Abstract
Juvenile hereditary hemochromatosis is a genetically heterogeneous disorder transmitted as an autosomal recessive trait. It is most often caused by mutations in the HJV gene and rarely in the HAMP gene. Hepcidin is considered to constitute a negative regulator of iron absorption, and its production is increased in inflammatory states and iron overload. We report the detection of a new mutation in the HAMP gene leading to juvenile hemochromatosis in 2 members of a Portuguese family. The mutation lies in the 5′-UTR (untranslated region) of the gene and creates a new initiation codon in the context of a Kozak sequence. We found no trace of hepcidin protein in the patients' urine, suggesting that ribosomes select the mutant initiation codon for translation. The decrease of hepcidin production would thus lead to increased iron absorption, resulting in iron deposition in parenchymal tissues. Phlebotomy therapy of the 2 patients resulted in impressive clinical improvement. (Blood. 2004;104: 2181-2183)
Introduction
Familial hemochromatosis is a clinically and genetically heterogeneous disease.1 A severe form of familial hemochromatosis is juvenile hemochromatosis (JH) that occurs in patients before the age of 30. Two genes responsible for JH have been described. Attention first focused on the HAMP gene, which codes for the peptide hepcidin.2,3 Transgenic mouse models indicate that hepcidin constitutes the prominent-negative regulator of iron absorption in mammals.4 Mutations in the HAMP gene were found in affected members of 3 families with JH.5,6 The second gene involved in JH was isolated on chromosome 1 and was called hemojuvelin (HJV).7
We describe a family of Portuguese origin in which 2 members with JH were found to be homozygous for a novel mutation in the 5′-UTR (untranslated region) of the HAMP gene.
Study design
Case report
A 29-year-old Portuguese man was diagnosed with insulin-dependent diabetes mellitus and severe heart failure. He exhibited skin hyperpigmentation, hepatosplenomegaly, and hypogonadism. The cardiac ejection fraction was 20%. Laboratory findings were as follows: aspartate aminotransferase (AST), 63 U/L; alanine aminotransferase (ALT), 76 U/L; hemoglobin (Hb), 126 g/L; thrombocytes, 99 × 109/L; serum iron (Fe), 41 μM; transferrin saturation, 100%; and serum ferritin, 1981 μg/L. Ultrasonography showed diffuse hepatomegaly and splenomegaly; liver biopsy showed severe diffuse hepatocellular siderosis and cirrhosis. The histologic iron index (score determined with method of Deugnier et al8 ) was highly elevated, score 49/29 = 1.68 (normal < 0.2). Hypogonadotrophic hypogonadism was diagnosed from the following laboratory findings: testosterone, 0.18 μg/mL (normal range, 3-8 μg/mL); follicle-stimulating hormone (FSH), less than 0.4 IU/L (normal range, 2.7-6.8 IU/L); and luteinizing hormone (LH), less than 0.3 IU/L (normal range, 2.5-6.8 IU/L). Juvenile hemochromatosis was suspected.
The patient was treated for 11 months by phlebotomy (400 mL per week) and erythropoietin 6000 UI intravenously once weekly. Ferritin decreased to 1187 μg/L, but transferrin saturation remained at 100%, and AST and ALT values did not vary. The clinical signs improved significantly.
The patient's 24-year-old sister had no clinical findings, but laboratory tests showed increased transaminases and iron overload (Fe = 52 μM, transferrin saturation = 100%, and ferritin = 810 μg/L). She was treated for 7 months by phlebotomy, and, at reevaluation, transferrin saturation was 50% and ferritin was 40 μg/L. Liver biopsy performed at that time showed moderate predominant periportal hepatocellular siderosis without cirrhosis; Deugnier score was elevated, 24/24 = 1.
The proband's parents had normal transferrin saturation and normal ferritin values.
Methods
Urine samples from the proband and his sister were analyzed for the presence of hepcidin after extraction of cationic peptides by Western blot with the use of rabbit anti-human hepcidin antibody9 ; normal values include 10 to 200 ng hepcidin/mg creatinine (detection limit, 2 ng hepcidin/mg creatinine).
All family members included in this study provided written consent to perform DNA studies on their blood samples. DNA was extracted from peripheral white blood cells (WBCs) and screened for C282Y and H63D mutations of the HFE1 gene.10 The HAMP gene was sequenced according to standard procedures.11 RNA was extracted from liver biopsies and analyzed by reverse transcriptase-polymerase chain reaction (RT-PCR) and real-time PCR with primers specific for the HAMP gene and abl gene as control (HAMP: P1 = 5′-GCACT GAGCT CCCAG ATCTG; P2 = 5′-CTACG TCTTG CAGCA CATCC C; abl: P1 = 5′-CCCAA CCTTT TCGTT GCACT GT; P2 = CGGCT CTCGG AGGAG ACGTA GA).11
Results and discussion
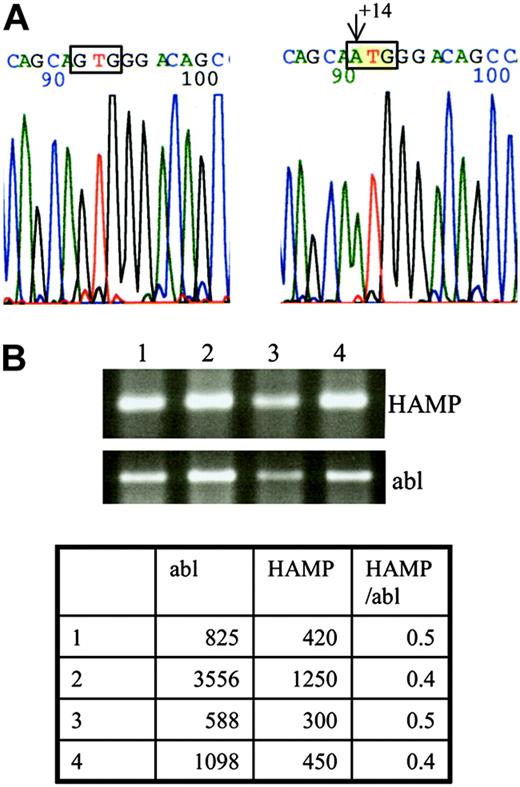
By screening the HFE1 gene we only found heterozygosity for the H63D mutation in the proband, his sister, and their mother. Sequencing of the hepcidin gene of the proband showed an undescribed homozygous G → A mutation at position +14 of the 5′-untranslated region (5′-UTR), relative to the Cap site of the mRNA (Figure 1A). On testing by restriction analysis (BsrDI and BtsI), this mutation was not found among 200 chromosomes from Portuguese subjects without iron overload. The patient's sister was homozygous for the same mutation, whereas both parents and a cousin were found to be heterozygous, with no signs of iron overload.
DNA and RNA analysis. (A) DNA sequencing of the HAMP gene led to identification of a new mutation at position +14 from the Cap site of the mRNA. Left side: wild-type nucleotide G at position +14; right side: mutated nucleotide A. (B) RNA was extracted from a biopsy obtained from the proband's liver and analyzed by RT-PCR for the presence of HAMP cDNA. The ethidium bromide stained agarose gel shows similar expression of the patient's HAMP cDNA (lane 4) compared with cDNA obtained from normal (lanes 1-2) and cirrhotic livers (lane 3). RT-PCR for abl was performed in parallel to ensure that equal quantities of cDNA were used for each PCR. Real-time PCR was performed for a more accurate determination of HAMP cDNA levels. The table shows only minor variations of the HAMP/abl ratios between the different samples (1-2, normal liver; 3, cirrhotic liver; 4, proband's liver).
DNA and RNA analysis. (A) DNA sequencing of the HAMP gene led to identification of a new mutation at position +14 from the Cap site of the mRNA. Left side: wild-type nucleotide G at position +14; right side: mutated nucleotide A. (B) RNA was extracted from a biopsy obtained from the proband's liver and analyzed by RT-PCR for the presence of HAMP cDNA. The ethidium bromide stained agarose gel shows similar expression of the patient's HAMP cDNA (lane 4) compared with cDNA obtained from normal (lanes 1-2) and cirrhotic livers (lane 3). RT-PCR for abl was performed in parallel to ensure that equal quantities of cDNA were used for each PCR. Real-time PCR was performed for a more accurate determination of HAMP cDNA levels. The table shows only minor variations of the HAMP/abl ratios between the different samples (1-2, normal liver; 3, cirrhotic liver; 4, proband's liver).
The proband's hepcidin cDNA quantities were found similar to those obtained from 5 control liver biopsies (normal and cirrhotic), (Figure 1B). No trace of hepcidin was found in the proband's or his sister's urine.
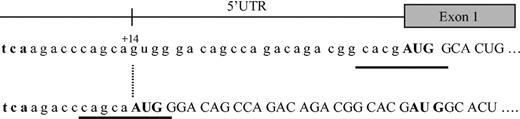
The JH in this 29-year-old man and his sister is thus secondary to a new mutation of the HAMP gene. This mutation creates a new initiation codon at position +14 of the 5′-UTR, which does not seem to affect the level of RNA produced, but induces a shift of the reading frame and the generation of an abnormal protein (Figure 2). This protein is probably unstable or otherwise degraded, because it was not found on bidimensional protein gel electrophoresis in a region predicted by the SWISS-2DPAGE database12 after nucleotide-protein sequence translation. Interestingly, the new AUG site seems to suppress the normal HAMP mRNA translation, because no hepcidin was found in the urine of either homozygous patient. Most eukaryotic mRNAs are translated by a scanning mechanism in which the small ribosomal subunit enters the mRNA at its 5′ end and migrates linearly in the 3′ direction until an AUG codon is encountered. Whether the ribosome then stops scanning and initiates translation depends on the presence of certain flanking nucleotides, the so-called Kozak sequence: GCCRCC AUG G.13 In our patient the newly created AUG site lies indeed in such a sequence with a G at position -3 and +4 (Figure 2). However, the normal AUG is located at a relatively short distance, ie, 25 nucleotides downstream. Given that the minimal distance between 2 adjacent ribosomes in the same mRNA strand is approximately 80 nucleotides,14 it is unlikely that the normal AUG is functional. Several examples of upstream AUG codons created by polymorphisms or mutations are reported in the literature, eg, β-thalassemia, coagulation factor XII, apolipoprotein(a).15-17 In each of these cases translation of the corresponding normal protein was diminished by the new out-of-frame initiation site.
Map of the 5′-UTR region of the HAMP gene. The normal hepcidin peptide is derived from the C-terminus of an 84 amino acid prepropeptide, encoded by a 0.4-kb mRNA, generated from 3 exons of a 2.5-kb gene on chromosome 19. The G → A mutation at position +14 creates a new AUG codon, which leads to a shift of the reading frame, inhibition of the synthesis of the normal hepcidin protein, and probably generation of a new abnormal protein instead. Kozak sequences are underlined. The initiation codon AUG is given in bold.
Map of the 5′-UTR region of the HAMP gene. The normal hepcidin peptide is derived from the C-terminus of an 84 amino acid prepropeptide, encoded by a 0.4-kb mRNA, generated from 3 exons of a 2.5-kb gene on chromosome 19. The G → A mutation at position +14 creates a new AUG codon, which leads to a shift of the reading frame, inhibition of the synthesis of the normal hepcidin protein, and probably generation of a new abnormal protein instead. Kozak sequences are underlined. The initiation codon AUG is given in bold.
The heterozygous state for this mutation, alone or in association with H63D of the HFE1 gene, does not lead to any iron overload, as shown by the normal iron status observed in the proband's parents.
The clinical phenotype of the proband was severe. This probably underlines the deleterious effect of complete transferrin saturation in JH secondary to hepcidin deficiency and the important total iron burden. In fact, although the patient underwent weekly phlebotomies, resulting in a decrease in ferritin levels, because hepcidin not only controls iron absorption in the intestine but also iron release from macrophages, we speculate that its absence in our patient greatly contributed to the disappearance of the iron storage function of tissue macrophages subsequently leading to massive iron deposition in parenchymal tissues and to complete serum transferrin saturation.
At diagnosis the proband exhibited anemia, and platelet counts were found decreased at multiple controls, probably secondary to hypersplenism (moderate portal hypertension secondary to liver cirrhosis). For that reason, he did not tolerate weekly bleeding and erythropoietin was added.
In the patient's sister, clinical manifestations were not severe. There are 2 possible explanations: (1) family screening allowed earlier diagnosis (24 versus 29 years in her brother) and (2) abundant and regular menstruations from the age of 12 years partially depleted iron overload. However, because cases described in the literature do not show a significant difference in the phenotype between men and women, we cannot exclude the existence of modifier genes affecting penetrance of juvenile hemochromatosis in this family.
Prepublished online as Blood First Edition Paper, June 15, 2004; DOI 10.1182/blood-2004-01-0332.
Supported by the Fondation pour la Recherche en Hématologie A. and P. Miescher (T.M. and P.B.), an unrestricted grant from VIFOR International, St Gallen, Switzerland (T.M. and P.B.), and a grant from La Fondation pour la Recherche Médicale (P.A.M.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal