F3 cells expressing WT Bcr-Abl (IC50 = 14 nM) with approximately 25-fold higher potency than imatinib mesylate (IC50 = 350 nM). In addition, AP23464 displayed excellent in vitro effectiveness against all tested Bcr-Abl kinase domain mutants except T315I. Specifically, AP23464 IC50 values were in the range of 8 to 26 nM for Ba/F3 cells expressing mutants Q252H, Y253F, M351T, and H396P (Figure 4; Table 1) and slightly higher for E255K (IC50 = 94 nM). In comparison, the reported IC50 range for treatment of these mutants with imatinib mesylate is 930 to 4400 nM.20 Proliferation of parental Ba/F3 cells (IC50 > 8 μM) and Ba/F3 cells expressing Bcr-Abl T315I (IC50 > 9 μM) was not significantly inhibited by AP23464.
As summarized earlier, we used both cellular and biochemical assays to test the specificity and potency of AP23464. The results of the 2 cellular assays were in excellent agreement (Table 1). Taken together, the similar AP23464 IC50 values for inhibiting cellular proliferation (IC50 range, 8-94 nM; T315I > 9 μM), cellular Bcr-Abl tyrosine phosphorylation (IC50 range, 9-57 nM; T315I > 5 μM), GST-Abl autophosphorylation (IC50 range, 31-110 nM; T315I > 5 μM), and GST-Abl kinase peptide substrate phosphorylation (IC50 range, 6-14 nM; T315I > 5 μM) suggest that cell killing is tightly linked to blockade of Bcr-Abl kinase activity and that the inhibitor enters cells efficiently.
Src-family kinases and Abl kinase share considerable homology, and several Src inhibitors of varying chemical classes have been found to be effective against the Abl kinase.35-38 We20 and others38-40 have investigated pyrido-[2,3-d]pyrimidine-type small molecule kinase inhibitors against cells expressing Bcr-Abl with clinically relevant kinase domain mutations. In our studies, PD180970 displayed potent inhibitory activity against WT Bcr-Abl (IC50 = 25 nM; imatinib mesylate IC50 = 300 nM), activation loop mutant H396P (IC50 = 15 nM), and mutant M351T (IC50 = 45 nM).20 Inhibition of nucleotide binding loop mutants Q252H, Y253F, and E255K by PD180970 was weaker, spanning an IC50 range of 120 to 140 nM. PD180970 was not effective against Bcr-Abl mutant T315I. Recent studies have identified pyrido-[2,3-d]pyrimidine analogues PD166326 and SKI DV-MO16 as more potent Bcr-Abl inhibitors compared with PD180970.40 These inhibitors are also capable of inhibiting several of the Bcr-Abl kinase domain mutants, except T315I. Major drawbacks of the pyrido-[2,3-d]pyrimidine family of inhibitors include limited solubility and a poor pharmacokinetic profile.
One particularly attractive feature of AP23464 and similar compounds is the potential for dual inhibition of Src and Abl. Bcr-Abl activates Src family kinases, which are in turn able to induce phosphorylation of Bcr-Abl and promote its interaction with Grb2.41-43 Also, up-regulation of the Src-related kinase Lyn has been observed in imatinib mesylate–resistant K562 cells.44,45 Several compounds designed as Src kinase inhibitors are also effective against Bcr-Abl. SKI-606, a 4-anilino-3-quinolinecarbonitrile, was highly effective against K562 cells in vitro and caused K562 xenograft reduction in nude mice.36
Warmuth et al37 examined the effects of the Src/Abl inhibitors PP1 and CGP76030 on growth and survival of cells expressing WT- or imatinib mesylate–resistant Bcr-Abl kinase. Both inhibitors blocked cellular WT Bcr-Abl tyrosine phosphorylation, albeit only at very high concentrations (PP1, 25 μM; CGP76030, 5 μM). Both inhibitors failed to suppress Bcr-Abl tryosine phosphorylation in cells expressing Bcr-Abl kinase domain mutant T315I, yet still significantly reduced cell growth and survival. The researchers suggest that the observed efficacy against cells expressing Bcr-Abl mutant T315I may be attributable to inhibition of Src family tryosine kinases and/or Akt kinase.37 In the present study, treatment of cells expressing Bcr-Abl mutant T315I with the highly potent Src and Abl kinase inhibitor AP23464 efficiently inhibited Src family kinase signaling (Figure 6B) but not Bcr-Abl signaling (Figure 6A). Importantly, inhibition of Src family kinase signaling had no discernible effect on proliferation of cells expressing Bcr-Abl mutant T315I, consistent with our assertion that blocking this pathway alone is not sufficient to kill T315I cells. Thus, inhibition of kinase(s) other than, or in addition to, Src family kinases may account for the previously reported activity of Src inhibitors PP1 and CGP76030 against Bcr-Abl/T315I cells.
Crystallographic studies comparing the Abl kinase domain in complex with imatinib mesylate or PD173955 (a close analog of PD180970) have provided a structural basis for the specificity of imatinib mesylate for Abl and for assessing the role of individual mutations in conferring imatinib mesylate resistance.21,22 The activation loop of imatinib mesylate can assume a closed/inactive conformation or an open/active conformation. Imatinib mesylate binds to the inactive form of Abl and induces massive shifts in the conformation of the highly flexible nucleotide binding loop (P-loop). Thus, despite the high degree of homology between Src and Abl, the unique tertiary structure of the inactive conformation of Abl accounts for the inability of imatinib mesylate to inhibit Src family kinases. Mutations Q252H, Y253F, and E255K, located within the nucleotide binding loop (P-loop), probably prevent this region from undergoing the conformation shifts required for imatinib mesylate binding. In addition, tyrosine 253 makes van der Waals contact with imatinib mesylate. Specifically, Thr-315 forms a direct hydrogen bond and makes van der Vaals contact with imatinib mesylate. The T315I mutation eliminates a crucial hydrogen bond and places a steric blockade within the imatinib mesylate binding site. M351T is located in the C-terminal loop, near the base of the activation loop and distant from the imatinib mesylate binding site. Met-351 is also a site of interaction with the SH2 domain. The M351T mutation may impair imatinib mesylate binding by restricting flexibility of the activation loop and/or by interrupting contacts with the SH2 domain that favor the inactive conformation of Bcr-Abl.46 H396P, a mutation within the activation loop, probably destabilizes the inactive, imatinib mesylate–accessible conformation of Bcr-Abl.
A 3-D molecular model of AP23464 complexed to Abl kinase in its ATP-binding, active conformation was constructed (Figure 7B) and compared with previously described X-ray crystallographic structures of Abl kinase complexed to imatinib mesylate (Figure 7C), and PD173955 (Figure 7D). The chemical structures of these 3 inhibitors (Figure 7A) vary significantly as do their comparative modes of binding. AP23464 is predicted to bind to the Abl kinase domain in a distinct, but partially overlapping manner, compared with imatinib mesylate and PD173955 (compare Figure 7B-D). The 3-hydroxyphenethyl group of AP23464 extends into a large hydrophobic specificity pocket that is also important for imatinib mesylate and PD173955 binding. The side chain of Abl kinase residue Thr-315 modulates access to this binding site, and the bulkier Ile-315 effectively blocks access to AP23464 as well as imatinib mesylate and PD173955. Consequently, all 3 inhibitors are unable to effectively bind Bcr-Abl mutant T315I. Unlike imatinib mesylate, AP23464 is predicted to bind only to the ATP-binding active conformation of Abl. Hence, AP23464 binding is not predicted to require an induced-fit mechanism involving extensive conformational changes within the P-loop. Binding of either imatinib mesylate or PD173955 to Abl kinase requires induced-fit conformational changes within the P-loop (residues Leu-248 to Tyr-257); however, discrete differences exist between each of the 2 complexes. Overall, comparison of the predicted 3-D molecular modeling of AP23464 and X-ray structures of both imatinib mesylate and PD173955 provides a basis to correlate with Abl kinase mutant inhibition results. In this study, AP23464 was found to be capable of binding Bcr-Abl P-loop and activation loop mutants as similarly described for pyrido-[2,3d]pyrimidines PD173955 (Figure 7D) and PD166326.21,22,47 Finally, a 3-D overlay of AP23464, imatinib mesylate, and PD17355 (Figure 7E) illustrates the varying chemical space these inhibitors can use to bind Abl kinase and provides further insight into their differential inhibitory properties against the mutants investigated in this study.
Single agent imatinib mesylate therapy induces complete cytogenetic remission with much greater frequency than any approach except allogeneic stem cell transplantation. However, a recent study shows that almost all patients have residual Bcr-Abl positivity when examined by reverse transcriptase–polymerase chain reaction (RT-PCR).48 This suggests that imatinib mesylate monotherapy may be incapable of eradicating the disease. One possibility is that cells harboring imatinib mesylate–refractory Bcr-Abl mutations persist at extremely low frequencies but gain a selective advantage on prolonged treatment with imatinib mesylate. AP23464 and other inhibitors that might be capable of eliminating these rare clones could prove to be useful in combination with imatinib mesylate. These data obtained in this study support the development and clinical evaluation of AP23464 and related compounds, either alone or in combination with imatinib mesylate.
Prepublished online as Blood First Edition Paper, July 15, 2004; DOI 10.1182/blood-2004-05-1851.
Supported by grants from the National Cancer Institute, the Leukemia and Lymphoma Society, the Doris Duke Charitable Foundation, and the Howard Hughes Medical Institute (B.J.D.).
Several of the authors (R.P., J.A.K., V.M.R., H.T., C.A.M., R.S.B., Y.W., R.S., W.C.S., D.D., T.C., and T.K.S.) are employed by ARIAD Pharmaceuticals Inc, whose potential product was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We dedicate this paper to Professor Heinz G. Floss on the occasion of his 70th birthday.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal