Abstract
Folate metabolism plays an essential role in DNA synthesis and methylation processes. Deviations in the flux of folate due to genetic variation could result in selective growth and genomic instability and affect susceptibility to various cancers including lymphoma. To test this hypothesis, genetic polymorphisms in the folate metabolic pathway were investigated using DNA from a population-based case-control study of non-Hodgkin lymphoma (NHL) conducted in the San Francisco Bay Area between 1988 and 1995. The polymorphisms examined and haplotypes generated included thymidylate synthase (TYMS 28-bp triple repeat [3R] → double repeat [2R], 1494del6, IVS6 -68C>T, 1122A>G, and 1053C>T); 5,10-methylenetetrahydrofolate reductase (MTHFR 677C>T and 1298A>C); serine hydroxymethyltransferase (SHMT1 C1420T); reduced folate carrier (RFC G80A); and methionine synthase (MTR A2756G), making the present study the largest and most comprehensive to date to evaluate associations between genetic polymorphisms in folatemetabolizing genes and NHL risk. The TYMS 6 base pair (bp)-6bp- (homozygous for 6bp deletion), IVS6 -68C>T, and 1053C>T genotypes (all in complete linkage disequilibrium) were all inversely associated with NHL (TYMS; odds ratio [OR] = 0.57; 0.34-0.94), particularly with diffuse large cell lymphoma (DLCL; OR = 0.29; 0.10-0.82). Further, the MTR 2756AG/GG and the MTHFR 677TT genotypes were associated with increased risk for NHL (OR = 1.3; 0.99-1.7) and follicular lymphoma (FL; OR = 1.8; 0.98-3.1), respectively. We did not observe any significant differences in genotype frequencies of the SHMT1 and RFC polymorphisms between the cases and controls. The associations of DLCL and FL with TYMS 1494del6 and MTHFR 677TT genotypes, respectively, suggest that folate metabolism may play an important role in the pathogenesis of specific subtypes of NHL. (Blood. 2004;104: 2155-2162)
Introduction
Folate is an important nutrient required for DNA synthesis and methylation and its deficiency has been associated with a number of malignancies.1-3 We have previously shown that a triple tandem repeat (3R) in the promoter region of the thymidylate synthase gene (TYMS 3R → 2R) and variant alleles in the cytosolic serine hydroxymethyltransferase (SHMT1 1420C>T); methionine synthase (MTR 2756A>G); and 5,10-methylenetetrahydrofolate reductase (MTHFR 677C>T and 1298A>C) genes in the folate metabolic pathway were associated with a reduced risk of adult acute lymphocytic leukemia (ALL).4,5 Others have shown similar relationships with childhood ALL and also that folate supplementation of the diet of pregnant mothers decreased the risk of childhood ALL.6,7 In addition to ALL, these same folate genetic polymorphisms have been implicated in the development of lymphoma.8,9 For example, in agreement with our previous findings in adult ALL, the TYMS 3R, SHMT1 1420T,9 MTHFR 677T,8 and MTR 2756G alleles10 were reported to have been inversely associated with lymphoma. It has been hypothesized that this protection results from decreased uracil misincorporation in DNA, reduced DNA double-strand breaks, and subsequent genetic lesions.
TYMS plays a crucial role in the balance of deoxynucleotides for DNA synthesis. The dynamic role of TYMS in DNA metabolism has led to its potential as a target for chemotherapeutic regimens. A double (2R) or triple (3R) 28-base pair (bp) repeat sequence in the promoter enhancer region of the TYMS gene influences protein expression in cancer cells, with higher efficiency conferred by the 3R allele.11,12 This enhanced expression can increase the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP), minimizing both the level of uracil that may be incorporated into DNA and resultant DNA double-strand breaks. Furthermore, a 6-bp deletion in the 3′-untranslated region (3′UTR) of TYMS has been described (1494del6)13 that has been associated with a decreased risk of spina bifida,14 although its functional relevance is unknown.
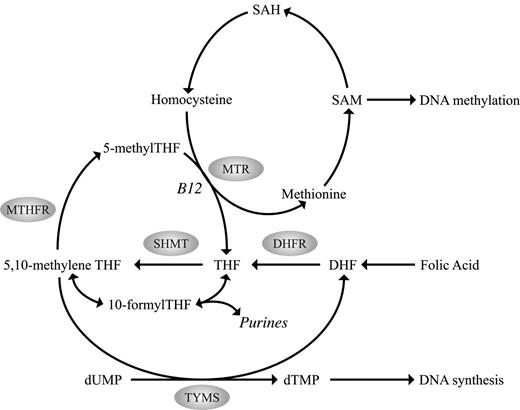
SHMT catalyzes the reversible conversion of tetrahydrofolate to 5,10-methylenetetrahydrofolate (methyleneTHF) providing 1-carbon units for S-adenosylmethionine (SAM), purine, and thymidine synthesis (Figure 1). The SHMT1 1420C>T polymorphism may influence the flux of folate toward thymidylate synthesis involving protein-protein interactions with TYMS. MTHFR reduces methyleneTHF to 5-methyltetrahydrofolate (5-methylTHF), which serves as a methyl donor for SAM production. The MTHFR 677C>T polymorphism, associated with reduced MTHFR enzyme activity and plasma folate levels and resultant hypomethylation, shifts pools of methyleneTHF from DNA methylation toward purine and DNA synthesis (Figure 1). Interaction between the MTHFR 677C>T and a common polymorphism in the reduced folate carrier gene (RFC 80G>A) has recently been reported.15 The reduced folate carrier facilitates the transport of 5-methylTHF from the circulation to peripheral cells. The G-to-A transition at nucleotide 80 replaces an arginine with a histidine in the protein. Although it is unknown whether this change alters folate transport, the 80AA variant has been associated with higher plasma folate levels. Further, an increased risk of having a child with spina bifida has been associated with the maternal 80GG genotype.16 A similar association in risk of spina bifida has been reported for the MTR 2756G allele.17 MTR is a cobalamin-dependent enzyme that catalyzes the methyl transfer from homocysteine to methionine, thus playing a critical role in maintaining adequate intracellular SAM levels for DNA methylation. The MTR 2756A>G polymorphism converts an aspartic acid to a glycine residue and has been predicted to alter enzyme activity that may affect DNA methylation processes, yet its function remains controversial.
Overview of the human folate metabolic pathway. SAM indicates S-adenosylmethionine; SAH, S-adenosylhomocysteine; DHF, dihydrofolate; DHFR, dihydrofolate reductase; THF, tetrahydrofolate; SHMT, serine hydroxymethyltransferase; 5,10-methyleneTHF, 5,10-methylenetetrahydrofolate; MTHFR, 5,10-methylenetetrahydrofolate reductase; 5-methylTHF, 5-methyltetrahydrofolate; 10-formylTHF, 10-formyltetrahydrofolate; MTR, methionine synthase; TYMS, thymidylate synthase; dTMP, deoxythymidine monophosphate; B12, vitamin B12; and dUMP, deoxyuridine monophosphate.
Overview of the human folate metabolic pathway. SAM indicates S-adenosylmethionine; SAH, S-adenosylhomocysteine; DHF, dihydrofolate; DHFR, dihydrofolate reductase; THF, tetrahydrofolate; SHMT, serine hydroxymethyltransferase; 5,10-methyleneTHF, 5,10-methylenetetrahydrofolate; MTHFR, 5,10-methylenetetrahydrofolate reductase; 5-methylTHF, 5-methyltetrahydrofolate; 10-formylTHF, 10-formyltetrahydrofolate; MTR, methionine synthase; TYMS, thymidylate synthase; dTMP, deoxythymidine monophosphate; B12, vitamin B12; and dUMP, deoxyuridine monophosphate.
Given the importance of folate in cancer risk, chromosomal integrity, and immune function, we studied whether polymorphisms in the folate-metabolizing and transport genes TYMS, MTHFR, SHMT1, MTR, and RFC influence non-Hodgkin lymphoma (NHL) risk. We examined the relationship between these polymorphisms involved in folate metabolism and NHL using DNA isolated from biospecimens collected from participants in a large case-control study of NHL in the San Francisco Bay Area.
Patients, materials, and methods
Study population
In a study conducted in the San Francisco Bay Area between 1988 and 1995, NHL patients between 21 and 74 years of age who were residents of 1 of the 6 Bay Area counties at the time of initial NHL diagnosis were identified by the Northern California Cancer Center's rapid case ascertainment. There were 1593 eligible patients (284 HIV positive) who completed in-person interviews (72% response rate). Random-digit dial was used to identify control participants and was supplemented by random sampling of the Health Care Financing Administration files for participants aged 65 years or older. Controls were frequency matched to patients by age within 5 years, sex, and county of residence. There were 2515 (78% response rate) eligible control participants (111 HIV positive) who completed in-person interviews. The racial/ethnic mix of the population included in these analyses was self-reported as white Hispanic (6%), white non-Hispanic (84%), black (4%), Asian (5%), and other (1%). This distribution of race did not differ by case-control status (Kruskal-Wallis, P = .71). Because of the small number of nonwhite participants and the potential for population stratification, results are presented for white participants only. Additional details have been published in earlier manuscripts.18-21
Biologic samples
Participants who had no history of chemotherapy within the past 3 months and met other requirements, including no current use of blood thinners or having had a Port-a-Cath in place, were eligible to participate in the laboratory portion of the study. Among eligible participants, blood specimens were obtained from 62% of patients and 62% of controls. Study protocols were approved by the University of California, San Francisco (UCSF) Committee on Human Research; participants provided written informed consent prior to interview, and collection of blood specimens was performed in accordance with the Declaration of Helsinki. The blood was processed using Ficoll-Paque separation and the lymphocytes were cryopreserved in liquid nitrogen. Personal identifiers were removed from all samples sent by UCSF investigators to the lab in Berkeley for DNA isolation of 458 cases and 812 control participants. DNA was isolated using a modified QIAamp DNA Blood Maxi Kit protocol (QIAgen, Santa Clarita, CA) and was quantified using PicoGreen dsDNA Quantitation kits (Molecular Probes, Eugene, OR) according to the manufacturers' specifications.
Histopathology
NHL histologic subtype and grade were rereviewed by an expert pathologist for 97% of all NHL study patients and classified using the Working Formulation (NHL Classification Project). To better reflect the Revised European American Lymphoma (REAL) Classification established in 1994 after the study recruitment was complete, Working Formulation diffuse large cell lymphoma (DLCL) and immunoblastic lymphoma were combined for the DLCL subtype and Working Formulation follicular small, mixed, and large cell lymphomas were combined for the follicular lymphoma (FL) subtype.
Genotyping
Genotyping was performed using Assays-by-Design supplied by Applied Biosystems (ABI; Foster City, CA). Reactions were performed with the following protocol on a GeneAmp polymerase chain reaction (PCR) 9700 or 7700 ABI Sequence Detection System: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. A post-PCR plate read on the 7700 was used to determine genotype. Probes and primer sets used for the TYMS 1494del6, TYMS IVS6 -68C>T, TYMS 1053C>T, TYMS 1122A>G, MTHFR 677C>T, MTHFR 1298A>C, SHMT1 1420C>T, MTR 2756A>G, and RFC 80G>A polymorphisms are listed in Table 1. The TYMS 3R → 2R 28-bp repeat was detected using a protocol previously described,5 and primer sets for the detection of this polymorphism are listed in Table 1. Linkage analysis for the TYMS and MTHFR polymorphisms was investigated using the Cocaphase program.22
Primers and TaqMan probes used for folate polymorphisms
SNP (RS no.), probe/primer . | 5′-3′ sequence . |
|---|---|
| TYMS 3R → 2R | |
| F | GTGGCTCCTGCGTTTCCCCC |
| R | CCGCGCCATGCCTGTGGCCGGCTCGGAGC |
| TYMS 1494del6 (16430) | |
| F | AGCTGAGTAACACCATCGATCATG |
| R | GGACGAATGCAGAACACTTCTTTA |
| AAGTTA | VIC-TGGTTATGAACTTTAAAGTT |
| Deletion | 6FAM-TGGTTATGAACTTTATAGTTG |
| TYMS IVS6 -68 C>T (1059394) | |
| F | TGCCATAATTGTACGACCTGTTGT |
| R | AGAACTTTGTTGATCACATCCTGTGT |
| C | VIC-CTCATGTCCGTGAAAC |
| T | 6FAM-CTCATGTCCATGAAAC |
| TYMS 1053C>T (699517) | |
| F | GAAATGGCTGTTTAGGGTGCTTT |
| R | CCCAACCCCTAAAGACTGACAATA |
| C | VIC-AAGGAGCTCGAAGGA |
| T | 6FAM-CAAAGGAGCTTGAAGG |
| TYMS 1122A>G (2790) | |
| F | GGATGCCGAGGTAAAAGTTCTTT |
| R | GATAGGTCACGGACAGATTTTTGA |
| A | VIC-CCTAGTTCCTTTTTCTTT |
| G | 6FAM-CCTAGTTCCTTCTTCTT |
| MTHFR 677C>T (1801133) | |
| F | CGGTGCATGCCTTCACAA |
| R | CTGACCTGAAGCACTTGAAGGA |
| C | VIC-TGATGAAATCGGCTCC |
| T | 6FAM-ATGATGAAATCGACTCC |
| MTHFR 1298A>C (1801131) | |
| F | CCCGAGAGGTAAAGAACAAAGACTT |
| R | GGAGGAGCTGCTGAAGATGTG |
| A | VIC-CAAAGACACTTTCTTC |
| C | 6FAM-AGACACTTGCTTCACT |
| SHMTI 1420C>T (1979277) | |
| F | CAGAGCCACCCTGAAAGAGTTC |
| R | AGTGGGCCCGCTCCTTTA |
| T | VIC-CGCCTCTTTCTTC |
| C | 6FAM-CGCCTCTCTCTTC |
| MTR 2756A>G (1805087) | |
| F | GAATACTTTGAGGAAATCATGGAAGA |
| R | TCTGTTTCTACCACTTACCTTGAGAGACT |
| A | VIC-AGACAGGACCATTATG |
| G | 6FAM-ACAGGGCCATTATG |
| RFC 80G>A (1051266) | |
| F | GGCCTGACCCCGAGCT |
| R | AGCCGTAGAAGCAAAGGTAGCA |
| G | VIC-CACGAGGCGCCGC |
| A | 6FAM-CGAGGTGCCGCCAG |
SNP (RS no.), probe/primer . | 5′-3′ sequence . |
|---|---|
| TYMS 3R → 2R | |
| F | GTGGCTCCTGCGTTTCCCCC |
| R | CCGCGCCATGCCTGTGGCCGGCTCGGAGC |
| TYMS 1494del6 (16430) | |
| F | AGCTGAGTAACACCATCGATCATG |
| R | GGACGAATGCAGAACACTTCTTTA |
| AAGTTA | VIC-TGGTTATGAACTTTAAAGTT |
| Deletion | 6FAM-TGGTTATGAACTTTATAGTTG |
| TYMS IVS6 -68 C>T (1059394) | |
| F | TGCCATAATTGTACGACCTGTTGT |
| R | AGAACTTTGTTGATCACATCCTGTGT |
| C | VIC-CTCATGTCCGTGAAAC |
| T | 6FAM-CTCATGTCCATGAAAC |
| TYMS 1053C>T (699517) | |
| F | GAAATGGCTGTTTAGGGTGCTTT |
| R | CCCAACCCCTAAAGACTGACAATA |
| C | VIC-AAGGAGCTCGAAGGA |
| T | 6FAM-CAAAGGAGCTTGAAGG |
| TYMS 1122A>G (2790) | |
| F | GGATGCCGAGGTAAAAGTTCTTT |
| R | GATAGGTCACGGACAGATTTTTGA |
| A | VIC-CCTAGTTCCTTTTTCTTT |
| G | 6FAM-CCTAGTTCCTTCTTCTT |
| MTHFR 677C>T (1801133) | |
| F | CGGTGCATGCCTTCACAA |
| R | CTGACCTGAAGCACTTGAAGGA |
| C | VIC-TGATGAAATCGGCTCC |
| T | 6FAM-ATGATGAAATCGACTCC |
| MTHFR 1298A>C (1801131) | |
| F | CCCGAGAGGTAAAGAACAAAGACTT |
| R | GGAGGAGCTGCTGAAGATGTG |
| A | VIC-CAAAGACACTTTCTTC |
| C | 6FAM-AGACACTTGCTTCACT |
| SHMTI 1420C>T (1979277) | |
| F | CAGAGCCACCCTGAAAGAGTTC |
| R | AGTGGGCCCGCTCCTTTA |
| T | VIC-CGCCTCTTTCTTC |
| C | 6FAM-CGCCTCTCTCTTC |
| MTR 2756A>G (1805087) | |
| F | GAATACTTTGAGGAAATCATGGAAGA |
| R | TCTGTTTCTACCACTTACCTTGAGAGACT |
| A | VIC-AGACAGGACCATTATG |
| G | 6FAM-ACAGGGCCATTATG |
| RFC 80G>A (1051266) | |
| F | GGCCTGACCCCGAGCT |
| R | AGCCGTAGAAGCAAAGGTAGCA |
| G | VIC-CACGAGGCGCCGC |
| A | 6FAM-CGAGGTGCCGCCAG |
RS indicates reference SNP.
Quality control procedures
Methodology verification of TaqMan genotyping assays was made by running 96 Coriell samples of known genotypes (as posted on the National Cancer Institute Single Nucleotide Polymorphism 500 [NCI SNP 500] website23 ) for each of the MTHFR 677C>T, MTHFR 1298A>C, TYMS 1053C>T, and TYMS 1122A>G loci. Verification of other TaqMan assays was performed either by direct sequencing or using standard restriction fragment length polymorphism analysis. For added quality control, 5% of samples were selected at random for repeat analysis using our standard TaqMan genotyping protocol. Four independent control samples were included and analyzed on each 96-well plate.
Statistical analyses
Analyses were restricted to HIV-negative white Hispanics and white non-Hispanics using unconditional logistic regression to obtain odds ratios (ORs). Initially, we used univariate analyses to examine the possible association between each polymorphism and NHL risk. We also investigated possible interactions between genes by including multiplicative terms in a logistic regression model. The presence of multiplicative interaction was tested using a likelihood ratio statistic comparing (nested) models with and without the interaction terms. Note, that a model with the null hypothesis of interaction rejected (ie, P value of likelihood ratio test < .05) suggests that the OR for lymphoma comparing alleles in one gene (eg, heterozygous to homozygous mutant) differ by genotype of the other gene in the model. We then treated the number of mutant variants as continuous variables and performed trend analyses, again using logistic regression. All statistical tests were 2-sided and results were considered significant for P of .05 or less; note, we are only controlling the type I error rate one test at a time and have not controlled the family-wise error rate for multiple tests. Results are referred to as borderline significant for P values greater than .05 and less than or equal to .10. In our analyses, we focused on TYMS because of the previous findings of an association with leukemia and lymphoma and also because it plays a crucial role in the balance of deoxynucleotides for DNA synthesis.
Results
Although many demographic characteristics of the study population have been described previously,19,24 selected characteristics of participants in the main study are presented for the reader's convenience in Table 2. Genotype distributions among controls were in conformity with Hardy-Weinberg equilibrium (HWE) with the exception of the TYMS 1494del6, MTR 2756A>G, and RFC 80G>A polymorphisms. However, upon restricting the analysis to whites (white Hispanics and white non-Hispanics), all genotype frequencies were consistent with HWE expectations. Thus, the genotype distributions among all white NHL cases and controls for the TYMS 3R → 2R, TYMS 1494del6, MTHFR 677C>T, MTHFR 1298A>C, SHMT1 1420C>T, MTR 2756A>G, and RFC 80G>A polymorphisms are shown in Table 3.
Demographic characteristics of population-based HIV-negative NHL patients and control participants (San Francisco Bay Area, CA)
Characteristic . | Cases, N = 1304 . | Controls, N = 2402 . |
|---|---|---|
| Age, y* | 56.2 ± 13.1 | 52.3 ± 14.4 |
| Sex, no. (%) | ||
| Men | 725 (56) | 1566 (65) |
| Women | 579 (44) | 836 (35) |
| Race, no. (%) | ||
| White Hispanic | 108 (8) | 168 (7) |
| White non-Hispanic | 1013 (78) | 1944 (81) |
| Black/African American | 67 (5) | 132 (6) |
| Asian/Pacific Islander | 95 (7) | 119 (5) |
| Other | 21 (2) | 39 (2) |
| Education, no. (%) | ||
| Fewer than 12 y | 155 (12) | 157 (7) |
| 12 y | 379 (29) | 532 (22) |
| 13 y to fewer than 16 y | 297 (23) | 608 (25) |
| 16 y or more | 472 (36) | 1104 (46) |
Characteristic . | Cases, N = 1304 . | Controls, N = 2402 . |
|---|---|---|
| Age, y* | 56.2 ± 13.1 | 52.3 ± 14.4 |
| Sex, no. (%) | ||
| Men | 725 (56) | 1566 (65) |
| Women | 579 (44) | 836 (35) |
| Race, no. (%) | ||
| White Hispanic | 108 (8) | 168 (7) |
| White non-Hispanic | 1013 (78) | 1944 (81) |
| Black/African American | 67 (5) | 132 (6) |
| Asian/Pacific Islander | 95 (7) | 119 (5) |
| Other | 21 (2) | 39 (2) |
| Education, no. (%) | ||
| Fewer than 12 y | 155 (12) | 157 (7) |
| 12 y | 379 (29) | 532 (22) |
| 13 y to fewer than 16 y | 297 (23) | 608 (25) |
| 16 y or more | 472 (36) | 1104 (46) |
There were 395 HIV-positive participants (284 cases, 111 controls) and 5 participants with HIV status unknown.
Age at diagnosis for patients and at interview for controls, mean ± standard deviation.
TYMS, MTHFR, SHMT1, MTR, and RFC genotype frequencies, odds ratios (ORs), and 95% confidence intervals (Cls) in NHL cases and controls using wild types as the referent groups
. | Control, no. (%) . | All NHL cases . | . | . | DLCL . | . | . | FL . | . | . | Other . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype . | . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | ||||||||
| TYMS 3R → 2R | |||||||||||||||||||||
| 3R/3R | 209 (29) | 97 (29) | 1.1 (.77-1.6) | .53 | 29 (27) | 1.0 (.55-1.8) | .99 | 33 (27) | 1.2 (.66-2.1) | .56 | 35 (33) | 1.2 (.68-2.1) | .53 | ||||||||
| 3R/2R | 346 (48) | 170 (51) | 1.2 (.85-1.7) | .31 | 55 (52) | 1.1 (.67-1.9) | .62 | 67 (55) | 1.5 (.86-2.5) | .16 | 48 (46) | 1.0 (.58-1.7) | .99 | ||||||||
| 2R/2R | 158 (22) | 65 (20) | 1.0 | NA | 22 (21) | 1.0 | NA | 21 (17) | 1.0 | NA | 22 (21) | 1.0 | NA | ||||||||
| 3R/3R vs 3R/2R | .94 (.70-1.3) | .71 | .87 (.54-1.4) | .58 | .81 (.52-1.3) | .37 | 1.2 (.76-1.9) | .43 | |||||||||||||
| SNA | 18 | 5 | 2 | 1 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS 1494del6 | |||||||||||||||||||||
| 6bp+6bp+ | 335 (46) | 166 (50) | 1.0 | NA | 58 (54) | 1.0 | NA | 62 (51) | 1.0 | NA | 47 (45) | 1.0 | NA | ||||||||
| 6bp+6bp− | 310 (43) | 144 (43) | .94 (.71-1.2) | .64 | 45 (42) | .85 (.56-1.3) | .46 | 51 (42) | .89 (.59-1.3) | .56 | 48 (46) | 1.1 (.72-1.7) | .65 | ||||||||
| 6bp−6bp− | 82 (11) | 23 (7) | .57 (.34-.94) | .02 | 4 (4) | .29 (.10-.82) | .01 | 9 (7) | .59 (.28-1.2) | .16 | 10 (10) | .87 (.42-1.8) | .71 | ||||||||
| −/− vs +/− | .61 (.37-1.0) | .05 | .34 (.12-.96) | .04 | .67 (.32-1.4) | .29 | .79 (.38-1.6) | .52 | |||||||||||||
| SNA | 4 | 4 | 1 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS IVS6 - 68C>T | |||||||||||||||||||||
| CC | 335 (46) | 168 (50) | 1.0 | NA | 58 (54) | 1.0 | NA | 62 (51) | 1.0 | NA | 48 (46) | 1.0 | NA | ||||||||
| CT | 307 (42) | 143 (43) | .92 (.71-1.2) | .59 | 46 (43) | .86 (.57-1.3) | .50 | 50 (41) | .88 (.58-1.3) | .53 | 47 (45) | 1.1 (.69-1.6) | .76 | ||||||||
| TT | 83 (11) | 22 (7) | .53 (.32-.88) | .01 | 4 (4) | .28 (.10-.80) | .01 | 9 (7) | .59 (.28-1.2) | .15 | 9 (9) | .76 (.36-1.6) | .47 | ||||||||
| TT vs CT | .57 (.34-.95) | .03 | .32 (.11-.92) | .03 | .67 (.31-1.4) | .29 | .71 (.33-1.5) | .37 | |||||||||||||
| SNA | 6 | 4 | 0 | 1 | 3 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS 1053C>T | |||||||||||||||||||||
| CC | 335 (46) | 166 (49) | 1.0 | NA | 58 (54) | 1.0 | NA | 62 (51) | 1.0 | NA | 46 (43) | 1.0 | NA | ||||||||
| CT | 310 (43) | 144 (43) | .94 (.71-1.2) | .64 | 45 (42) | .83 (.55-1.3) | .41 | 51 (42) | .89 (.59-1.3) | .56 | 48 (45) | 1.1 (.73-1.7) | .59 | ||||||||
| TT | 83 (11) | 26 (8) | .63 (.39-1.0) | .05 | 5 (5) | .34 (.13-.90) | .02 | 9 (7) | .59 (.28-1.2) | .15 | 12 (11) | 1.0 (.53-2.1) | .88 | ||||||||
| TT vs CT | .67 (.41-1.1) | .10 | .41 (.16-1.1) | .07 | .66 (.31-1.4) | .27 | .93 (.47-1.8) | .84 | |||||||||||||
| SNA | 3 | 1 | 0 | 0 | 1 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS 1122A>G | |||||||||||||||||||||
| AA | 465 (64) | 217 (65) | 1.0 | NA | 71 (66) | 1.0 | NA | 81 (67) | 1.0 | NA | 65 (61) | 1.0 | NA | ||||||||
| AG | 228 (31) | 105 (31) | .99 (.74-1.3) | .93 | 33 (31) | .94 (.56-1.5) | .81 | 35 (29) | .88 (.57-1.3) | .56 | 37 (35) | 1.2 (.75-1.8) | .50 | ||||||||
| GG | 35 (5) | 12 (4) | .73 (.37-1.4) | .37 | 3 (3) | .56 (.17-1.9) | .90 | 5 (4) | .82 (.31-2.1) | .69 | 4 (4) | .82 (.28-2.4) | .14 | ||||||||
| GG vs AG | .74 (.37-1.5) | .40 | .59 (.17-2.0) | .40 | .93 (.34-2.5) | .89 | .70 (.23-2.1) | .53 | |||||||||||||
| SNA | 3 | 3 | 1 | 1 | 1 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| MTHFR 677C>T | |||||||||||||||||||||
| CC | 288 (40) | 122 (37) | 1.0 | NA | 37 (35) | 1.0 | NA | 44 (36) | 1.0 | NA | 41 (39) | 1.0 | NA | ||||||||
| CT | 350 (48) | 160 (48) | 1.1 (.81-1.4) | .60 | 51 (48) | 1.1 (.72-1.8) | .58 | 55 (45) | 1.0 (.67-1.6) | .90 | 54 (51) | 1.1 (.70-1.7) | .72 | ||||||||
| TT | 84 (12) | 52 (16) | 1.5 (.97-2.2) | .07 | 19 (18) | 1.8 (.96-3.2) | .06 | 23 (19) | 1.8 (1.0-3.1) | .04 | 10 (10) | .84 (.40-1.7) | .63 | ||||||||
| SNA | 9 | 3 | 1 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| MTHFR 1298A>C | |||||||||||||||||||||
| AA | 341 (47) | 178 (53) | 1.0 | NA | 59 (56) | 1.0 | NA | 63 (52) | 1.0 | NA | 56 (53) | 1.0 | NA | ||||||||
| AC | 310 (43) | 128 (38) | .79 (.60-1.0) | .09 | 38 (36) | .71 (.46-1.1) | .12 | 49 (40) | .86 (.57-1.3) | .45 | 41 (39) | .80 (.52-1.2) | .32 | ||||||||
| CC | 71 (10) | 27 (8) | .73 (.45-1.2) | .19 | 9 (8) | .73 (.35-1.5) | .41 | 10 (8) | .76 (.37-1.6) | .46 | 8 (8) | .69 (.31-1.5) | .34 | ||||||||
| SNA | 9 | 4 | 2 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| SHMT1 1420C>T | |||||||||||||||||||||
| CC | 355 (49) | 150 (45) | 1.0 | NA | 47 (44) | 1.0 | NA | 61 (50) | 1.0 | NA | 42 (40) | 1.0 | NA | ||||||||
| CT | 302 (41) | 140 (42) | 1.1 (.83-1.4) | .51 | 46 (43) | 1.1 (.74-1.8) | .53 | 45 (37) | .87 (.57-1.3) | .50 | 49 (47) | 1.4 (.88-2.1) | .16 | ||||||||
| TT | 72 (10) | 43 (13) | 1.4 (.92-2.2) | .11 | 14 (13) | 1.5 (.77-2.8) | .24 | 16 (13) | 1.3 (.70-2.4) | .40 | 13 (13) | 1.5 (.78-3.0) | .21 | ||||||||
| SNA | 2 | 4 | 1 | 0 | 3 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| MTR 2756A>G | |||||||||||||||||||||
| AA | 489 (67) | 201 (61) | 1.0 | NA | 63 (60) | 1.0 | NA | 76 (63) | 1.0 | NA | 62 (59) | 1.0 | NA | ||||||||
| AG/GG | 242 (33) | 129 (39) | 1.3 (.99-1.7) | .06 | 42 (40) | 1.3 (.88-2.0) | .16 | 44 (37) | 1.2 (.78-1.7) | .44 | 43 (41) | 1.4 (.92-2.1) | .11 | ||||||||
| SNA | 0 | 7 | 3 | 2 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| RFC 80G>A | |||||||||||||||||||||
| GG | 266 (36) | 109 (32) | 1.0 | NA | 34 (32) | 1.0 | NA | 37 (30) | 1.0 | NA | 38 (36) | 1.0 | NA | ||||||||
| GA | 331 (45) | 158 (48) | 1.2 (.87-1.6) | .31 | 55 (51) | 1.3 (.82-2.0) | .26 | 57 (48) | 1.2 (.79-1.9) | .35 | 46 (44) | .97 (.61-1.5) | .91 | ||||||||
| AA | 132 (18) | 67 (20) | 1.2 (.86-1.8) | .26 | 18 (17) | 1.1 (.58-2.0) | .83 | 28 (23) | 1.5 (.89-2.6) | .12 | 21 (20) | 1.1 (.63-2.0) | .71 | ||||||||
| SNA | 2 | 3 | 1 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
. | Control, no. (%) . | All NHL cases . | . | . | DLCL . | . | . | FL . | . | . | Other . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype . | . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | ||||||||
| TYMS 3R → 2R | |||||||||||||||||||||
| 3R/3R | 209 (29) | 97 (29) | 1.1 (.77-1.6) | .53 | 29 (27) | 1.0 (.55-1.8) | .99 | 33 (27) | 1.2 (.66-2.1) | .56 | 35 (33) | 1.2 (.68-2.1) | .53 | ||||||||
| 3R/2R | 346 (48) | 170 (51) | 1.2 (.85-1.7) | .31 | 55 (52) | 1.1 (.67-1.9) | .62 | 67 (55) | 1.5 (.86-2.5) | .16 | 48 (46) | 1.0 (.58-1.7) | .99 | ||||||||
| 2R/2R | 158 (22) | 65 (20) | 1.0 | NA | 22 (21) | 1.0 | NA | 21 (17) | 1.0 | NA | 22 (21) | 1.0 | NA | ||||||||
| 3R/3R vs 3R/2R | .94 (.70-1.3) | .71 | .87 (.54-1.4) | .58 | .81 (.52-1.3) | .37 | 1.2 (.76-1.9) | .43 | |||||||||||||
| SNA | 18 | 5 | 2 | 1 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS 1494del6 | |||||||||||||||||||||
| 6bp+6bp+ | 335 (46) | 166 (50) | 1.0 | NA | 58 (54) | 1.0 | NA | 62 (51) | 1.0 | NA | 47 (45) | 1.0 | NA | ||||||||
| 6bp+6bp− | 310 (43) | 144 (43) | .94 (.71-1.2) | .64 | 45 (42) | .85 (.56-1.3) | .46 | 51 (42) | .89 (.59-1.3) | .56 | 48 (46) | 1.1 (.72-1.7) | .65 | ||||||||
| 6bp−6bp− | 82 (11) | 23 (7) | .57 (.34-.94) | .02 | 4 (4) | .29 (.10-.82) | .01 | 9 (7) | .59 (.28-1.2) | .16 | 10 (10) | .87 (.42-1.8) | .71 | ||||||||
| −/− vs +/− | .61 (.37-1.0) | .05 | .34 (.12-.96) | .04 | .67 (.32-1.4) | .29 | .79 (.38-1.6) | .52 | |||||||||||||
| SNA | 4 | 4 | 1 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS IVS6 - 68C>T | |||||||||||||||||||||
| CC | 335 (46) | 168 (50) | 1.0 | NA | 58 (54) | 1.0 | NA | 62 (51) | 1.0 | NA | 48 (46) | 1.0 | NA | ||||||||
| CT | 307 (42) | 143 (43) | .92 (.71-1.2) | .59 | 46 (43) | .86 (.57-1.3) | .50 | 50 (41) | .88 (.58-1.3) | .53 | 47 (45) | 1.1 (.69-1.6) | .76 | ||||||||
| TT | 83 (11) | 22 (7) | .53 (.32-.88) | .01 | 4 (4) | .28 (.10-.80) | .01 | 9 (7) | .59 (.28-1.2) | .15 | 9 (9) | .76 (.36-1.6) | .47 | ||||||||
| TT vs CT | .57 (.34-.95) | .03 | .32 (.11-.92) | .03 | .67 (.31-1.4) | .29 | .71 (.33-1.5) | .37 | |||||||||||||
| SNA | 6 | 4 | 0 | 1 | 3 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS 1053C>T | |||||||||||||||||||||
| CC | 335 (46) | 166 (49) | 1.0 | NA | 58 (54) | 1.0 | NA | 62 (51) | 1.0 | NA | 46 (43) | 1.0 | NA | ||||||||
| CT | 310 (43) | 144 (43) | .94 (.71-1.2) | .64 | 45 (42) | .83 (.55-1.3) | .41 | 51 (42) | .89 (.59-1.3) | .56 | 48 (45) | 1.1 (.73-1.7) | .59 | ||||||||
| TT | 83 (11) | 26 (8) | .63 (.39-1.0) | .05 | 5 (5) | .34 (.13-.90) | .02 | 9 (7) | .59 (.28-1.2) | .15 | 12 (11) | 1.0 (.53-2.1) | .88 | ||||||||
| TT vs CT | .67 (.41-1.1) | .10 | .41 (.16-1.1) | .07 | .66 (.31-1.4) | .27 | .93 (.47-1.8) | .84 | |||||||||||||
| SNA | 3 | 1 | 0 | 0 | 1 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| TYMS 1122A>G | |||||||||||||||||||||
| AA | 465 (64) | 217 (65) | 1.0 | NA | 71 (66) | 1.0 | NA | 81 (67) | 1.0 | NA | 65 (61) | 1.0 | NA | ||||||||
| AG | 228 (31) | 105 (31) | .99 (.74-1.3) | .93 | 33 (31) | .94 (.56-1.5) | .81 | 35 (29) | .88 (.57-1.3) | .56 | 37 (35) | 1.2 (.75-1.8) | .50 | ||||||||
| GG | 35 (5) | 12 (4) | .73 (.37-1.4) | .37 | 3 (3) | .56 (.17-1.9) | .90 | 5 (4) | .82 (.31-2.1) | .69 | 4 (4) | .82 (.28-2.4) | .14 | ||||||||
| GG vs AG | .74 (.37-1.5) | .40 | .59 (.17-2.0) | .40 | .93 (.34-2.5) | .89 | .70 (.23-2.1) | .53 | |||||||||||||
| SNA | 3 | 3 | 1 | 1 | 1 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| MTHFR 677C>T | |||||||||||||||||||||
| CC | 288 (40) | 122 (37) | 1.0 | NA | 37 (35) | 1.0 | NA | 44 (36) | 1.0 | NA | 41 (39) | 1.0 | NA | ||||||||
| CT | 350 (48) | 160 (48) | 1.1 (.81-1.4) | .60 | 51 (48) | 1.1 (.72-1.8) | .58 | 55 (45) | 1.0 (.67-1.6) | .90 | 54 (51) | 1.1 (.70-1.7) | .72 | ||||||||
| TT | 84 (12) | 52 (16) | 1.5 (.97-2.2) | .07 | 19 (18) | 1.8 (.96-3.2) | .06 | 23 (19) | 1.8 (1.0-3.1) | .04 | 10 (10) | .84 (.40-1.7) | .63 | ||||||||
| SNA | 9 | 3 | 1 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| MTHFR 1298A>C | |||||||||||||||||||||
| AA | 341 (47) | 178 (53) | 1.0 | NA | 59 (56) | 1.0 | NA | 63 (52) | 1.0 | NA | 56 (53) | 1.0 | NA | ||||||||
| AC | 310 (43) | 128 (38) | .79 (.60-1.0) | .09 | 38 (36) | .71 (.46-1.1) | .12 | 49 (40) | .86 (.57-1.3) | .45 | 41 (39) | .80 (.52-1.2) | .32 | ||||||||
| CC | 71 (10) | 27 (8) | .73 (.45-1.2) | .19 | 9 (8) | .73 (.35-1.5) | .41 | 10 (8) | .76 (.37-1.6) | .46 | 8 (8) | .69 (.31-1.5) | .34 | ||||||||
| SNA | 9 | 4 | 2 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| SHMT1 1420C>T | |||||||||||||||||||||
| CC | 355 (49) | 150 (45) | 1.0 | NA | 47 (44) | 1.0 | NA | 61 (50) | 1.0 | NA | 42 (40) | 1.0 | NA | ||||||||
| CT | 302 (41) | 140 (42) | 1.1 (.83-1.4) | .51 | 46 (43) | 1.1 (.74-1.8) | .53 | 45 (37) | .87 (.57-1.3) | .50 | 49 (47) | 1.4 (.88-2.1) | .16 | ||||||||
| TT | 72 (10) | 43 (13) | 1.4 (.92-2.2) | .11 | 14 (13) | 1.5 (.77-2.8) | .24 | 16 (13) | 1.3 (.70-2.4) | .40 | 13 (13) | 1.5 (.78-3.0) | .21 | ||||||||
| SNA | 2 | 4 | 1 | 0 | 3 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| MTR 2756A>G | |||||||||||||||||||||
| AA | 489 (67) | 201 (61) | 1.0 | NA | 63 (60) | 1.0 | NA | 76 (63) | 1.0 | NA | 62 (59) | 1.0 | NA | ||||||||
| AG/GG | 242 (33) | 129 (39) | 1.3 (.99-1.7) | .06 | 42 (40) | 1.3 (.88-2.0) | .16 | 44 (37) | 1.2 (.78-1.7) | .44 | 43 (41) | 1.4 (.92-2.1) | .11 | ||||||||
| SNA | 0 | 7 | 3 | 2 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
| RFC 80G>A | |||||||||||||||||||||
| GG | 266 (36) | 109 (32) | 1.0 | NA | 34 (32) | 1.0 | NA | 37 (30) | 1.0 | NA | 38 (36) | 1.0 | NA | ||||||||
| GA | 331 (45) | 158 (48) | 1.2 (.87-1.6) | .31 | 55 (51) | 1.3 (.82-2.0) | .26 | 57 (48) | 1.2 (.79-1.9) | .35 | 46 (44) | .97 (.61-1.5) | .91 | ||||||||
| AA | 132 (18) | 67 (20) | 1.2 (.86-1.8) | .26 | 18 (17) | 1.1 (.58-2.0) | .83 | 28 (23) | 1.5 (.89-2.6) | .12 | 21 (20) | 1.1 (.63-2.0) | .71 | ||||||||
| SNA | 2 | 3 | 1 | 0 | 2 | ||||||||||||||||
| Total | 731 | 337 | 108 | 122 | 107 | ||||||||||||||||
DLCL indicates diffuse large cell lymphoma; FL, follicular lymphoma; NA, not applicable; and SNA, samples not amplified.
Consistent with previous reports, the variant allele frequencies in the control group for the TYMS 3R → 2R and 1494del6 polymorphisms were 46.0% and 32.5%, respectively.13,25 Two percent of controls and 0.6% of cases possessed at least one allele encompassing between four and nine 28-bp repeats and were therefore excluded from further analysis of the TYMS 5′UTR 28-bp repeat. In a univariate analysis, the TYMS 3R → 2R 28-bp repeat polymorphism did not significantly affect the risk of NHL in this population. Univariate analysis of the TYMS 1494del6 polymorphism revealed that the homozygous variant (6bp-6bp-) genotype was inversely associated with NHL (OR = 0.57; 0.34-0.94), particularly with DLCL (OR = 0.29; 0.10-0.82). Simultaneously including both polymorphisms in the model did not alter the magnitude of the OR for the association between the 1494 6bp-6bp- genotype and NHL risk by more than 10% (data not shown). Furthermore, the TYMS 3R → 2R and TYMS 1494del6 alleles were in relatively strong linkage disequilibrium among controls (D′ = 0.90).
In the model containing terms representing multiplicative interaction between the genes, interaction was observed between the TYMS 3R → 2R 28-bp repeat and 1494del6 polymorphisms (likelihood ratio test P = .01); the estimated odds ratio for the 2R allele was influenced by the presence or absence of the 6-bp deletion in the 3′UTR (Table 4). Specifically, the combined TYMS 2R2R/6bp+6bp+ genotype was associated with a 1.5-fold increased risk of NHL compared with the 3R3R/6bp+6bp+ genotype (OR = 1.5; 0.87-2.6). However, in the presence of a single copy of the 6bp- allele, an inverse association with NHL was observed (OR = 0.41; 0.20-0.87). This odds ratio was further reduced among those with 2 copies of the 6bp- allele (OR = 0.25; 0.03-2.1), although the confidence interval was wide.
Interaction between TYMS 3R → 2R and 1494del6 polymorphisms in all NHL cases and controls
. | Controls, no. . | AII NHL cases, no. . | OR (95% CI) . | OR (95% CI)* . |
|---|---|---|---|---|
| At TYMS 6bp+6bp+ | ||||
| 3R3R | 77 | 26 | 1.0 (referent) | 1.0 |
| 3R2R | 151 | 86 | 1.7 (1.0-2.8) | 1.7 (1.0-2.8) |
| 2R2R | 102 | 52 | 1.5 (.87-2.6) | 1.5 (.87-2.6) |
| At TYMS 6bp+6bp− | ||||
| 3R3R | 90 | 52 | 1.0 (referent) | 1.7 (.97-3.0) |
| 3R2R | 168 | 79 | .81 (.53-1.3) | 1.4 (.83-2.3) |
| 2R2R | 46 | 11 | .41 (.20-.87) | .71 (.32-1.6) |
| At TYMS 6bp−6bp− | ||||
| 3R3R | 42 | 17 | 1.0 (referent) | 1.2 (.58-2.5) |
| 3R2R | 27 | 4 | .37 (.11-1.2) | .44 (.14-1.4) |
| 2R2R | 10 | 1 | .25 (.03-2.1) | .30 (.04-2.5) |
. | Controls, no. . | AII NHL cases, no. . | OR (95% CI) . | OR (95% CI)* . |
|---|---|---|---|---|
| At TYMS 6bp+6bp+ | ||||
| 3R3R | 77 | 26 | 1.0 (referent) | 1.0 |
| 3R2R | 151 | 86 | 1.7 (1.0-2.8) | 1.7 (1.0-2.8) |
| 2R2R | 102 | 52 | 1.5 (.87-2.6) | 1.5 (.87-2.6) |
| At TYMS 6bp+6bp− | ||||
| 3R3R | 90 | 52 | 1.0 (referent) | 1.7 (.97-3.0) |
| 3R2R | 168 | 79 | .81 (.53-1.3) | 1.4 (.83-2.3) |
| 2R2R | 46 | 11 | .41 (.20-.87) | .71 (.32-1.6) |
| At TYMS 6bp−6bp− | ||||
| 3R3R | 42 | 17 | 1.0 (referent) | 1.2 (.58-2.5) |
| 3R2R | 27 | 4 | .37 (.11-1.2) | .44 (.14-1.4) |
| 2R2R | 10 | 1 | .25 (.03-2.1) | .30 (.04-2.5) |
Likelihood ratio test for interaction: P = .01.
OR and 95% CI computed using 3R3R/6bp+6bp+ as referent.
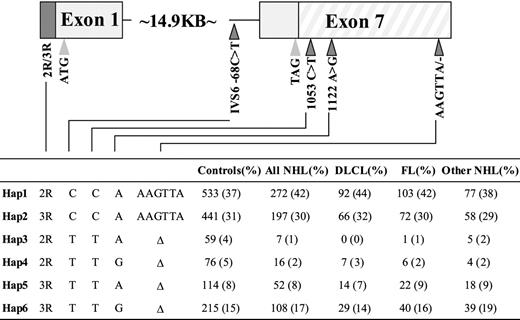
To investigate the relationship between TYMS polymorphisms and NHL risk in more detail, we evaluated 2 additional polymorphisms located in the 3′UTR of exon 7, TYMS 1053C>T and 1122A>G, and an additional polymorphism, IVS6 -68C>T, located in intron 6, 68 bp upstream of exon 7 (Figure 2). These polymorphisms were identified using the Ensembl Genome Browser by the Wellcome Trust Sanger Institute.26
White TYMS polymorphisms and haplotype frequencies. The first and last exons of TYMS and the relative positions of the polymorphisms that were investigated are depicted. Thin lines indicate introns; boxes, exons (3′UTR is striped, 3R → 2R is dark fill, coding is light fill); dark triangles, polymorphisms; and light triangles, start and stop codons. Haplotype frequencies were determined using the Cocaphase program.
White TYMS polymorphisms and haplotype frequencies. The first and last exons of TYMS and the relative positions of the polymorphisms that were investigated are depicted. Thin lines indicate introns; boxes, exons (3′UTR is striped, 3R → 2R is dark fill, coding is light fill); dark triangles, polymorphisms; and light triangles, start and stop codons. Haplotype frequencies were determined using the Cocaphase program.
Genotype distributions and risk estimates obtained in our analyses of the IVS6 -68C>T and 1053C>T polymorphisms were nearly identical to those observed for 1494del6 (Table 3). Specifically, reduced risk estimates for NHL were found for homozygous variants IVS6 -68TT (OR = 0.53; 0.32-0.88) and 1053TT (OR = 0.63; 0.39-1.0) that were even greater for DLCL (OR = 0.28; 0.10-0.80 and OR = 0.34; 0.13-0.90, respectively). No significant differences in genotype frequencies were observed between the cases and controls for the TYMS 1122A>G polymorphism.
Cocaphase analysis of all 5 TYMS polymorphisms revealed that 6 common haplotypes accounted for more than 99% of the estimated haplotypes among whites and that the IVS6 -68C>T, 1053C>T, and 1494del6 polymorphisms were in complete linkage disequilibrium (D′ = 1.0; Figure 2). There was also linkage disequilibrium between these 3 polymorphisms and 1122A>G (D′ = 0.99) and to a lesser extent with the 28-bp repeat (D′ = 0.36). The haplotypes that contained the 5′UTR 2R and 3′UTR 6bp- alleles, Hap3 and Hap4, were associated with significantly reduced risk for NHL (OR = 0.23, P < .001; and OR = 0.41, P = .02, respectively; Table 5). Subgroup analysis showed that Hap3 also was associated with reduced risk estimates for DLCL (OR = 0.01; P < .001) and FL (OR = 0.13; P = .01).
TYMS haplotypes and NHL risk
. | AII NHL . | . | DLCL . | . | FL . | . | Other NHL . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | OR . | P . | OR . | P . | OR . | P . | OR . | P . | ||||
| Hap1 | 1.00 | .23 | 1.00 | .15 | 1.00 | .37 | 1.00 | .66 | ||||
| Hap2 | 0.88 | .46 | 0.88 | .33 | 0.85 | .71 | 0.92 | .81 | ||||
| Hap3 | 0.23 | < .001 | 0.01 | < .001 | 0.13 | .01 | 0.54 | .23 | ||||
| Hap4 | 0.41 | .02 | 0.50 | .16 | 0.41 | .13 | 0.36 | .08 | ||||
| Hap5 | 0.90 | .51 | 0.72 | .13 | 0.98 | .95 | 1.09 | .99 | ||||
| Hap6 | 0.99 | .73 | 0.80 | .59 | 0.96 | .90 | 1.24 | .32 | ||||
. | AII NHL . | . | DLCL . | . | FL . | . | Other NHL . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | OR . | P . | OR . | P . | OR . | P . | OR . | P . | ||||
| Hap1 | 1.00 | .23 | 1.00 | .15 | 1.00 | .37 | 1.00 | .66 | ||||
| Hap2 | 0.88 | .46 | 0.88 | .33 | 0.85 | .71 | 0.92 | .81 | ||||
| Hap3 | 0.23 | < .001 | 0.01 | < .001 | 0.13 | .01 | 0.54 | .23 | ||||
| Hap4 | 0.41 | .02 | 0.50 | .16 | 0.41 | .13 | 0.36 | .08 | ||||
| Hap5 | 0.90 | .51 | 0.72 | .13 | 0.98 | .95 | 1.09 | .99 | ||||
| Hap6 | 0.99 | .73 | 0.80 | .59 | 0.96 | .90 | 1.24 | .32 | ||||
See Figure 2 for details of haplotypes.
In the control population, variant allele frequencies for MTHFR 677C>T and 1298A>C were 36.0% and 31.5%, consistent with previous studies in white populations.27,28 In the main effects models that evaluated these polymorphisms individually, a 1.5-fold increased risk estimate for NHL was observed among those with the MTHFR 677TT genotype compared with MTHFR 677CC (OR = 1.5; 0.97-2.2), whereas the MTHFR 1298A>C genotype frequencies were similar between the cases and controls. In a Cocaphase analysis, the 2 MTHFR polymorphisms were in linkage disequilibrium (D′ = 0.98). Three haplotypes accounted for almost 100% of the estimated haplotypes (Table 6). HapB (677C/1298C) was associated with a borderline decrease in the NHL risk estimate (OR = 0.88; P = .07). In addition, HapC (677T/1298A) was associated with borderline increased risk for DLCL (OR = 1.2; P = .08) and for FL (OR = 1.3; P = .09).
MTHFR haplotype frequencies and odds ratios (ORs) in NHL cases and controls
. | . | . | . | AII NHL . | . | . | DLCL . | . | . | FL . | . | . | Other NHL . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | 677C>T . | 1298A>C . | Controls, no. (%) . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | ||||||||
| HapA | C | A | 477 (33) | 220 (33) | 1.00 | .98 | 67 (32) | 1.00 | .69 | 74 (30) | 1.00 | .41 | 79 (38) | 1.00 | .19 | ||||||||
| HapB | C | C | 449 (31) | 182 (27) | 0.88 | .07 | 56 (26) | 0.89 | .16 | 69 (28) | 0.99 | .36 | 57 (27) | 0.77 | .23 | ||||||||
| HapC | T | A | 515 (36) | 264 (40) | 1.11 | .08 | 89 (42) | 1.23 | .08 | 101 (41) | 1.30 | .09 | 74 (35) | 0.87 | .89 | ||||||||
. | . | . | . | AII NHL . | . | . | DLCL . | . | . | FL . | . | . | Other NHL . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | 677C>T . | 1298A>C . | Controls, no. (%) . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | No. (%) . | OR . | P . | ||||||||
| HapA | C | A | 477 (33) | 220 (33) | 1.00 | .98 | 67 (32) | 1.00 | .69 | 74 (30) | 1.00 | .41 | 79 (38) | 1.00 | .19 | ||||||||
| HapB | C | C | 449 (31) | 182 (27) | 0.88 | .07 | 56 (26) | 0.89 | .16 | 69 (28) | 0.99 | .36 | 57 (27) | 0.77 | .23 | ||||||||
| HapC | T | A | 515 (36) | 264 (40) | 1.11 | .08 | 89 (42) | 1.23 | .08 | 101 (41) | 1.30 | .09 | 74 (35) | 0.87 | .89 | ||||||||
Since the MTHFR 677C>T and 1298A>C polymorphisms are linked, we estimated both the unadjusted ORs (Table 3) as well as including both polymorphisms in a regression model to estimate the adjusted odds ratios. Model results revealed no association between either polymorphism and NHL or DLCL (data not shown). However, upon stratification by NHL histologic subtype, the MTHFR 677T allele was associated with borderline increased risk of FL for both 677CT (adjusted OR = 1.8; 0.92-3.4) and 677TT (adjusted OR = 1.8; 0.98-3.1) genotypes. Combined heterozygosity of MTHFR 677CT/1298AC did not modify the risk estimates.
The frequencies of the polymorphic SHMT1 1420T, MTR 2756G, and RFC 80A alleles in the controls were 30.5%, 19.0%, and 40.5%, respectively, and within the range of previous reports (Table 3).5,15,29 Due to the low frequency of the MTR 2756GG homozygous variants, ORs were computed for the MTR 2756AG/GG genotypes combined. In a main effects model, a small increased risk estimate for NHL was associated with the MTR 2756AG/GG genotype when MTR 2756AA was the referent group (OR = 1.3; 0.99-1.7). Stratification by subtype did not significantly alter risk estimates.
No interactions were detected between the TYMS, MTHFR, SHMT1, MTR, and RFC genes and risk of NHL. There was insufficient power to explore interactions by NHL subtype.
Discussion
In all, 10 polymorphisms were examined and haplotypes were generated for TYMS and MTHFR, making the present study the largest and most comprehensive to date to evaluate the association between genetic polymorphisms in folate-metabolizing genes and NHL risk. This study has provided further evidence that folate, particularly the DNA synthesis pathway, may play a role in risk of NHL. Whereas we observed similar genotype frequencies for the TYMS 3R → 2R polymorphism in our cases and controls, evaluation of the TYMS 1494del6 polymorphism revealed that those who harbored 2 copies of the 6-bp deletion had a 43% reduced NHL risk estimate compared with nondeletion homozygotes (6bp+6bp+). This effect was even greater for DLCL where the 6bp-6bp- genotype was associated with a 70% reduced risk estimate. Furthermore, we detected significant statistical interaction between the TYMS 3R → 2R and TYMS 1494del6 polymorphisms (P = .01). Namely, among those without the 6-bp deletion (6bp+6bp+), the 2R allele was associated with a 50% to 70% increased risk estimate for NHL. Yet, when estimating risk across TYMS 1494del6 genotype strata, an inverse association was apparent among those with the 2R allele compared with the 3R3R variant. However, estimates were based on few subjects (TYMS 2R2R/6bp-6bp-, 1 case and 10 controls; 3R2R/6bp-6bp-, 4 cases and 27 controls) and were imprecise.
The functional effect of the 1494 6-bp deletion is unknown, particularly with regard to how it may interact with the 28-bp tandem repeat in the 5′UTR. It is possible that this deletion in the 3′UTR may influence the cellular distribution, stability, and translational efficiency of TYMS mRNA, which may have a positive effect on TYMS enzyme production. The 28-bp tandem repeat sequence in the 5′UTR of the TYMS gene acts as a cis-enhancing element that affects TYMS expression. The 3R3R genotype, when compared with the 2R alleles, has been associated with enhanced mRNA translation efficiency,12 elevated intratu-moral TYMS mRNA30 and protein expression levels,12 and lower response rates and survival benefit from 5-fluorouracil-based chemotherapy.30,31 Its inhibition results in depletion of deoxythymidine triphosphate (dTTP) pools followed by thymineless death or, in some instances, high levels of uracil misincorporation in DNA followed by extensive repair by uracil DNA glycosylase and subsequent double-strand breaks in DNA. These double-strand breaks promote chromosomal instability, translocations, and aberrations that may contribute to increased cancer risk. We have previously shown an inverse association between the 3R allele and risk of adult ALL,5 and a similar association has recently been reported for lymphoma where a 60% increased odds ratio was associated with the 2R allele.8 The inconsistency between the elevated lymphoma risk estimate associated with the TYMS 2R allele reported by Hishida et al8 but not observed in the present study may reflect vast differences in allele frequencies found in Asian and white populations. The Hishida et al8 study was composed of a Japanese population where the 2R allele frequency was 14%, whereas in the present study of white Hispanics and white non-Hispanics, the 2R allele frequency was 46%.
Inverse associations between 2 of the less frequent TYMS haplotypes and NHL were also detected. In particular, TYMS Hap3 and 4 (Figure 2) accounted for only 4% and 5% of control and 1% and 2% of case haplotypes, respectively. Functional studies will be needed to discern if any of the tightly linked IVS6 -68C>T, 1053C>T, and 1494del6 polymorphisms are relevant to disease susceptibility and whether they are linked to another yet unknown susceptibility allele. Both the 1053C>T and 1494del6 polymorphisms occur in the 3′UTR and it may be that presence of the variant alleles result in enhanced mRNA stability and/or translational efficiency leading to increased fidelity of DNA synthesis and repair. Polymorphisms in UTRs may modulate 5′UTR-3′UTR interactions and the binding of associated regulatory factors.32 Additional studies will be necessary to explore the function of these and the TYMS 3R → 2R polymorphisms and their interaction. Nonetheless, given the tight linkage found, any one of these three 3′UTR polymorphisms may be used as a tag polymorphism in future association studies without the need to genotype all 3.
MTHFR plays a central role in directing the folate pool toward the remethylation of homocysteine to methionine at the expense of purine and DNA synthesis. The MTHFR 677C>T polymorphism results in a thermolabile enzyme with reduced activity that hinders DNA methylation while providing greater flux of folate for nucleotide synthesis. This may result in enhanced DNA repair by reducing levels of uracil that might otherwise be incorporated into DNA. However, reduced MTHFR enzyme activity also lowers availability of SAM required for methylation processes that can likewise promote hypomethylation of DNA,25,33 the effects of which are exacerbated under conditions of low folate status.34 The effects of the MTR 2756A>G polymorphism on homocysteine levels remain controversial,35,36 yet the variant 2756G allele has been inversely associated with a number of diseases.33,37,38 Previously, we reported an inverse association between the MTHFR 677T, MTHFR 1298C, and MTR 2756G alleles and ALL risk.4,5 Similar findings for MTHFR were reported for lymphoma by Matsuo et al9 where the MTHFR 677CC and 1298AA genotypes were associated with increased odds ratios in a study of 98 NHL patients, but these results were not corroborated in another study of 111 DLCL cases.39 Further, Matsuo's group9 reported that the MTR 2756 GG genotype conferred a 3.8-fold increased risk of lymphoma, whereas in another study of 151 NHL patients, the G allele was associated with a 60% reduced odds ratio.10
In the present study, positive associations were observed between the MTHFR 677TT genotype and FL and the MTR 2756AG/GG genotypes and overall NHL, suggesting that these polymorphisms may be modest predictors of disease risk. The MTHFR haplotype data corroborate the regression results showing that those who harbored the 677T allele (HapC) were at an increased risk of FL. The haplotype data also suggested that 677T was associated with increased risk for NHL and DLCL and that 1298C (HapB) may be inversely associated with NHL. However, larger studies are needed to provide sufficient power to assess these possible associations. These findings suggest that low folate and elevated homocysteine levels may increase FL risk. One possible mechanism may be that elevated homocysteine levels result in heightened global DNA hypomethylation that could potentially invoke chromosomal instability, reactivation of transposable elements, and/or loss of imprinting,40 factors that may contribute to increased lymphoma risk. The pathogenesis of FL involves the transformation of a germinal center-derived B cell with a t(14;18) chromosomal translocation resulting in a cell survival advantage through antiapoptotic mechanisms. Previous studies suggest that hypomethylation of the BCL-2 gene may be associated with this translocation event.41
One limitation of the study is that sample sizes were insufficient to evaluate the effect of these genetic polymorphisms among African American and Asian American participants. Other limitations include the lack of dietary information related to folate intake and possible selection bias associated with patients who were eligible for the laboratory portion of the study, although the latter is unlikely since no differences were evident between sex, race, education, mean age, or case-control status.42
In conclusion, the decreased risk estimates for NHL and particularly DLCL associated with the variant TYMS 1494del6, IVS6 -68C>T, and 1053C>T genotypes and the increased risk for FL associated with the low activity MTHFR 677TT genotype suggest that both DNA synthesis and repair and methylation processes may play important roles in the pathogenesis of specific NHL subtypes. Studies are currently underway to investigate the more specific role of TYMS in lymphoma risk. Furthermore, our findings on TYMS, MTHFR, and MTR polymorphisms and NHL risk are inconsistent with some previous reports, emphasizing the need for larger studies. The international consortium on lymphoma research, InterLymph, will be able to provide support for truly significant findings through much larger combined data sets.
Prepublished online as Blood First Edition Paper, June 15, 2004; DOI 10.1182/blood-2004-02-0557.
Supported by RO1 grant numbers CA104682 (M.T.S., principal investigator [PI]), CA45614 (E.A.H., PI), CA89745 (E.A.H., PI), and CA66529 (E.A.H., PI) from the National Cancer Institute and Center Grant P30ES01896 (M.T.S., PI) from the National Institute of Environmental Health Sciences and the National Foundation for Cancer Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful for the pathology review provided by Dr Ronald F. Dorfman at Stanford University.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal