Abstract
von Willebrand disease (VWD) type 1 is difficult to diagnose because of bleeding variability and low heritability of von Willebrand factor (VWF) levels. We compared a bleeding severity score and bleeding times to candidate gene haplotypes within pedigrees of 14 index cases, using a covariance components model for multivariate traits (Mendel: QTL Association). These pedigrees included 13 affected and 40 unaffected relatives, as defined by plasma ristocetin cofactor (VWF:RCo) levels. The bleeding severity score was derived from a detailed history. Donors were genotyped using a primer extension method, and 9 candidate genes were selected for analysis. VWF:RCo levels had the strongest influence on bleeding severity score and bleeding time. ITGA2 haplotype 2 (807C) and ITGA2B haplotype 1 (Ile843) were each associated with increased bleeding severity scores (P < .01 and P < .01, respectively). GP6 haplotype b (Pro219) was also associated with increased scores (P = .03) after adjustment for donor age. No association was observed with 6 other candidate genes, GP1BA, ITGB3, VWF, FGB, IL6, or TXA2R. Increased plasma VWF:Ag levels were associated with VWF haplotype 1 (–1793G; P = .02). These results establish that genetic differences in the adhesion receptor subunits α2, αIIb, and GPVI can influence the phenotype of VWD type 1.
Introduction
In arterial flow, the initial transient arrest of platelets on collagen requires von Willebrand factor (VWF) acting as a molecular bridge between collagen and the glycoprotein Ib (GPIb) complex.1 Next, the direct binding of platelet α2β1 and platelet GPVI to collagen anchors the platelet more firmly to the matrix and facilitates signal transduction that leads to an activated, procoagulant platelet monolayer.2 Genetic variation in any of the responsible receptors might have an impact on platelet function in vivo, particularly in mild forms of von Willebrand disease (VWD) type 1, which accounts for at least 60% of all cases of VWD.3
Three criteria must be satisfied to make the correct diagnosis. First, the bleeding history must be mainly mucocutaneous; second, the inheritance is usually autosomal dominant; and third, laboratory findings must demonstrate reduced levels of otherwise functionally normal VWF, evident by concomitant reduction of VWF ristocetin cofactor activity (VWF:RCo) and VWF antigen (VWF:Ag) with a normal VWF multimeric pattern.3 A positive bleeding history since childhood with symptoms observed in at least 2 different sites is considered the most important criterion. The true incidence is difficult to determine with accuracy because the diagnosis is not always straightforward, especially in patients with very mild defects and no apparent family history. Genetic follow-up will often identify mutations that inhibit the synthesis/secretion or enhance the clearance of VWF.4 VWF multimer assays will typically demonstrate decreased levels of all multimer sizes. Quite often, individuals with very similar or identical VWF levels will exhibit only modest, if any, bleeding tendency. These exceptions to the paradigm have confounded both the diagnosis and prognostic predictability in VWD type 1.
We selected haplotypes of 5 platelet glycoprotein genes that have already been implicated in risk for thrombosis or bleeding or both. These are (Table 1): GPIbα (GP1BA)5,6 , GPVI (GP6),7 integrin α2 (ITGA2),8 integrin αIIb (ITGA2B),9 and integrin β3 (ITGB3).10
Candidate glycoprotein genes
Gene (GenBank accession no.) and haplotype . | Other designation . | SNP . |
|---|---|---|
| ITGA2 (NT_023081) | ||
| 1 | 807T; Bgl II (+); Ase I (neg) | 2915969A; 2916690G |
| 2 | 807C; Bgl II (neg); Ase I (+) | 2915969G; 2916690A |
| 3 | 807C; Bgl II (neg); Ase I (neg) | 2915969G; 2916690G |
| - 92C | None | 2850525C |
| - 92G | None | 2850525G |
| - 52C | None | 2850565C |
| - 52T | None | 2850565T |
| GP6 (NT_011225) | ||
| a | Ser219 | 13039T |
| b | Pro219 | 13039C |
| GPIBA (NT_010823) | ||
| 1 | HPA-2a; Thr145 | 757C |
| 2 | HPA-2b; Met145 | 757T |
| - 5T | None | 271T |
| - 5C | None | 271C |
| ITGB3 (NT_010833) | ||
| 1 | HPA-1a; P1A1; Leu33 | 29519T |
| 2 | HPA-1b; P1A2; Pro33 | 29519C |
| ITGA2B (M33320) | ||
| 1 | HPA-3a; Baka; Ile843 | 10893T |
| 2 | HPA-3b; Bakb; Ser843 | 10893G |
| VWF (NT_009731) | ||
| 1 | - 1793G | - 1793G |
| 2 | - 1793C | - 1793C |
| IL6 (AF372214) | ||
| 1 | - 174G | 1510G |
| 2 | - 174C | 1510C |
| FGB (NM_005141) | ||
| 1 | - 455G | - 455G |
| 2 | - 455A | - 455A |
| TXA2R (NM_001060) | ||
| 1 | 924C | 1915C |
| 2 | 924T | 1915T |
Gene (GenBank accession no.) and haplotype . | Other designation . | SNP . |
|---|---|---|
| ITGA2 (NT_023081) | ||
| 1 | 807T; Bgl II (+); Ase I (neg) | 2915969A; 2916690G |
| 2 | 807C; Bgl II (neg); Ase I (+) | 2915969G; 2916690A |
| 3 | 807C; Bgl II (neg); Ase I (neg) | 2915969G; 2916690G |
| - 92C | None | 2850525C |
| - 92G | None | 2850525G |
| - 52C | None | 2850565C |
| - 52T | None | 2850565T |
| GP6 (NT_011225) | ||
| a | Ser219 | 13039T |
| b | Pro219 | 13039C |
| GPIBA (NT_010823) | ||
| 1 | HPA-2a; Thr145 | 757C |
| 2 | HPA-2b; Met145 | 757T |
| - 5T | None | 271T |
| - 5C | None | 271C |
| ITGB3 (NT_010833) | ||
| 1 | HPA-1a; P1A1; Leu33 | 29519T |
| 2 | HPA-1b; P1A2; Pro33 | 29519C |
| ITGA2B (M33320) | ||
| 1 | HPA-3a; Baka; Ile843 | 10893T |
| 2 | HPA-3b; Bakb; Ser843 | 10893G |
| VWF (NT_009731) | ||
| 1 | - 1793G | - 1793G |
| 2 | - 1793C | - 1793C |
| IL6 (AF372214) | ||
| 1 | - 174G | 1510G |
| 2 | - 174C | 1510C |
| FGB (NM_005141) | ||
| 1 | - 455G | - 455G |
| 2 | - 455A | - 455A |
| TXA2R (NM_001060) | ||
| 1 | 924C | 1915C |
| 2 | 924T | 1915T |
Quantitative differences in platelet α2β1 have been correlated with inheritance of 3 major ITGA2 haplotypes. Haplotype 1 (807T/1648G) is associated with highest levels of α2β1; haplotype 3 (807C/1648A), with intermediate to high levels; and haplotype 2 (807C/1648G), with lowest levels.8 A correlation has been found between haplotype 1 (807T; high receptor density) and risk for arterial thrombosis in younger men with a history of myocardial infarction,11,12 women who are heavy smokers,13 patients with diabetic retinopathy,14 and younger patients with stroke.15 These 3 haplotypes can be defined by 2 single nucleotide polymorphisms (SNPs) within intron 7 depicted in Table 1 (Bgl II and Ase I sites).
Two major GP6 haplotypes have been defined. Haplotype a (T13254) encodes a serine at residue 219, whereas haplotype b (C13254) encodes a proline.7 A recent report by Joutsi-Korhonen et al16 presents biologic evidence that platelets homozygous for haplotype b exhibit a decreased tendency toward thrombosis on a collagen surface in flowing whole blood or in the Platelet Function Analyzer-100 (PFA-100; Dade-Behring International, Düdingen, Switzerland). On the other hand, homozygosity for haplotype b was associated with risk for myocardial infarction, particularly among older women (≥ 60 years old) who were smokers and carried the fibrinogen Bβ–455A haplotype (higher fibrinogen level).7
Polymorphism of the αIIb gene is responsible for the expression of the Bak (HPA-3) alloantigen system. The allele Ile843 confers the Baka (HPA-3a) alloantigen determinant (frequency 0.616), whereas Ser843 represents the Bakb (HPA-3b) epitope. An initial indication that HPA-3 is related to mortality after stroke17 was followed by larger studies that did not find a risk association in either stroke18 or coronary artery disease.19 However, Reiner et al9 found that homozygosity for the Ser843 allele was associated with an approximately 5-fold increased risk of ischemic stroke among subgroups of women who carried a diagnosis of hypertension or diabetes (OR = 4.51; 95% CI, 1.01-20.13) or had elevated plasma homocysteine levels (OR = 5.94; 95% CI, 1.53-23.05).
We included haplotypes of 4 other genes that directly or indirectly influence thrombotic potential. These include 2 major haplotypes of the promoter regions of the VWF gene itself (VWF),20 the fibrinogen-B gene (FBG),21 and the interleukin 6 gene (IL6),22 and 2 major haplotypes of the thromboxane A2 receptor (TXA2R).23
Haplotype differences in each of these 9 genes can potentially influence the efficiency of hemostasis, and our objective is to analyze the association of these haplotypes with risk for bleeding in VWD type 1, through an analysis of index cases and their family members.
Patients, materials, and methods
Criteria for enrollment of VWD patients and healthy individuals
This study was organized in 14 different families from the Milan area, part of the 154 European families already enrolled in the study entitled “Molecular and Clinical Markers for Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD)” under the Fifth Framework Program of the European Community (Coordinators: Prof I. R. Peake and F. Rodeghiero). The criteria used in this study were the same as those applied to the entire population of European VWD. Patients with VWD type 1 were classified according to the previous definitions of the Scientific Standardization Committee (SSC) on VWF of the International Society on Thrombosis and Haemostasis (ISTH).24 By these criteria, all index cases previously diagnosed as VWD type 1 in 12 different European centers were eligible for enrollment if their family showed at least 2 affected individuals and additional nonaffected members.
This portion of the project was approved by the local Institutional Review Board of the University Hospital of Milan. After individuals signed an informed consent, 14 index cases of VWD type 1 and 18 affected and 38 unaffected relatives were enrolled between April and October 2002 on the basis of a previous diagnosis of VWD type 1 according to the records of the Angelo Bianchi Bonomi Hemophilia Thrombosis Center of Milan.
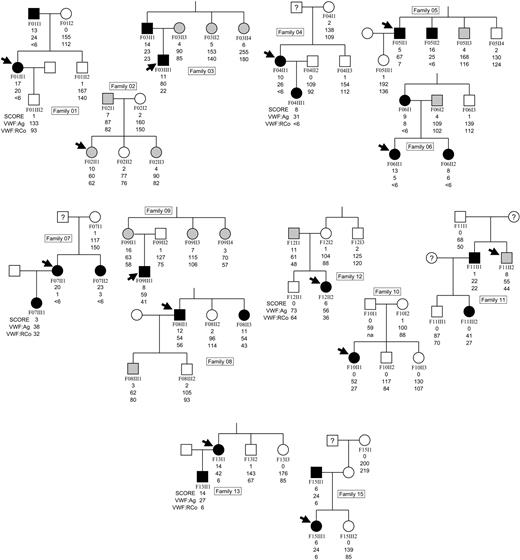
According to the enrollment criteria of the European project, all DNA and plasma samples derived from VWD type 1 families and controls were indicated by progressive letter-numerical codes as follows: P03 (partner 3, Milan), F00 (family number from 1-14), I-IV (generation levels in Roman numbers, from I-IV), and 00 (individuals in numbers, from 1-10). These codes are reported in the pedigree charts (Figure 1).
Pedigrees for 14 families affected by VWD. Black symbols indicate affected individuals (VWF: RCo ≤ 45 U/dL, if blood group non-O; ≤ 35 U/dL, if blood group O); gray symbols, individuals with bleeding severity scores ≥ 3; and white symbols, otherwise healthy individuals. Arrows indicate index cases within each pedigree.
Pedigrees for 14 families affected by VWD. Black symbols indicate affected individuals (VWF: RCo ≤ 45 U/dL, if blood group non-O; ≤ 35 U/dL, if blood group O); gray symbols, individuals with bleeding severity scores ≥ 3; and white symbols, otherwise healthy individuals. Arrows indicate index cases within each pedigree.
In this blinded study, the subjects were recruited, interviewed, assigned a bleeding severity score, and tested for all laboratory and clinical parameters in Milan. DNA samples were then coded without knowledge of these data or the identification of the family members and their relationships, and the genotyping and statistical analysis were performed at The Scripps Research Institute (TSRI). Normal unrelated donor pools consisted of age- and gender-matched individuals from either the Milan area (Milan controls, n = 52) or white, non-Hispanic donors recruited through the General Clinical Research Center (GCRC) of Scripps Clinic and Green Hospital, La Jolla (white non-Hispanic controls, n = 145).
Standardized criteria to evaluate bleeding history
A bleeding history was derived from detailed questionnaires, and a score was compiled, as described.4 This questionnaire was administered to all the affected and nonaffected members (including index cases) of the different families from the same well-trained hematologist who was attending the outpatient care unit at the Angelo Bianchi Bonomi Hemophilia Thrombosis Center. To avoid bias during the interview, the hematologist was not aware at that moment which patient would be considered the index case. The severity of bleeding episodes was ranked from 0 to 3, as shown in Table 2, in each of 11 bleeding manifestation categories: epistaxis, cutaneous symptomatic bleeding, bleeding from minor wounds, oral cavity bleeding, gastrointestinal bleeding, bleeding associated with tooth extraction, surgery, postpartum hematoma, muscle hematoma, hemarthrosis, and menorrhagia. The total bleeding score is the numerical sum of the scores for each category and was calculated in the laboratory area independently by another hematologist.
The bleeding severity scale in patients with VWD
. | Severity, by symptoms and therapy . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Bleeding category . | 0 . | 1 . | 2 . | 3 . | |||
| Epistaxis | Absent | < 10 episodes/y | 10 episodes/y | Desmopressin or FVIII/VWF conc | |||
| < 10 min/episode | < 10 min/episode | ||||||
| No therapy | No therapy or local therapy | ||||||
| Cutaneous bleeding | Absent | < 10 episodes/y | > 10 episodes/y | Desmopressin or FVIII/VWF conc | |||
| No therapy | No therapy or local therapy | ||||||
| Bleeding from minor wounds | Absent | < 10 episodes/y | > 10 episodes/y | Desmopressin or FVIII/VWF conc | |||
| < 5 min/episode | > 5 min/episode | ||||||
| No therapy | No therapy or local therapy | ||||||
| Oral cavity bleeding | Absent | After minimal trauma only | Spontaneous | Desmopressin or FVIII/VWF conc | |||
| No therapy | No therapy or local therapy | ||||||
| Gastrointestinal bleeding | Absent | 1 bleeding episode | > 1 bleeding episode | Desmopressin or FVIII/VWF conc | |||
| No therapy | No therapy or local therapy | ||||||
| Bleeding after tooth extraction | Absent | Sometimes | Always | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | Local therapy | ||||||
| Bleeding after surgery | Absent | Minor bleeding | Major bleeding | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | Local therapy | ||||||
| Postpartum hemorrhage | Absent | Minor bleeding | Major bleeding | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | Local therapy | ||||||
| Muscle hematomas | Absent | After major trauma | After minor trauma | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | No therapy or local therapy | ||||||
| Hemarthrosis | Absent | After major trauma | After minor trauma | Desmopressin or FVIII/VWF conc | |||
| No therapy | Local therapy | ||||||
| Menorrhagia | Absent | No therapy or local therapy | Birth control pills | Desmopressin or FVIII/VWF conc | |||
. | Severity, by symptoms and therapy . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Bleeding category . | 0 . | 1 . | 2 . | 3 . | |||
| Epistaxis | Absent | < 10 episodes/y | 10 episodes/y | Desmopressin or FVIII/VWF conc | |||
| < 10 min/episode | < 10 min/episode | ||||||
| No therapy | No therapy or local therapy | ||||||
| Cutaneous bleeding | Absent | < 10 episodes/y | > 10 episodes/y | Desmopressin or FVIII/VWF conc | |||
| No therapy | No therapy or local therapy | ||||||
| Bleeding from minor wounds | Absent | < 10 episodes/y | > 10 episodes/y | Desmopressin or FVIII/VWF conc | |||
| < 5 min/episode | > 5 min/episode | ||||||
| No therapy | No therapy or local therapy | ||||||
| Oral cavity bleeding | Absent | After minimal trauma only | Spontaneous | Desmopressin or FVIII/VWF conc | |||
| No therapy | No therapy or local therapy | ||||||
| Gastrointestinal bleeding | Absent | 1 bleeding episode | > 1 bleeding episode | Desmopressin or FVIII/VWF conc | |||
| No therapy | No therapy or local therapy | ||||||
| Bleeding after tooth extraction | Absent | Sometimes | Always | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | Local therapy | ||||||
| Bleeding after surgery | Absent | Minor bleeding | Major bleeding | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | Local therapy | ||||||
| Postpartum hemorrhage | Absent | Minor bleeding | Major bleeding | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | Local therapy | ||||||
| Muscle hematomas | Absent | After major trauma | After minor trauma | Desmopressin or FVIII/VWF conc | |||
| No therapy or local therapy | No therapy or local therapy | ||||||
| Hemarthrosis | Absent | After major trauma | After minor trauma | Desmopressin or FVIII/VWF conc | |||
| No therapy | Local therapy | ||||||
| Menorrhagia | Absent | No therapy or local therapy | Birth control pills | Desmopressin or FVIII/VWF conc | |||
Conc indicates concentrate.
Laboratory assays
Blood samples were drawn and processed as previously described.25 Venous blood for hemostasis tests was drawn in 0.125 mM citrate. For VWF multimeric analysis, blood was drawn in 6 mM EDTA (ethylenediaminetetraacetic acid). Blood samples were centrifuged at 3000g for 20 minutes to obtain platelet-poor plasma. For multimeric analysis, plasma was transferred to another tube and centrifuged at 40 000g for 20 minutes to remove residual platelets. All plasma samples were frozen in ethanol-dry ice and stored at –70° C until tested.
The bleeding time (BT) was measured by the Simplate II device (General Diagnostics, Morris Plane, NJ). The PFA-100 was not available in all participating European centers and was therefore not considered a substitutive assay for primary hemostasis in VWD. VWF antigen (VWF: Ag) was measured by enzyme-linked immunosorbent assay (ELISA) and ristocetin cofactor activity (VWF:RCo) was measured by aggregometry of formalin-fixed platelets, as described.25 After centrifugation at 3500g at 4° C for 10 minutes, supernatants were frozen at –80° C for VWF:Ag and VWF:RCo assays. Multimeric analysis was performed on low-resolution agarose gels. All FVIII/VWF measurements were expressed in international units (IU), with reference to a plasma pool standardized against the International Reference Preparation for FVIII/VWF-related activities.
Pedigrees of the families
The 14 family pedigrees are shown in Figure 1, with individuals indicated by a coded number (see “Criteria for enrollment of VWD patients and healthy individuals”). Index cases are noted by a bold horizontal arrow, and below each symbol, the values for (top to bottom) bleeding severity score, VWF antigen (VWF:Ag), and ristocetin cofactor activity (VWF:RCo) are indicated. Affected individuals are designated by black symbols and defined as those with a VWF:RCo level equal to or less than 45 U/dL, if blood group non-O, or equal to or less than 35 U/dL, if blood group O. Among these affected individuals, bleeding severity scores ranged from 3 to 23. Family members with bleeding severity scores greater than or equal to 3 but VWF:RCo levels above the cutoff level are indicated by gray symbols. Individuals with bleeding severity scores less than or equal to 2 (ie, below the affected donor range) and VWF:RCo levels above the cutoff level are considered normal and indicated by white symbols.
PCR amplification of the target polymorphic DNA fragment
DNA was isolated from citrate-anticoagulated whole blood using the DNeasy tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Fifty nanograms gDNA was sufficient for each polymerase chain reaction (PCR). PCR primer pairs were designed to amplify specific fragments of DNA encompassing each of SNP, as previously described.26
After polymerase extension of minisequencing primers, antidigoxygenin antibody conjugated to cyanine 5 (Cy5; Jackson ImmunoResearch Laboratories, West Grove, PA) and streptavidin-phycoerythrin conjugate (Molecular Probes, Eugene, OR) were added. After incubation for 15 minutes, slides were rinsed and scanned using a ScanArray 5000 (Packard Bioscience, Downers Grove, IL) confocal scanner equipped with 543-nm HeNe and 632-nm HeNe lasers to detect the signals of the labeled dideoxyribonucleotides (ddNTPs). Fluorescent signal data were quantified using Imagene image analysis software (Biodiscovery, Marina Del Ray, CA) and exported into Excel (Microsoft, Seattle, WA) for further analysis. Data collected from samples were compiled on an X-Y scatter plot along with control samples of known genotype. Genotypes were called based on data clustering.
Statistical analysis: covariance components model for multivariate traits
In the QTL Association model, genotypes at the candidate gene locus are treated as predictors modifying the quantitative trait.27
In our analyses, the QTL Association model was implemented using the software package MENDEL 5.0 (Dr Kenneth Lange, Dept of Human Genetics, UCLA).28 For each candidate gene, MENDEL assesses association by conducting a likelihood ratio test to determine whether the haplotype regression coefficients are significantly different from zero. Continuous variables, such as age, platelet count, plasma VWF:RCo levels, and plasma VWF:Ag levels are handled as covariates. The haplotypes associated with minimum or maximum values for each of the quantitative traits, bleeding time, plasma VWF:Ag levels, bleeding severity score, and adjusted bleeding severity score (score/age), were computed, and the significance of the association indicated by P. When necessary to achieve a normal distribution, quantitative traits were subjected to appropriate transformations, such as square root, and reciprocal or standardization by gender and checked for normality by the Kolmogorov-Smirnov test (SigmaStat 3.0; SYSTAT Software, Richmond, CA).
Results
VWD type 1 pedigrees
All the families were considered VWD type 1 at the time of enrollment because the diagnosis was based on previous laboratory analyses reported in the local database. When a diagnosis was performed again during this study, several affected individuals showed an apparent reduction of VWF:RCo versus VWF:Ag (F01, F03, F04, F05, F13, F14), whereas others (F06 and F07) were characterized by very low VWF measurements. All of these patients were selected among a large cohort of 150 VWD families according to the enrollment criteria of the European project (see “Criteria for enrollment of VWD patients and healthy individuals”) and they have been treated in the past with desmopressin (DDAVP), a drug than induces VWF release from endothelial cells (data not shown).
The gene haplotype frequencies among the 14 pedigree members, when considered as a single population, are not significantly different from those of a normal control group from Milan (Table 3).
Haplotype frequencies
Gene and haplotype . | Pedigree frequency . | M control frequency . | P* . | WNH control frequency . | P† . |
|---|---|---|---|---|---|
| ITGA2 | |||||
| 1 | 0.41 | 0.35 | .30 | 0.39 | .89 |
| 2 | 0.50 | 0.50 | 0.52 | ||
| 3 | 0.09 | 0.15 | 0.09 | ||
| - 92C | 0.82 | 0.85 | .66 | 0.88 | .08 |
| - 92G | 0.18 | 0.15 | 0.12 | ||
| - 52C | 0.63 | 0.59 | .63 | 0.65 | .77 |
| - 52T | 0.37 | 0.41 | 0.35 | ||
| GP6 | |||||
| a | 0.82 | 0.90 | .14 | 0.86 | .33 |
| b | 0.18 | 0.10 | 0.14 | ||
| GPIBA | |||||
| 1 | 0.86 | 0.91 | .35 | 0.92 | .02 |
| 2 | 0.14 | 0.09 | 0.08 | ||
| - 5T | 0.88 | 0.93 | .32 | 0.87 | .85 |
| - 5C | 0.12 | 0.07 | 0.13 | ||
| ITGB3 | |||||
| 1 | 0.86 | 0.79 | .22 | 0.83 | .59 |
| 2 | 0.14 | 0.21 | 0.17 | ||
| ITGA2B | |||||
| 1 | 0.57 | 0.55 | .87 | 0.59 | .72 |
| 2 | 0.43 | 0.45 | 0.41 | ||
| VWF | |||||
| 1 | 0.61 | 0.59 | .81 | 0.62 | .98 |
| 2 | 0.39 | 0.41 | 0.38 | ||
| IL6 | |||||
| 1 | 0.67 | 0.64 | .69 | 0.68 | .92 |
| 2 | 0.33 | 0.36 | 0.32 | ||
| FGB | |||||
| 1 | 0.74 | 0.73 | .94 | 0.78 | .37 |
| 2 | 0.26 | 0.27 | 0.22 | ||
| TXA2R | |||||
| 1 | 0.66 | 0.79 | .06 | 0.75 | .08 |
| 2 | 0.34 | 0.21 | 0.25 |
Gene and haplotype . | Pedigree frequency . | M control frequency . | P* . | WNH control frequency . | P† . |
|---|---|---|---|---|---|
| ITGA2 | |||||
| 1 | 0.41 | 0.35 | .30 | 0.39 | .89 |
| 2 | 0.50 | 0.50 | 0.52 | ||
| 3 | 0.09 | 0.15 | 0.09 | ||
| - 92C | 0.82 | 0.85 | .66 | 0.88 | .08 |
| - 92G | 0.18 | 0.15 | 0.12 | ||
| - 52C | 0.63 | 0.59 | .63 | 0.65 | .77 |
| - 52T | 0.37 | 0.41 | 0.35 | ||
| GP6 | |||||
| a | 0.82 | 0.90 | .14 | 0.86 | .33 |
| b | 0.18 | 0.10 | 0.14 | ||
| GPIBA | |||||
| 1 | 0.86 | 0.91 | .35 | 0.92 | .02 |
| 2 | 0.14 | 0.09 | 0.08 | ||
| - 5T | 0.88 | 0.93 | .32 | 0.87 | .85 |
| - 5C | 0.12 | 0.07 | 0.13 | ||
| ITGB3 | |||||
| 1 | 0.86 | 0.79 | .22 | 0.83 | .59 |
| 2 | 0.14 | 0.21 | 0.17 | ||
| ITGA2B | |||||
| 1 | 0.57 | 0.55 | .87 | 0.59 | .72 |
| 2 | 0.43 | 0.45 | 0.41 | ||
| VWF | |||||
| 1 | 0.61 | 0.59 | .81 | 0.62 | .98 |
| 2 | 0.39 | 0.41 | 0.38 | ||
| IL6 | |||||
| 1 | 0.67 | 0.64 | .69 | 0.68 | .92 |
| 2 | 0.33 | 0.36 | 0.32 | ||
| FGB | |||||
| 1 | 0.74 | 0.73 | .94 | 0.78 | .37 |
| 2 | 0.26 | 0.27 | 0.22 | ||
| TXA2R | |||||
| 1 | 0.66 | 0.79 | .06 | 0.75 | .08 |
| 2 | 0.34 | 0.21 | 0.25 |
Haplotype frequencies within the 14 pedigrees, including affected and unaffected individuals (n = 67).
M control indicates Milan controls (n = 52); WNH control, white non-Hispanic controls (n = 145).
P versus Milan controls by χ2 (Sigma Stat 3.0).
P versus white non-Hispanic controls by χ2 (Sigma Stat 3.0).
However, when compared to the white non-Hispanic control group, the frequencies of the GP1BA haplotypes 1 and 2 are significantly different (P = .02, χ2 analysis). The frequency of GP1BA haplotype 1 among the VWD pedigrees (0.86) is significantly lower than that seen in the control white non-Hispanic population (0.92; P = .02) and notably, albeit not significantly, lower than that of the Milan controls (0.91; P = .35). At the same time, the fact that haplotype frequencies can vary even among populations with comparable ethnic backgrounds reinforces the potential for type 1 errors that can be introduced in case-control studies. Such errors can be avoided in pedigree analyses. Furthermore, despite the decrease in GP1BA haplotype 1 among the VWD pedigrees, the statistical analysis described will show that GP1BA haplotypes are not associated with a more severe bleeding phenotype.
For purposes of comparison, the descriptive statistical analyses of the bleeding severity scores and BTs of pedigree members as a function of each of the candidate gene haplotypes are summarized in Table 4.
Descriptive statistical analysis
. | . | Score . | . | BT, min . | . | ||
|---|---|---|---|---|---|---|---|
| Gene . | No. . | Mean . | SD . | Mean . | SD . | ||
| ITGA2 | |||||||
| 11 | 10 | 7.9 | 8.4 | 11.4 | 10.0 | ||
| 12 | 18 | 6.5 | 4.8 | 9.4 | 8.4 | ||
| 13 | 5 | 0.8 | 1.3 | 6.9 | 3.3 | ||
| 22 | 14 | 6.3 | 5.0 | 7.1 | 2.9 | ||
| 23 | 19 | 4.0 | 5.4 | 6.9 | 4.8 | ||
| 33 | 1 | 0 | — | 6.5 | — | ||
| ITGA2-92 | |||||||
| cc | 42 | 6.1 | 6.5 | 8.6 | 7.3 | ||
| cg | 23 | 4.0 | 3.8 | 7.9 | 5.4 | ||
| gg | 2 | 7.5 | 4.9 | 6.8 | 1.8 | ||
| ITGA2-52 | |||||||
| cc | 25 | 5.7 | 4.3 | 7.1 | 5.1 | ||
| ct | 29 | 4.3 | 5.3 | 8.4 | 5.5 | ||
| tt | 13 | 7.3 | 8.3 | 10.3 | 10.4 | ||
| GP6 | |||||||
| aa | 56 | 4.7 | 4.9 | 7.6 | 5.8 | ||
| ab | 10 | 8.0 | 6.8 | 9.7 | 7.5 | ||
| bb | 1 | 23 | — | 30 | — | ||
| IL6 | |||||||
| 11 | 33 | 5.5 | 5.4 | 7.8 | 6.7 | ||
| 12 | 20 | 4.1 | 4.0 | 6.8 | 2.0 | ||
| 22 | 14 | 7.6 | 7.9 | 11.7 | 9.4 | ||
| GP1BA | |||||||
| 11 | 53 | 5.3 | 5.8 | 8.1 | 6.5 | ||
| 12 | 13 | 4.8 | 5.4 | 8.6 | 7.8 | ||
| 22 | 1 | 10 | — | 10 | — | ||
| GPIBA-5 | |||||||
| tt | 53 | 5.4 | 6.0 | 8.1 | 6.9 | ||
| tc | 14 | 5.5 | 4.5 | 8.9 | 5.3 | ||
| ITGB3 | |||||||
| 11 | 48 | 6.1 | 6.0 | 8.8 | 6.8 | ||
| 12 | 16 | 3.6 | 4.5 | 7.3 | 6.4 | ||
| 22 | 3 | 5.0 | 5.2 | 5.5 | 3.0 | ||
| ITGA2B | |||||||
| 11 | 25 | 7.8 | 6.6 | 10.3 | 9.6 | ||
| 12 | 31 | 4.5 | 4.8 | 7.4 | 4.2 | ||
| 22 | 11 | 2.4 | 4.1 | 6.2 | 2.0 | ||
| VWF | |||||||
| 11 | 25 | 6.3 | 6.5 | 9.5 | 7.9 | ||
| 12 | 27 | 5.5 | 5.7 | 7.9 | 6.7 | ||
| 22 | 15 | 3.9 | 4.0 | 6.9 | 2.9 | ||
| FGB | |||||||
| 11 | 35 | 5.9 | 6.0 | 8.1 | 6.4 | ||
| 12 | 24 | 5.5 | 5.9 | 8.0 | 6.5 | ||
| 22 | 8 | 3.3 | 3.3 | 8.7 | 5.1 | ||
| TXA2R | |||||||
| 11 | 33 | 5.7 | 6.2 | 7.2 | 6.1 | ||
| 12 | 26 | 4.8 | 5.4 | 9.4 | 6.8 | ||
| 22 | 8 | 6.3 | 4.6 | 7.7 | 3.8 | ||
. | . | Score . | . | BT, min . | . | ||
|---|---|---|---|---|---|---|---|
| Gene . | No. . | Mean . | SD . | Mean . | SD . | ||
| ITGA2 | |||||||
| 11 | 10 | 7.9 | 8.4 | 11.4 | 10.0 | ||
| 12 | 18 | 6.5 | 4.8 | 9.4 | 8.4 | ||
| 13 | 5 | 0.8 | 1.3 | 6.9 | 3.3 | ||
| 22 | 14 | 6.3 | 5.0 | 7.1 | 2.9 | ||
| 23 | 19 | 4.0 | 5.4 | 6.9 | 4.8 | ||
| 33 | 1 | 0 | — | 6.5 | — | ||
| ITGA2-92 | |||||||
| cc | 42 | 6.1 | 6.5 | 8.6 | 7.3 | ||
| cg | 23 | 4.0 | 3.8 | 7.9 | 5.4 | ||
| gg | 2 | 7.5 | 4.9 | 6.8 | 1.8 | ||
| ITGA2-52 | |||||||
| cc | 25 | 5.7 | 4.3 | 7.1 | 5.1 | ||
| ct | 29 | 4.3 | 5.3 | 8.4 | 5.5 | ||
| tt | 13 | 7.3 | 8.3 | 10.3 | 10.4 | ||
| GP6 | |||||||
| aa | 56 | 4.7 | 4.9 | 7.6 | 5.8 | ||
| ab | 10 | 8.0 | 6.8 | 9.7 | 7.5 | ||
| bb | 1 | 23 | — | 30 | — | ||
| IL6 | |||||||
| 11 | 33 | 5.5 | 5.4 | 7.8 | 6.7 | ||
| 12 | 20 | 4.1 | 4.0 | 6.8 | 2.0 | ||
| 22 | 14 | 7.6 | 7.9 | 11.7 | 9.4 | ||
| GP1BA | |||||||
| 11 | 53 | 5.3 | 5.8 | 8.1 | 6.5 | ||
| 12 | 13 | 4.8 | 5.4 | 8.6 | 7.8 | ||
| 22 | 1 | 10 | — | 10 | — | ||
| GPIBA-5 | |||||||
| tt | 53 | 5.4 | 6.0 | 8.1 | 6.9 | ||
| tc | 14 | 5.5 | 4.5 | 8.9 | 5.3 | ||
| ITGB3 | |||||||
| 11 | 48 | 6.1 | 6.0 | 8.8 | 6.8 | ||
| 12 | 16 | 3.6 | 4.5 | 7.3 | 6.4 | ||
| 22 | 3 | 5.0 | 5.2 | 5.5 | 3.0 | ||
| ITGA2B | |||||||
| 11 | 25 | 7.8 | 6.6 | 10.3 | 9.6 | ||
| 12 | 31 | 4.5 | 4.8 | 7.4 | 4.2 | ||
| 22 | 11 | 2.4 | 4.1 | 6.2 | 2.0 | ||
| VWF | |||||||
| 11 | 25 | 6.3 | 6.5 | 9.5 | 7.9 | ||
| 12 | 27 | 5.5 | 5.7 | 7.9 | 6.7 | ||
| 22 | 15 | 3.9 | 4.0 | 6.9 | 2.9 | ||
| FGB | |||||||
| 11 | 35 | 5.9 | 6.0 | 8.1 | 6.4 | ||
| 12 | 24 | 5.5 | 5.9 | 8.0 | 6.5 | ||
| 22 | 8 | 3.3 | 3.3 | 8.7 | 5.1 | ||
| TXA2R | |||||||
| 11 | 33 | 5.7 | 6.2 | 7.2 | 6.1 | ||
| 12 | 26 | 4.8 | 5.4 | 9.4 | 6.8 | ||
| 22 | 8 | 6.3 | 4.6 | 7.7 | 3.8 | ||
Genetic correlations
The results are depicted in Table 5. For each gene, the minimum haplotype is defined as that associated with lower values of the quantitative trait in question, whereas the maximum haplotype is that associated with higher values.
Candidate gene haplotype association: maximum likelihood estimates
Bleeding scores, by gene . | Minimum haplotype . | Estimate . | Maximum haplotype . | Estimate . | P . |
|---|---|---|---|---|---|
| Bleeding severity score | |||||
| ITGA2 | 3 | -0.5020 | 2 | 0.3718 | .01 |
| -92 | G | -0.0174 | C | 0.0174 | .84 |
| -52 | A | -0.0769 | G | 0.0769 | .25 |
| GP6 | a | -0.0855 | b | 0.0855 | .48 |
| GPIBA | 2 | -0.0718 | 1 | 0.0718 | .51 |
| -5 | T | -0.0417 | C | 0.0417 | .73 |
| ITGB3 | 2 | -0.1482 | 1 | 0.1482 | .11 |
| ITGA2B | 2 | -0.1698 | 1 | 0.1698 | .03 |
| VWF | 1 | -0.0584 | 2 | 0.0584 | .41 |
| IL6 | 2 | -0.0035 | 1 | 0.0035 | .96 |
| FGB | 2 | -0.0958 | 1 | 0.0958 | .23 |
| TBXA2R | 2 | -0.0088 | 1 | 0.0088 | .90 |
| Adjusted bleeding severity score, score/age | |||||
| ITGA2 | 3 | -0.4708 | 2 | 0.3374 | .01 |
| -92 | G | -0.0099 | C | 0.0099 | .93 |
| -52 | A | -0.0961 | G | 0.0961 | .28 |
| GP6 | a | -0.3312 | b | 0.3312 | .03 |
| GPIBA | 1 | -0.1263 | 2 | 0.1263 | .37 |
| -5 | C | -0.0564 | T | 0.0564 | .73 |
| ITGB3 | 2 | -0.0984 | 1 | 0.0984 | .40 |
| ITGA2B | 2 | -0.2544 | 1 | 0.2544 | .01 |
| VWF | 2 | -0.1120 | 1 | 0.1120 | .20 |
| IL6 | 1 | -0.0170 | 2 | 0.0170 | .85 |
| FGB | 2 | -0.0075 | 1 | 0.0075 | .94 |
| TBXA2R | 1 | -0.0509 | 2 | 0.0509 | .58 |
| Plasma VWF:Ag level | |||||
| ITGA2 | 1 | -0.1512 | 2 | 0.1364 | .37 |
| -92 | C | -0.1030 | G | 0.1030 | .37 |
| -52 | G | -0.0802 | A | 0.0802 | .39 |
| GP6 | b | -0.2522 | a | 0.2522 | .09 |
| GPIBA | 2 | -0.1868 | 1 | 0.1868 | .26 |
| -5 | C | -0.0307 | T | 0.0307 | .84 |
| ITGB3 | 1 | -0.0772 | 2 | 0.0772 | .52 |
| ITGA2B | 1 | -0.1572 | 2 | 0.1572 | .15 |
| VWF | 2 | -0.1939 | 1 | 0.1939 | .02 |
| IL6 | 2 | -0.0885 | 1 | 0.0885 | .38 |
| FGB | 2 | -0.0197 | 1 | 0.0197 | .84 |
| TBXA2R | 1 | -0.0480 | 2 | 0.0480 | .61 |
| Bleeding time | |||||
| ITGA2 | 3 | 0.2144 | 1 | -0.1321 | .35 |
| -92 | G | 0.0750 | C | -0.0750 | .48 |
| -52 | G | 0.0066 | A | -0.0066 | .94 |
| GP6 | a | 0.2329 | b | -0.2329 | .11 |
| GPIBA | 2 | 0.0661 | 1 | -0.0661 | .61 |
| -5 | T | 0.2418 | C | -0.2418 | .10 |
| ITGB3 | 2 | 0.1910 | 1 | -0.1910 | .07 |
| ITGA2B | 2 | 0.0673 | 1 | -0.0673 | .44 |
| VWF | 2 | 0.0482 | 1 | -0.0482 | .56 |
| IL6 | 1 | 0.1852 | 2 | -0.1852 | .12 |
| FGB | 1 | 0.0737 | 2 | -0.0737 | .40 |
| TBXA2R | 1 | 0.1132 | 2 | -0.1132 | .18 |
Bleeding scores, by gene . | Minimum haplotype . | Estimate . | Maximum haplotype . | Estimate . | P . |
|---|---|---|---|---|---|
| Bleeding severity score | |||||
| ITGA2 | 3 | -0.5020 | 2 | 0.3718 | .01 |
| -92 | G | -0.0174 | C | 0.0174 | .84 |
| -52 | A | -0.0769 | G | 0.0769 | .25 |
| GP6 | a | -0.0855 | b | 0.0855 | .48 |
| GPIBA | 2 | -0.0718 | 1 | 0.0718 | .51 |
| -5 | T | -0.0417 | C | 0.0417 | .73 |
| ITGB3 | 2 | -0.1482 | 1 | 0.1482 | .11 |
| ITGA2B | 2 | -0.1698 | 1 | 0.1698 | .03 |
| VWF | 1 | -0.0584 | 2 | 0.0584 | .41 |
| IL6 | 2 | -0.0035 | 1 | 0.0035 | .96 |
| FGB | 2 | -0.0958 | 1 | 0.0958 | .23 |
| TBXA2R | 2 | -0.0088 | 1 | 0.0088 | .90 |
| Adjusted bleeding severity score, score/age | |||||
| ITGA2 | 3 | -0.4708 | 2 | 0.3374 | .01 |
| -92 | G | -0.0099 | C | 0.0099 | .93 |
| -52 | A | -0.0961 | G | 0.0961 | .28 |
| GP6 | a | -0.3312 | b | 0.3312 | .03 |
| GPIBA | 1 | -0.1263 | 2 | 0.1263 | .37 |
| -5 | C | -0.0564 | T | 0.0564 | .73 |
| ITGB3 | 2 | -0.0984 | 1 | 0.0984 | .40 |
| ITGA2B | 2 | -0.2544 | 1 | 0.2544 | .01 |
| VWF | 2 | -0.1120 | 1 | 0.1120 | .20 |
| IL6 | 1 | -0.0170 | 2 | 0.0170 | .85 |
| FGB | 2 | -0.0075 | 1 | 0.0075 | .94 |
| TBXA2R | 1 | -0.0509 | 2 | 0.0509 | .58 |
| Plasma VWF:Ag level | |||||
| ITGA2 | 1 | -0.1512 | 2 | 0.1364 | .37 |
| -92 | C | -0.1030 | G | 0.1030 | .37 |
| -52 | G | -0.0802 | A | 0.0802 | .39 |
| GP6 | b | -0.2522 | a | 0.2522 | .09 |
| GPIBA | 2 | -0.1868 | 1 | 0.1868 | .26 |
| -5 | C | -0.0307 | T | 0.0307 | .84 |
| ITGB3 | 1 | -0.0772 | 2 | 0.0772 | .52 |
| ITGA2B | 1 | -0.1572 | 2 | 0.1572 | .15 |
| VWF | 2 | -0.1939 | 1 | 0.1939 | .02 |
| IL6 | 2 | -0.0885 | 1 | 0.0885 | .38 |
| FGB | 2 | -0.0197 | 1 | 0.0197 | .84 |
| TBXA2R | 1 | -0.0480 | 2 | 0.0480 | .61 |
| Bleeding time | |||||
| ITGA2 | 3 | 0.2144 | 1 | -0.1321 | .35 |
| -92 | G | 0.0750 | C | -0.0750 | .48 |
| -52 | G | 0.0066 | A | -0.0066 | .94 |
| GP6 | a | 0.2329 | b | -0.2329 | .11 |
| GPIBA | 2 | 0.0661 | 1 | -0.0661 | .61 |
| -5 | T | 0.2418 | C | -0.2418 | .10 |
| ITGB3 | 2 | 0.1910 | 1 | -0.1910 | .07 |
| ITGA2B | 2 | 0.0673 | 1 | -0.0673 | .44 |
| VWF | 2 | 0.0482 | 1 | -0.0482 | .56 |
| IL6 | 1 | 0.1852 | 2 | -0.1852 | .12 |
| FGB | 1 | 0.0737 | 2 | -0.0737 | .40 |
| TBXA2R | 1 | 0.1132 | 2 | -0.1132 | .18 |
Boldface indicates P < .05.
In the case of bleeding severity score, not adjusted for age (Table 5), ITGA2 haplotype 2 is most often associated with higher bleeding severity scores. Conversely, ITGA2 haplotype 3 is most often associated with lower bleeding scores. This particular association is statistically significant (P = .01) and as such is indicated in bold. These results can be interpreted to mean that haplotype 2 is associated with bleeding risk, whereas haplotype 3 is protective. A similar relationship was seen with ITGA2B haplotypes, where haplotype 1 (Ile843; Baka) was associated with risk for bleeding, whereas haplotype 2 (Ser843; Bakb) was protective (P = .03). None of the other 7 candidate gene haplotypes exhibited a statistically significant association with bleeding severity score.
When the bleeding severity score was adjusted for age (score/age; Table 5), the associations of ITGA2 and ITGA2B haplotypes remained statistically significant (P = .01 and .01, respectively). In addition, a statistically significant association was now apparent with GP6 haplotypes: Haplotype b (Pro219) was associated with risk for bleeding, whereas haplotype a (Ser219) was protective (P = .03). None of the other 6 candidate gene haplotypes exhibited a statistically significant association with score/age.
The VWF promoter haplotypes did not show an association with either measure of bleeding severity. On the other hand, with regard to plasma VWF:Ag levels, VWF promoter haplotype 1 (–1793G) was associated with higher levels (P = .02; Table 5).
With respect to BT (Table 5), there were no statistically significant association with any of the haplotypes of the 9 candidate genes.
The covariance components model used in this analysis also permits a simultaneous assessment of the impact of other parameters (covariates). In this manner, the influence of each parameter (VWF:RCo level, VWF:Ag level, platelet count, gender, and age) on bleeding severity score and BT, relative to candidate gene haplotypes, can be determined. A maximum likelihood estimate and standard error (SE) of the estimate are computed. Under the null hypothesis, the ratio of the absolute value of the estimate to the corresponding SE is approximately standard normal. A ratio of 2.0 or greater is arbitrarily accepted to indicate a significant effect, and a comparison of the ratios provides an indication of the relative impact of each parameter. In each comparison of parameter estimates, VWF:RCo and VWF:Ag gave essentially identical results, so these parameters will be referred to collectively as VWF level.
With regard to bleeding severity score (Table 6), parameter estimates clearly show that VWF level has the strongest impact on the score. This was an inverse effect, as reflected in the negative estimate, such that a decrease in score was associated with an increase in VWF level. ITGA2 haplotype 3 greater than ITGAB haplotype 2 each exerted an inverse effect on the score. Conversely, those parameters that were associated with an increase in score, in order of greatest impact, were ITGA2 haplotype 2 and ITGA2B haplotype 1. It is noteworthy that gender, platelet count, and GP6 haplotype had no significant impact on the score.
Parameter estimates for bleeding severity score
Parameter . | Estimate . | SE . | Ratio* . |
|---|---|---|---|
| VWF:RCo | -1.2521 | 0.1878 | 6.67 |
| VWF:Ag | -1.1248 | 0.1693 | 6.64 |
| ITGA2 haplotype 3 | -0.5020 | 0.1122 | 4.47 |
| ITGA2B haplotype 2 | -0.1698 | 0.0726 | 2.34 |
| Platelet count | -0.0017 | 0.0019 | 0.89 |
| GP6 haplotype a | -0.0855 | 0.1201 | 0.71 |
| Male | -0.0311 | 0.0929 | 0.33 |
| ITGA2 haplotype 2 | 0.3718 | 0.0935 | 3.98 |
| ITGA2B haplotype 1 | 0.1698 | 0.0726 | 2.34 |
| Age | 0.0098 | 0.0051 | 1.92 |
| ITGA2 haplotype 1 | 0.1302 | 0.0833 | 1.56 |
| GP6 haplotype b | 0.0855 | 0.1201 | 0.71 |
| Female | 0.0311 | 0.0929 | 0.33 |
Parameter . | Estimate . | SE . | Ratio* . |
|---|---|---|---|
| VWF:RCo | -1.2521 | 0.1878 | 6.67 |
| VWF:Ag | -1.1248 | 0.1693 | 6.64 |
| ITGA2 haplotype 3 | -0.5020 | 0.1122 | 4.47 |
| ITGA2B haplotype 2 | -0.1698 | 0.0726 | 2.34 |
| Platelet count | -0.0017 | 0.0019 | 0.89 |
| GP6 haplotype a | -0.0855 | 0.1201 | 0.71 |
| Male | -0.0311 | 0.0929 | 0.33 |
| ITGA2 haplotype 2 | 0.3718 | 0.0935 | 3.98 |
| ITGA2B haplotype 1 | 0.1698 | 0.0726 | 2.34 |
| Age | 0.0098 | 0.0051 | 1.92 |
| ITGA2 haplotype 1 | 0.1302 | 0.0833 | 1.56 |
| GP6 haplotype b | 0.0855 | 0.1201 | 0.71 |
| Female | 0.0311 | 0.0929 | 0.33 |
Absolute value (estimate)/SE.
These relationships held true when the bleeding severity score was adjusted for age of the individual (score/age; Table 7), except that the impact of GP6 haplotypes was now significant. The relative impact of the 3 protective candidate gene haplotypes of concern was ITGA2 haplotype 3 more than ITGA2B haplotype 2 more than GP6 haplotype a.
Parameter estimates for adjusted bleeding severity score (score/age)
Parameter . | Estimate . | SE . | Ratio* . |
|---|---|---|---|
| VWF:Ag | -1.0234 | 0.1703 | 6.01 |
| VWF:RCo | -1.1309 | 0.1913 | 5.91 |
| ITGA2 haplotype 3 | -0.4708 | 0.1499 | 3.14 |
| ITGA2B haplotype 2 | -0.2544 | 0.0871 | 2.92 |
| GP6 haplotype a | -0.3312 | 0.1545 | 2.14 |
| Platelet count | -0.0028 | 0.0019 | 1.47 |
| Male | -0.0203 | 0.0954 | 0.21 |
| ITGA2 haplotype 2 | 0.3374 | 0.1246 | 2.71 |
| ITGA2B haplotype 1 | 0.2544 | 0.0871 | 2.92 |
| GP6 haplotype b | 0.3312 | 0.1545 | 2.14 |
| ITGA2 haplotype 1 | 0.1334 | 0.1167 | 1.14 |
| Female | 0.0203 | 0.0954 | 0.21 |
Parameter . | Estimate . | SE . | Ratio* . |
|---|---|---|---|
| VWF:Ag | -1.0234 | 0.1703 | 6.01 |
| VWF:RCo | -1.1309 | 0.1913 | 5.91 |
| ITGA2 haplotype 3 | -0.4708 | 0.1499 | 3.14 |
| ITGA2B haplotype 2 | -0.2544 | 0.0871 | 2.92 |
| GP6 haplotype a | -0.3312 | 0.1545 | 2.14 |
| Platelet count | -0.0028 | 0.0019 | 1.47 |
| Male | -0.0203 | 0.0954 | 0.21 |
| ITGA2 haplotype 2 | 0.3374 | 0.1246 | 2.71 |
| ITGA2B haplotype 1 | 0.2544 | 0.0871 | 2.92 |
| GP6 haplotype b | 0.3312 | 0.1545 | 2.14 |
| ITGA2 haplotype 1 | 0.1334 | 0.1167 | 1.14 |
| Female | 0.0203 | 0.0954 | 0.21 |
Absolute value (estimate)/SE.
As noted, none of the candidate gene haplotypes exerted a significant influence on bleeding time. On the other hand, VWF level was again the single parameter most responsible for variation in BT (Table 8).
Parameter estimates for bleeding time
Parameter . | Estimate . | SE . | Ratio* . |
|---|---|---|---|
| VWF:RCo | -0.7875 | 0.2279 | 3.46 |
| VWF:Ag | -0.6093 | 0.2125 | 2.87 |
| Female | -0.0231 | 0.1171 | 0.20 |
| Platelet count | 0.0019 | 0.0023 | 0.83 |
| Male | 0.0231 | 0.1171 | 0.20 |
| Age | 0.0000 | 0.0066 | 0.00 |
Parameter . | Estimate . | SE . | Ratio* . |
|---|---|---|---|
| VWF:RCo | -0.7875 | 0.2279 | 3.46 |
| VWF:Ag | -0.6093 | 0.2125 | 2.87 |
| Female | -0.0231 | 0.1171 | 0.20 |
| Platelet count | 0.0019 | 0.0023 | 0.83 |
| Male | 0.0231 | 0.1171 | 0.20 |
| Age | 0.0000 | 0.0066 | 0.00 |
Absolute value (estimate)/SE.
Discussion
We have tried to correlate integrin haplotypes with bleeding tendency in a well-characterized group of families previously diagnosed as VWD type 1. These families were initially recruited according to predefined enrollment criteria applied to the 154 families enrolled by 12 hemophilia centers from 9 European countries. The aims of this European project were to determine the best clinical and molecular markers for diagnosis and management of previously diagnosed VWD type 1. Because several factors are known to interfere with VWF levels within the same VWD families and to influence their clinical behavior in terms of bleeding tendency, this pilot study on the role of adhesion receptor gene haplotypes on bleeding tendency in 14 families collected by one partner (P03, Milan) was approved by all of the other partners of this European project.
All 14 families with previously diagnosed VWD type 1 satisfied the criteria of enrollment, but several affected individuals showed an apparent reduction of VWF:RCo versus VWF:Ag (F01, F03, F04, F05, F13, F14), whereas others (F06 and F07) were characterized by very low VWF measurements. This is not surprising because the diagnosis of VWD has been improved in the last 10 years. Updated criteria for the classification of VWD type 1 have not yet been approved by the SSC of the ISTH. A panel of experts of this committee is now working on a proposal for an updated classification of VWD type 1 versus 2.
To better understand the influence of secondary gene polymorphisms on bleeding risk in VWD type 1, we elected to study pedigrees of index cases. Case-control studies can provide useful indirect evidence for the contribution of a genetic effect, and we have previously demonstrated in the initial case-control study of this kind that the ITGA2 SNP 807C is more common among individuals with VWD type 1.29 However, case-control studies suffer from a number of weaknesses that preclude the identification of many genetic associations.30 These include the introduction of type 1 errors resulting from hidden population stratification, the lack of a direct evaluation of familial transmission, and poor ability to discover novel genes involved in intrapopulation variation in disease risk. Contributing to these weaknesses are small but statistically significant differences in haplotype frequency that might be peculiar to the specific populations under study. One such frequency difference was seen for GP1BA haplotypes in the pedigrees that were the subject of this study (Table 3).
Family-based approaches, particularly those that involve large pedigrees, provide a more powerful approach to the study of genetic risk associations. The benefit of genetic linkage studies in pedigrees derives from the presence of case-parent triads and multiplex sibships. For studies of genetic risk, designs based on the genotyping of index cases and their parents are highly informative and minimize spurious conclusions that might result from hidden genetic structure, such as stratification or admixture that can afflict a case-control study.31 The underlying benefit of case-parent triad lies in the fact that transmission of alleles from parents to offspring follows mendelian probabilities and for each parental mating type, the mendelian genotype proportions persist among offspring until such time as the probands are studied. In addition, multiplex sibships will be enriched in frequency for a disease susceptibility allele and thereby create a stronger contrast to parents and unaffected sibs.32
The paradox of VWD type 1 is that bleeding risk does not correlate with plasma VWF levels.3 Among healthy donors, plasma VWF levels vary over a wide range, and 95% of all values fall within 50% and 200% of the mean. ABO blood type has a major influence on VWF levels, such that the average level for persons who are blood type O is 25% to 35% lower than that of persons who are not type O.33 This accounts for about 30% of the genetic variance of VWF.34 The SNP G–1793C in the VWF promoter also correlates with small changes in mean VWF level. Among blood group O donors, mean VWF levels are 77% for genotype CC, 86% for genotype GC, and 93% for genotype GG.20 Because of the high allele frequency of –1793G (0.65), it can contribute modestly to the variation in VWF levels. In our study, we confirm a positive influence of –1793G on plasma VWF:Ag and VWF:RCo. However, it appears that the impact of the VWF promoter haplotypes are not sufficiently important that they can be directly associated to variation in bleeding phenotype.
In an earlier case-control study,29 we showed that the 807C dimorphism of the integrin gene ITGA2 was associated with diminished platelet responsiveness to collagen in a high shear environment, reflected by a prolongation of the PFA-100 collagen/epinephrine-closure time. However, that study preceded the finding that there are 2 major haplotypes that share the 807C allele: haplotype 3 (807C/1648A) is associated with higher levels of the integrin, whereas haplotype 2 (807C/1648G) is associated with markedly reduced levels of this receptor.8 The current study refines our initial association study, demonstrating that haplotype 2 contributes to increased bleeding severity score and increased risk, whereas haplotype 3 is protective. The protective effect offered by the presence of haplotype 3 may not be explained exclusively by increased levels of the integrin and suggests that haplotype 3 may be associated with increased function of the integrin in addition to any effect on expression. The unique difference of haplotype 3 is its expression of the alloantigen HPA-5b (Bra) resulting from a lysine to glutamate substitution at residue 505. It remains to be determined whether this amino acid substitution has a significant influence on platelet function tests ex vivo. Aside from this, our results reinforce the need to distinguish the major haplotypes 2 and 3 of ITGA2, which share the 807C SNP, because each has a quantitatively and perhaps qualitatively opposite influence on the function of this integrin.
With regard to GP6 haplotypes, our results show a statistically significant, albeit weaker, association between haplotype b (Pro219) and risk for bleeding, which was only apparent when the bleeding severity score was adjusted for age. Conversely, haplotype a (Ser219) was protective. These findings are not consistent with the observation of Croft et al7 that homozygosity for haplotype b would be associated with risk for acute myocardial infarction, but are consistent with the observations of Joutsi-Korhonen et al16 that haplotype a confers increased reactivity to the receptor.
The involvement of ITGA2 and GP6 haplotypes in risk for bleeding in mild VWD is certainly not a coincidence. Although some have argued that one or the other of GPVI or integrin α2β1 is the more important for platelet adhesion and signaling on collagens, it is more likely that the concerted activity of both is essential for optimum platelet function. Receptor cooperation is supported by several observations. Mice genetically deficient in GPVI exhibit a modest abnormality in hemostatic function35 whereby platelet adhesion and thrombus formation persist, albeit with some loss of thrombus stability. At the same time, similar results occur with mice that are deficient in α2β1,36 where adhesion and thrombus formation are moderately impaired but not completely eliminated. Consistent with this theme, 2 receptor models of platelet adhesion to collagen and resultant platelet activation emphasize the integrated cross-talk between these 2 receptors. Lastly, mouse strain differences in platelet aggregation by collagen have been recently correlated with a difference in expression of integrin α2β1, but not other prominent receptors, such as GPVI, GPIbα or integrin αIIbβ3.37
Polymorphism of ITGA2B is responsible for the expression of the Bak (HPA-3) alloantigen system.38 This polymorphism lies adjacent to the binding region of the murine monoclonal PMI-1 antibody, which inhibits platelet adhesion and spreading on certain substrata.39 Adenosine diphosphate (ADP) stimulation of platelets results in a fibrinogen-dependent increase in binding of the PMI-1 antibody, and peptides containing arginine-glycine-aspartate (RGD) also reversibly increase the binding of this antibody to cells and to purified glycoprotein αIIbβ3.40 Thus, the αIIb region encompassing HPA-3 and PMI.1 epitopes may participate in adhesive functions through postreceptor occupancy events. The study of Reiner et al9 is consistent with this hypothesis, wherein homozygosity for the ITGA2B haplotype 2 (HPA-3b allele) was associated with an approximately 5-fold increased risk of ischemic stroke among subgroups of women who carried a diagnosis of hypertension or diabetes (OR = 4.51; 95% CI, 1.01-20.13) or had elevated plasma homocysteine levels (OR = 5.94; 95% CI, 1.53-23.05). Our findings in a different clinical setting complement the observation of Reiner et al,9 showing that the complementary haplotype 1 would decrease the tendency to thrombosis and thus increase risk for bleeding in VWD type 1.
The results of our study document that, despite its variability and poor heritability, the level of VWF, measured as antigen or ristocetin cofactor, remains the single most important parameter associated with bleeding severity and bleeding times in families of VWD type 1 index cases. Our results also provide unique confirmation of the importance of ITGA2, GP6, and ITGA2B haplotypes in disease outcome, demonstrating that genetically controlled attenuation of certain adhesion receptors (whether through expression or activity) can influence risk for morbidity in clinical settings where hemostasis is compromised. In contrast, a number of other glycoprotein haplotypes were not predictive, namely, those represented by the GP1BA, integrin ITGB3, VWF, FGB, IL6, and TXA2R genes.
Prepublished online as Blood First Edition Paper, June 29, 2004; DOI 10.1182/blood-2004-01-0349.
Supported by grant R01 HL46979 from the National Heart, Lung and Blood Institute (T.J.K.). The General Clinical Research Center (GCRC) of Scripps Clinic is supported by National Institutes of Health grant M01 RR00833. The work in Milan (L.B., M.T.C., A.B.F.) was supported by a grant from the European Community QLRT 1999-30387 for the project entitled “Molecular and Clinical Markers for Diagnosis and Management of type 1 von Willebrand Disease” (A.B.F.). I.R.P. is the Scientific Coordinator of this European project.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This is manuscript 16315-MEM from TSRI. We thank Drs Kenneth Lange and Eric Sobel (Department of Human Genetics, UCLA School of Medicine) for their excellent genetic and statistical counseling throughout this study. We also thank Drs ClaudiaMistretta and Francesca Giannello for assistance in interviewing patients and calculating bleeding scores. We acknowledge the comments received from all of the other partners of the European project entitled “Molecular and Clinical Markers for Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD): Ann Gooedeve (P01, Sheffield, United Kingdom), Drs Francesco Rodeghiero and Giancarlo Castaman (P02, Vicenza, Italy), Dr Javier Battle (P04, La Coruna, Spain), Drs Dominique Meyer, Mazurier, Goodemand (P05, Paris and Lille, France), Dr Jeroen Eikenboom (P06, Leiden, The Netherlands), Drs Reinhard Schneppenheim and Ulrich Budde (P07, Hamburg, Germany), Dr Jorgen Ingerslev (P08, Aarhus, Denmark), Drs Zdena Vorlova and David Habart (P09, Prague, Czech Republic), Dr Lars Holmberg and Stefan Lethagen (P10, Malmo, Sweden), Dr John Pasi (P11, Leicester, United Kingdom), and Dr Frank Hill (P12, Birmingham, United Kingdom).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal