Abstract
Notch proteins comprise a family of transmembrane receptors. Ligand activation of Notch releases the intracellular domain of the receptor that translocates to the nucleus and regulates transcription through the DNA-binding protein RBP-Jκ. Previously, it has been shown that the Notch4 intracellular region (N4IC) can inhibit endothelial sprouting and angiogenesis. Here, N4IC deletion mutants were assessed for their ability to inhibit human microvascular endothelial cell (HMEC) sprouting with the use of a quantitative endothelial sprouting assay. Deletion of the ankyrin repeats, but not the RAM (RBP-Jκ associated module) domain or C-terminal region (CT), abrogated the inhibition of fibroblast growth factor 2 (FGF-2)- and vascular endothelial growth factor (VEGF)-induced sprouting by Notch4, whereas the ankyrin repeats alone partially blocked sprouting. The ankyrin repeats were also the only domain required for up-regulation of RBP-Jκ-dependent gene expression. Interestingly, enforced expression of the ankyrin domain alone was sufficient to up-regulate some, but not all, RBP-Jκ-dependent genes. Although N4IC reduced VEGF receptor-2 (VEGFR-2) and vascular endothelial (VE)-cadherin expression, neither of these events is necessary and sufficient to explain N4IC-mediated inhibition of sprouting. A constitutively active RBP-Jκ mutant significantly inhibited HMEC sprouting but not as strongly as N4IC. Thus, Notch4-induced inhibition of sprouting requires the ankyrin repeats and appears to involve RBP-Jκ-dependent and -independent signaling. (Blood. 2004;104:1760-1768)
Introduction
Notch proteins are a highly conserved family of transmembrane receptors involved in intercellular signaling that regulate cell fate.1 Notch interacts with ligands presented on neighboring cells, triggering a 2-step proteolytic cleavage of the receptor that releases its C-terminal intracellular region (NIC).2,3 NIC is then capable of translocating to the nucleus and up-regulating the transcription of target genes.4,5 As a result of this signaling mechanism, enforced expression of the intracellular domain of the receptor provides constitutive Notch activity.6,7
In the nucleus, NIC regulates transcription through association with the DNA-binding protein RBP-Jκ (also known as CBF1, KBF2, or CSL). The primary gene targets of RBP-Jκ include members of the hairy and enhancer of split (HES) and hairy related transcription factor (HRT) families of basic-helix-loop-helix transcriptional repressors. In the absence of NIC, RBP-Jκ actively represses transcription by way of recruitment of a corepressor complex.8 Nuclear translocation of NIC leads to dissociation of repressor proteins from RBP-Jκ and formation of a coactivator complex.9-13
RBP-Jκ-independent Notch signaling also exists. Studies on loss-of-function mutants indicate that the activity of Supressor of Hairless (Su(H)) and Lag-1-RBP-Jκ homologs in Drosophila and Caenorhabditis elegans, respectively, do not account for all observed Notch functions.14-18 There is also growing support for the existence of RBP-Jκ-independent Notch signaling in mammals.19-22
Four mammalian Notch homologs have been identified to date and include Notch1, Notch2, Notch3, and Notch4.23-28 The intracellular region of each homolog is composed of several discrete domains, including a RAM (RBP-Jκ-associated module) domain that has high affinity for RBP-Jκ and 6 ankyrin repeats that also bind RBP-Jκ as well as other components of the transcriptional coactivator complex.9,13,29-31 The function of the region C-terminal to the ankyrin repeats is not well defined but includes a proline-glutamate-serine-threonine (PEST) motif that is involved in protein turnover.32,33
Functionally, Notch receptors and ligands are necessary for vascular development. Targeted deletion of Notch1 causes embryonic lethality because of defects in blood vessel development, and this phenotype is enhanced in Notch1/Notch4 double knock-out mice.34,35 Paradoxically, similar vascular abnormalities occur in transgenic mice expressing a constitutively active Notch4 mutant in the endothelium, demonstrating the need for critical regulation of Notch activity during vascular development.36 In these various mutant mice, the primary vascular plexus forms normally, but there is a failure to properly remodel this immature network, implicating a role for Notch in angiogenesis, the process of developing new blood vessels from the existing vasculature in response to various factors such as vascular endothelial growth factor (VEGF, also VEGF-A) and fibroblast growth factor-2 (FGF-2).37-39
The regulatory role Notch4 plays in endothelial cells during angiogenesis is of particular interest because this receptor is primarily expressed in the vascular endothelium of embryonic and adult mammals.27,40,41 Enforced expression of the Notch4 intracellular domain (N4IC) inhibits VEGF- and FGF-2-induced endothelial sprouting in 3-dimensional fibrin gels.42 Activated Notch4 or Notch1, as well as a downstream Notch effector, HRT1, have each been shown to down-regulate VEGF receptor 2 (VEGFR-2) gene expression, which may provide one explanation of how Notch signals inhibit endothelial network formation.43,44 However, the critical Notch4 domains required for its antiangiogenic activity have not been defined.
In the present study, the requirement of individual Notch4 domains to inhibit endothelial sprouting was investigated with the use of a quantitative assay.42,45,46 Activated Notch4 was confirmed to inhibit human microvascular endothelial cell (HMEC) sprouting in response to FGF-2 and VEGF, as shown previously.42 Inhibition of endothelial sprouting by Notch4 requires the ankyrin repeats, but not the RAM domain or C-terminal region (CT) as demonstrated by the expression of Notch4 mutants deleted in these individual domains. In parallel, enforced expression of only the ankyrin repeats of Notch4 partially inhibited sprouting. Similarly, activation of RBP-Jκ independently of Notch only partially inhibited endothelial sprouting. Deletion of the ankyrin repeats, but not the RAM domain or CT, abolished Notch4 induction of RBP-Jκ-dependent gene expression. Taken together, our findings indicate that Notch4-induced inhibition of endothelial sprouting requires the ankyrin repeats and likely involves signaling through both RBP-Jκ-dependent and -independent pathways.
Materials and methods
Cell culture
HMECs immortalized by the SV40 T antigen were provided by the Centers for Disease Control and Prevention (Atlanta, GA).47 Cells were cultured in MCDB medium (Sigma, St Louis, MO) supplemented with 10% heat-inactivated fetal calf serum (FCS; HyClone, Logan, UT), 10 ng/mL epidermal growth factor (Sigma), and 50 U/mL penicillin and 50 μg/mL streptomycin (Gibco, Gaithersburg, MD) (HMEC medium). Cells were maintained at 37°C in 5% CO2.
Plasmid constructs and gene transfer
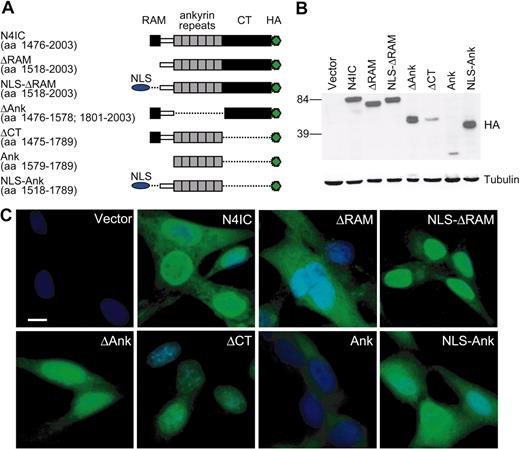
The N4IC construct, described previously, contains a C-terminal hemagglutinin (HA) epitope tag and includes amino acids (aa's) 1476 to 2003 of the 2003 aa full-length Notch4.42 The N4IC deletion mutants were constructed by polymerase chain reaction (PCR) with the use of N4IC as a template and also include C-terminal HA-tags. cDNAs were inserted into the LNCX retroviral vector, in which expression is controlled by the cytomegalovirus (CMV) immediate early enhancer/promoter. The N4IC mutants (Figure 1A) include constructs (1) lacking the entire RAM domain (ΔRAM; encodes aa 1518-2003), (2) lacking the RAM and N-terminally fused with an SV40-derived nuclear localization signal (NLS; NLS-ΔRAM; encodes aa 1518-2003), (3) lacking all 6 ankyrin repeats (ΔAnk encodes aa's 1476-1578 and 1801-2003), (4) lacking the C-terminal region (ΔCT; aa's 1475-1789), (5) composed of only the 6 ankyrin repeats (Ank; encodes aa's 1579-1789), and (6) composed of the 6 ankyrin repeats plus additional upstream sequence and fused with an N-terminal SV40 NLS (NLS-Ank; encodes aa's 1518-1789). The NLS sequence used encodes the amino acid sequence DPKKKRKV. N4IC was also cloned into the MSCV-IRES-YFP (MIY) retroviral vector, as was RBP-VP16, a constitutively active RBP-Jκ. In the MIY vector, gene expression is controlled by the murine stem cell virus long terminal repeats (LTRs). RBP-VP16 has an N-terminal FLAG-tag and was constructed by PCR amplification of the 3′ region of the mouse RBP-VP16 cDNA (gift of E. Manet, Institut National de la Santé et de la Recherche Médicale [INSERM], Lyon, France).48 This PCR product, which includes the coding region for the VP16 transactivation domain, was digested with AflII and ligated to the corresponding AflII site of the cDNA for FLAG-RBP-Jκ, which itself was derived from the RBP-2N isoform of human RBP-Jκ (gift of R. Schmid, University of Ulm, Germany).49 The 4xRBP-Jκ luciferase plasmid includes 4 copies of an RBP-Jκ binding element cloned into pGL2pro (Promega, Madison, WI), an SV40 promoter-driven firefly luciferase plasmid (gift of S.D. Hayward, Johns Hopkins School of Medicine, Baltimore, MD).50 The HRT2 luciferase comprises a 10-kb fragment of the mouse HRT2 promoter cloned into pGL3basic (Promega), a promoterless firefly luciferase vector (gift of E.N. Olson, University of Texas Southwestern Medical Center, Dallas).51
Deletion of the RAM domain inhibits Notch4 nuclear localization. (A) Structure diagrams of the HA epitope-tagged Notch4 intracellular region (N4IC) and related deletion constructs. The amino acid (aa) numbers from the 2003 residue human Notch4 protein that are included in each mutant are indicated in parentheses. NLS indicates nuclear localization signal. (B) Expression of the N4IC constructs in HMECs as detected by immunoblotting with anti-HA antibody. (C) Expression and subcellular localization of the N4IC constructs as detected by immunofluorescent staining of transduced HMEC lines with anti-HA antibody. Nuclei are counterstained with DAPI. Original magnification, × 400; bar = 10 μm.
Deletion of the RAM domain inhibits Notch4 nuclear localization. (A) Structure diagrams of the HA epitope-tagged Notch4 intracellular region (N4IC) and related deletion constructs. The amino acid (aa) numbers from the 2003 residue human Notch4 protein that are included in each mutant are indicated in parentheses. NLS indicates nuclear localization signal. (B) Expression of the N4IC constructs in HMECs as detected by immunoblotting with anti-HA antibody. (C) Expression and subcellular localization of the N4IC constructs as detected by immunofluorescent staining of transduced HMEC lines with anti-HA antibody. Nuclei are counterstained with DAPI. Original magnification, × 400; bar = 10 μm.
HMECs were transduced with the empty vector control or vector with a cDNA insert as described previously.52 Polyclonal HMEC lines were isolated by selection in 300 μg/mL G418 (Gibco) for the LNCX constructs and by sorting cells for yellow fluorescence protein (YFP) expression using a fluorescence activated cell sorter (FACS) 440 (Becton Dickinson [BD], San Jose, CA) for the MIY constructs.
Immunoblotting
Total cellular extracts were prepared from confluent cell monolayers and stored at -80°C until use. Total protein (40 μg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Primary antibodies used included a mouse anti-HA epitope monoclonal antibody (1:4000 dilution; Sigma), the M5 mouse anti-FLAG epitope monoclonal antibody (1:1000 dilution; Sigma), and a mouse anti-α-tubulin monoclonal antibody (1:5000 dilution; Sigma). The secondary antibody was horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Bio-Rad Laboratories, Hercules, CA).
Immunofluorescence
Transduced HMEC lines were cultured overnight on chamber slides (BD), fixed with 4% paraformaldehyde (Fisher Scientific, Suwannee, GA) for 15 minutes and then permeabilized with cold methanol (Fisher Scientific) for 3 minutes. Nonspecific binding was blocked by incubation with phosphate-buffered saline (PBS), 5% goat serum. Cells were stained with the mouse anti-HA monoclonal primary antibody (1:100 dilution) for 1 hour and then for 30 minutes with an AlexaFluor 488-conjugated goat anti-mouse IgG secondary antibody (1:500 dilution; Molecular Probes, Eugene, OR). Nuclei were counterstained with 4′,6′diamidino-2-phenylindole (DAPI) for 5 minutes, and coverslips were mounted with 50% glycerol. Slides were viewed through a 40× Neofluor objective (numerical objective 0.75) using a Zeiss Axioplan II Imaging inverted microscope (Carl Zeiss, Toronto, Canada), and images were captured with a 1350EX cooled charge-coupled device (CCD) digital camera (QImaging, Burnaby, BC, Canada) using Northern Eclipse software (Empix Imaging, Mississauga, ON, Canada).
Endothelial sprouting assay
Endothelial sprouting was assessed as previously described.42,45,46 Briefly, microcarrier beads coated with gelatin were seeded with HMEC lines at a ratio of approximately 200 HMECs/bead and embedded in fibrin gels in 96-well plates (∼ 50 beads/well). Fibrin gels were supplemented either with FGF-2 (15 ng/mL) or VEGF165 (30 ng/mL) or with no angiogenic factor. The overlying medium contained either MCDB + 2% fetal bovine serum (FBS) alone (basal medium) or was supplemented with FGF-2 (15 ng/mL), or VEGF165 (30 ng/mL). After 3 days of incubation with daily medium changes, the number of capillary-like tubes formed was quantitated by counting the number of tubelike structures more than 150 μm in length per microcarrier bead (sprouts/bead), counting all beads in every well. Images were captured with a Nikon Coolpix 950 camera (Nikon, Tokyo, Japan) through a 10× objective lens (numerical aperture 0.25).
Transient transfection and luciferase assays
Transient transfection of luciferase reporter plasmids was carried out by electroporation. Transduced HMEC lines were grown to approximately 80% confluence and then trypsinized and resuspended in HMEC medium. Cells (1.5 × 106/transfection) were pelleted at 200 g for 5 minutes, washed with PBS, pelleted as previous, and then resuspended in 0.4 mL electroporation buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 137 mM sodium chloride, 5 mM potassium chloride, 0.7 mM sodium phosphate, 6 mM d-glucose, pH 7.0 53) containing luciferase reporter plasmid DNA. The cell-DNA mixture was transferred to a 4-mm gap electroporation cuvette (Bio-Rad), left for 10 minutes at room temperature, and then electroporated at a fixed capacitance of 900 μF and 200 V using a Bio-Rad Gene Pulser II instrument. For each transfection, 2.5 μg 4xRBP-Jκ-binding promoter luciferase plus 1 μg RL-CMV (Promega) or 5 μg HRT2 promoter luciferase plus 1 μg RL-CMV were used. The RL-CMV reporter contains the renilla luciferase cDNA expressed under control of the CMV immediate early enhancer/promoter and serves as a normalization control for transfection efficiency. After electroporation, the cells were left for 10 minutes at room temperature before plating in prewarmed (37°C) HMEC medium. The medium was changed 24 hours later, and cells were harvested for assay 48 hours after transfection. Lysis and dual-luciferase reporter assays were performed according to the manufacturer's recommendations (Promega) with luminescence measured on a Tropix tube luminometer (BIO/CAN Scientific, Mississauga, ON, Canada). Luminescence values of mock transfections were subtracted from sample luminescence readings to give the net firefly and net renilla luciferase units. The net firefly units divided by the net renilla units determined the relative luciferase units (RLUs).
RNA isolation and RT-PCR
Total RNA was isolated from confluent cell monolayers with use of TRIzol Reagent (Invitrogen, Carlsbad, CA). First strand cDNA was synthesized with use of 50-μL reactions containing 2.5 μg RNA and 200 units SuperScript II reverse transcriptase (Invitrogen). Following RNAse H (2 U/reaction) (Invitrogen) treatment, PCR reactions were performed, and amplicons were resolved on Tris acetate ethylenediaminetetraacetic acid (TAE) agarose gels. No PCR products were detected in the negative control reactions performed without reverse transcriptase (RT). Following are the primers and reaction conditions used for amplification: HRT1, sense 5′-ggagaggcgccgctgtagtta-3′ and antisense 5′-caagggcgtgcgcgtcaaagta-3′ primers, 57°C annealing temperature, and 28 reaction cycles; HRT2, sense 5′-tgagcataggattccgagagtgc-3′ and 5′-antisense gaaggacagagggaagctgtgtg-3′ primers, 57°C annealing temperature, and 28 reaction cycles; HRT3, sense 5′-cactggtgggacaggattctttg-3′ and antisense 5′-gtaagcagccgaccctgtaggac-3′ primers, 57°C annealing temperature, and 30 reaction cycles; HES1, sense 5′-aggcggacattctggaaatg-3′ and 5′-antisense cggtacttccccagcacactt-3′ primers, 55°C annealing temperature, and 30 reaction cycles; HES4, sense 5′-caccgcaagtcctccaag-3′ and antisense 5′-tcacctccgccagacact-3′ primers, 53°C annealing temperature, and 30 reaction cycles; FGFR-1, sense 5′-agctccatattggacatc-3′ and antisense 5′-tatgatgctccaggtggc-3′ primers, 54°C annealing temperature, and 24 reaction cycles; VEGFR-2, sense 5′-agccctgtgcgctcaactgtc-3′ and antisense 5′-aagagaaacactaggcaaacc-3′ primers, 55°C annealing temperature, and 30 reaction cycles. GAPDH (glyceraldehyde-3-phosphate dehydrogenase), sense 5′-cccatcaccatcttccag-3′ and antisense 5′-atgaccttgcccacagcc-3′ primers, 55°C annealing temperature, and 22 reaction cycles.
Migration assay
Statistics
Results were analyzed by analysis of variance (ANOVA) to ascertain differences between groups, followed by a Tukey test for multiple comparisons.
Results
Subcellular localization of Notch4 mutants
A series of deletion mutants of the human Notch4 intracellular region (N4IC) were constructed to determine the functional relevance of specific domains. The Notch4 deletion mutants included constructs lacking either the RAM domain (ΔRAM), all 6 ankyrin repeats (ΔAnk), or the C-terminal region (ΔCT), as well as a construct consisting of the ankyrin repeats alone (Ank) (Figure 1A). Each construct was C-terminally tagged with an HA epitope and stably expressed in HMECs, as confirmed by immunoblotting (Figure 1B).
Because activated Notch functions primarily by modulating transcription at promoter regulatory sites, it was important to ensure that each mutant localized to the nucleus. It has previously been reported that the RAM domain is required for nuclear targeting of Notch4 in mouse mammary epithelial cells.54 As seen in Figure 1C, mutants lacking the RAM domain, including the ΔRAM and Ank constructs, localized predominantly to the cytoplasm in HMECs. Conversely, the intact N4IC, as well as the ΔAnk and ΔCT deletion mutants, were all predominantly expressed in the nucleus with varying degrees of cytoplasmic staining. Expression of the ΔCT and Ank mutants in HMECs were notably reduced compared with the other constructs as indicated by the immunoblotting (Figure 1B). Fusion of an SV40-derived NLS sequence to the N-terminus of ΔRAM and Ank restored the ability of these mutants to localize to the nucleus (Figure 1C). The NLS-ΔRAM mutant targeted to the nucleus with minimal cytoplasmic staining. In contrast, the NLS-Ank protein, although showing increased nuclear localization, still showed significant cytoplasmic distribution. To focus on Notch signaling in the nucleus, all subsequent experiments were carried out with the NLS-ΔRAM and NLS-Ank constructs rather than ΔRAM and Ank.
Notch4-induced inhibition of endothelial sprouting requires the ankyrin repeats
Notch4 is predominantly expressed in the vascular endothelium,28,34,41 and constitutive activation of Notch4 in endothelial cells blocks angiogenesis.36,42 Using an in vitro model of angiogenesis in which endothelial cells coated on microcarrier beads are induced to form cellular sprouts within a 3-dimensional fibrin gel, we have previously shown that N4IC inhibits serum-induced sprouting as well as that induced by FGF-2 and VEGF.42 However, the structural elements of Notch4 that are required for regulating endothelial cell morphogenesis during vascular remodeling remain unknown. Figure 2A shows phase-contrast micrographs of microcarrier beads coated with either control cells or cells expressing activated Notch4 and incorporated into FGF-2-containing fibrin gels. Whereas extensive sprouting is observed in the control cells, there is minimal sprouting in the HMECs expressing N4IC (HMEC-N4IC). Transmission electron microscopy confirmed previous findings that this assay of morphogenesis mimics endothelial tube formation, because we detected the formation of lumina at the base of the sprouts (Figure 2B).46 As quantitated in Figure 2C, mutants lacking either the RAM motif or the CT domain blocked endothelial sprouting as effectively as N4IC in response to all stimuli. Conversely, deletion of the Notch4 ankyrin repeats abrogated the ability of the receptor to inhibit endothelial sprouting. To determine whether the ankyrin repeats alone were sufficient for Notch4 function, HMECs sprouting in cells expressing the ankyrin repeats targeted to the nucleus (NLS-Ank) were determined. Sprouting of HMEC-NLS-Ank was significantly less than control (P < .001), but significantly greater than N4IC-expressing endothelial cells (P < .001) for all stimuli (Table 1). Therefore, although the ankyrin repeats appear necessary for Notch4-induced inhibition of endothelial sprouting, this domain is only partially sufficient to inhibit sprouting in and of itself.
The ankyrin repeats are required for Notch4-mediated inhibition of endothelial sprouting. (A) Phase-contrast micrographs of microcarrier beads seeded with HMECs transduced with N4IC or empty vector control and stimulated with FGF-2. Arrows indicate capillary-like sprouts of sufficient length to be counted after 3 days of stimulation. Original magnification, × 100. (B) Transmission electron micrographs of sectioned fibrin gels containing sprouting HMECs. Panel i demonstrates the base of a sprout forming a lumen that excludes the fibrin gel (original magnification, × 9000; bar = 5 μm. Panel ii demonstrates another lumen formed by HMECs. Arrowheads point to adherens-like junctions and the arrow points to a coated pit (original magnification, × 54 000; bar = 1 μm. Electron microscopy was performed on a Philips 400 transmission electron microscope, and images were photographed with the built-in 3550 × objective (i) and the 21 500 × objective (ii). Images were scanned on an Epson Perfection scanner using Photoshop Element. (C) Quantitation of sprouting for the transduced cell lines after 3 days of stimulation with basal medium or medium supplemented with FGF-2 or VEGF. The number of sprouts per microcarrier bead (sprouts/bead) were counted and graphed as means + SD. Data are from a single experiment done in triplicate. The relative sprouting patterns are representative of at least 4 separate experiments.
The ankyrin repeats are required for Notch4-mediated inhibition of endothelial sprouting. (A) Phase-contrast micrographs of microcarrier beads seeded with HMECs transduced with N4IC or empty vector control and stimulated with FGF-2. Arrows indicate capillary-like sprouts of sufficient length to be counted after 3 days of stimulation. Original magnification, × 100. (B) Transmission electron micrographs of sectioned fibrin gels containing sprouting HMECs. Panel i demonstrates the base of a sprout forming a lumen that excludes the fibrin gel (original magnification, × 9000; bar = 5 μm. Panel ii demonstrates another lumen formed by HMECs. Arrowheads point to adherens-like junctions and the arrow points to a coated pit (original magnification, × 54 000; bar = 1 μm. Electron microscopy was performed on a Philips 400 transmission electron microscope, and images were photographed with the built-in 3550 × objective (i) and the 21 500 × objective (ii). Images were scanned on an Epson Perfection scanner using Photoshop Element. (C) Quantitation of sprouting for the transduced cell lines after 3 days of stimulation with basal medium or medium supplemented with FGF-2 or VEGF. The number of sprouts per microcarrier bead (sprouts/bead) were counted and graphed as means + SD. Data are from a single experiment done in triplicate. The relative sprouting patterns are representative of at least 4 separate experiments.
Statistical analysis of the effect of Notch 4 mutants on HMEC sprouting
Comparisons of each N4IC construct to empty vector control, P . | . | . | . | . | . | Comparison of N4IC vs NLS-Ank, P . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | N4IC . | NLS-ΔRAM . | ΔAnk . | ΔCT . | NLS-Ank . | . | |||||
| Medium | < .001 | < .001 | < .05 | < .001 | < .001 | < .001 | |||||
| FGF-2 | < .001 | < .001 | < .05 | < .001 | < .001 | < .001 | |||||
| VEGF | < .001 | < .001 | < .05 | < .001 | < .001 | < .001 | |||||
Comparisons of each N4IC construct to empty vector control, P . | . | . | . | . | . | Comparison of N4IC vs NLS-Ank, P . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | N4IC . | NLS-ΔRAM . | ΔAnk . | ΔCT . | NLS-Ank . | . | |||||
| Medium | < .001 | < .001 | < .05 | < .001 | < .001 | < .001 | |||||
| FGF-2 | < .001 | < .001 | < .05 | < .001 | < .001 | < .001 | |||||
| VEGF | < .001 | < .001 | < .05 | < .001 | < .001 | < .001 | |||||
Table shows multiple comparisons of different cell lines receiving a given growth factor stimulus.
Induction of RBP-Jκ-dependent gene expression by Notch4 requires the ankyrin repeats
Activated Notch translocates to the nucleus and associates with the DNA-binding protein RBP-Jκ, thereby derepressing and/or coactivating the transcription of genes belonging to the HES and HRT families of basic helix-loop-helix factors.55 Growing evidence suggests Notch may also signal through RBP-Jκ-independent pathways.19-22 To determine the ability of the various Notch4 mutants to derepress/coactivate RBP-Jκ-dependent signaling, 2 distinct reporter plasmids containing promoters with RBP-Jκ-binding sites were used. In the first assay, HMEC-N4IC mutant cell lines were transiently transfected with a reporter construct containing 4 copies of an RBP-Jκ-binding element upstream of a minimal SV40 promoter driving the firefly luciferase gene (4xRBP-Jκ luciferase).50 N4IC activation of the RBP-Jκ-dependent promoter-reporter (5.1-fold up-regulation) was abolished by deletion of the ankyrin domain, whereas mutants lacking the RAM (5.9-fold up-regulation) or CT (5.5-fold up-regulation) domains were fully able to activate the RBP-Jκ-dependent promoter (Figure 3A). However, although the ankyrin repeats alone appeared to slightly activate the 4xRBP-Jκ promoter (1.7-fold up-regulation), these results did not achieve statistical significance (P = .9).
Notch4 induction of RBP-Jκ-dependent gene expression requires the ankyrin repeats. Reporter assays using a reporter construct with 4 copies of a RBP-Jκ-binding element upstream of an SV40 promoter-driven firefly luciferase gene (4xRBP-Jκ luciferase) (A) or a HRT2 promoter-driven firefly luciferase gene (HRT2 luciferase) (B). Reporter plasmids were electroporated into the HMEC-N4IC mutant cell lines along with a CMV promoter-driven renilla luciferase plasmid used as a normalization control for transfection efficiency. Cell lysates were harvested 48 hours after electroporation, and the relative luciferase units (RLUs) were determined as the ratio of firefly-derived luminescence over renilla-derived luminescence. Data are means + SD for a single experiment done in triplicate. Fold increases are reported for each N4IC construct cell line as compared with the empty vector control cell line. *P < .01 and **P < .001 for sample means compared with the empty vector control. The relative RLU patterns are representative of at least 3 separate experiments. (C) RT-PCR was performed using single-stranded cDNA reverse-transcribed from total RNA isolated from the HMEC-N4IC mutant cell lines. PCR amplifications were done with primers specific for fragments of the HRT1-3, HES1, HES4, and GAPDH cDNA sequences. Quantitation of 4 independent experiments was performed by densitometry of HRT2 (D) and HES4 (E) with levels normalized to GAPDH expression. *P < .05 and **P < .01 for sample means compared with the empty vector control.
Notch4 induction of RBP-Jκ-dependent gene expression requires the ankyrin repeats. Reporter assays using a reporter construct with 4 copies of a RBP-Jκ-binding element upstream of an SV40 promoter-driven firefly luciferase gene (4xRBP-Jκ luciferase) (A) or a HRT2 promoter-driven firefly luciferase gene (HRT2 luciferase) (B). Reporter plasmids were electroporated into the HMEC-N4IC mutant cell lines along with a CMV promoter-driven renilla luciferase plasmid used as a normalization control for transfection efficiency. Cell lysates were harvested 48 hours after electroporation, and the relative luciferase units (RLUs) were determined as the ratio of firefly-derived luminescence over renilla-derived luminescence. Data are means + SD for a single experiment done in triplicate. Fold increases are reported for each N4IC construct cell line as compared with the empty vector control cell line. *P < .01 and **P < .001 for sample means compared with the empty vector control. The relative RLU patterns are representative of at least 3 separate experiments. (C) RT-PCR was performed using single-stranded cDNA reverse-transcribed from total RNA isolated from the HMEC-N4IC mutant cell lines. PCR amplifications were done with primers specific for fragments of the HRT1-3, HES1, HES4, and GAPDH cDNA sequences. Quantitation of 4 independent experiments was performed by densitometry of HRT2 (D) and HES4 (E) with levels normalized to GAPDH expression. *P < .05 and **P < .01 for sample means compared with the empty vector control.
With the use of a second RBP-Jκ-dependent reporter comprising a 10-kb fragment of the mouse HRT2 promoter driving the firefly luciferase gene (HRT2 luciferase),51 similar results were seen. N4IC activated the HRT2 promoter (4.3-fold up-regulation), in a manner dependent on the ankyrin repeats but not the RAM motif (4.0-fold up-regulation of HRT2 luciferase with the ΔRAM mutant) (Figure 3B). ΔCT also up-regulated the HRT2 promoter (2.9-fold up-regulation), although activation was less than that induced by N4IC (P < .001), suggesting that the CT domain likely contributes in a differential manner to derepression/coactivation of individual RBP-Jκ-dependent genes. Again, the ankyrin repeats alone slightly activated the HRT2 promoter, but this was not statistically significant (P = .9).
As an independent measure of the ability of the Notch4 mutants to activate distinct RBP-Jκ-dependent promoters, mRNA levels of different members of the HRT and HES families of genes were analyzed. The HRT genes were the primary focus because their expression has been established in the mammalian vasculature.56-58 As well, N4IC can induce HRT1 expression in cultured human endothelial cells.43 HES1 was chosen because it has been the most extensively studied of all known Notch effectors.55 HES4 was examined in addition to HES1 and the HRTs to determine whether there were any general differences in N4IC-mediated effects on HES family members compared with HRT family members. N4IC up-regulated all analyzed transcripts as determined by RT-PCR (Figure 3C). Specifically, HRT1 and HRT2 were strongly induced, whereas only slight induction of HRT3, HES1, and HES4 were detected. The ΔRAM and ΔCT mutants also induced each of the HRT and HES transcripts. Conversely, the ΔAnk mutant was incapable of inducing expression of any of the transcripts, verifying the requirement of the ankyrin repeats for RBP-Jκ derepression/coactivation in HMECs. Interestingly, NLS-Ank up-regulated HRT1, HRT2, and HES1 mRNA, but had no effect on expression of HRT3 or HES4 (Figure 3C). Quantitation of mRNA induction from 4 independent experiments verified that NLS-Ank up-regulated HRT2 but not HES4 mRNA, providing evidence for potentially different mechanisms of activation of different Notch-dependent promoters (Figure 3D-E).
Notch4-mediated inhibition of endothelial sprouting is independent of VEGFR-2 levels
The Notch signaling pathway down-regulates VEGF receptor-2 (VEGFR-2) mRNA expression in human capillary endothelial cells and human umbilical vein endothelial cells and reduces the responsiveness of these cells to VEGF-induced proliferation.43,44 We aimed to determine whether changes in VEGFR-2 expression were responsible for Notch4-mediated inhibition of sprouting in HMECs. In agreement with published results for other endothelial cell types, enforced expression of activated Notch4 down-regulated VEGFR-2 mRNA expression in HMECs (data not shown). However, expression of the FGF receptor 1 was not altered at the mRNA level (data not shown). Whether Notch also inhibits VEGFR-2 protein expression has not been reported. When examined by immunoblotting VEGFR-2 protein was down-regulated by N4IC as well as the NLS-ΔRAM construct, but the ΔCT construct had no effect (Figure 4A-B). These results indicate that the inhibition of endothelial sprouting by Notch4 is not solely dependent on the down-regulation of VEGFR-2 expression.
Notch4-induced inhibition of endothelial sprouting is independent of VEGFR-2 expression. (A) Total protein harvested from each of the Notch mutant cell lines was assayed for expression of VEGFR-2 protein by immunoblotting. Immunoblotting for α-tubulin demonstrates equivalent loading of total protein. Results are representative of 5 separate experiments and quantitation of the 3 experiments was performed by densitometry with VEGFR-2 levels normalized to α-tubulin (B). *P < .01 for sample means compared with the empty vector control.
Notch4-induced inhibition of endothelial sprouting is independent of VEGFR-2 expression. (A) Total protein harvested from each of the Notch mutant cell lines was assayed for expression of VEGFR-2 protein by immunoblotting. Immunoblotting for α-tubulin demonstrates equivalent loading of total protein. Results are representative of 5 separate experiments and quantitation of the 3 experiments was performed by densitometry with VEGFR-2 levels normalized to α-tubulin (B). *P < .01 for sample means compared with the empty vector control.
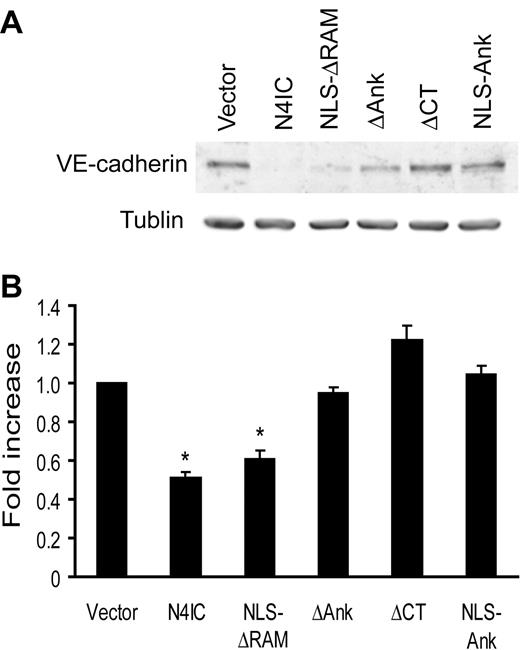
Inhibition of endothelial sprouting by Notch4 is independent of VE-cadherin expression
Recently, we have shown that enforced expression of N4IC causes an endothelial-to-mesenchymal transformation in various endothelial cell types, including HMECs.59 This phenotypic switch of endothelial cells is defined by the down-regulation of vascular endothelial (VE)-cadherin and up-regulation of smooth muscle α-actin (SMA) expression. Because lack of VE-cadherin disrupts angiogenesis, VE-cadherin protein levels in the HMEC-N4IC mutant cell lines were compared to determine whether endothelial transdifferentiation correlated with inhibition of sprouting.60 As detected by immunoblotting, only N4IC and NLS-ΔRAM down-regulated VE-cadherin (Figure 5A-B). ΔCT did not alter VE-cadherin expression, but fully inhibited endothelial cell sprouting. NLS-Ank, which partially inhibits sprouting, was similarly unable to regulate VE-cadherin. Collectively, these results strongly suggest that Notch4-initiated inhibition of endothelial sprouting includes events in addition to VE-cadherin down-regulation.
Inhibition of endothelial sprouting by Notch4 is independent of VE-cadherin expression. (A) Total protein harvested from each of the Notch mutant cell lines was assayed for expression of VE-cadherin protein by immunoblotting. Immunoblotting for α-tubulin demonstrates equivalent loading of total protein. Results are representative of 3 separate experiments, and quantitation of the 3 experiments was performed by densitometry with VE-cadherin levels normalized to α-tubulin (B). *P < .001 for sample means compared with the empty vector control.
Inhibition of endothelial sprouting by Notch4 is independent of VE-cadherin expression. (A) Total protein harvested from each of the Notch mutant cell lines was assayed for expression of VE-cadherin protein by immunoblotting. Immunoblotting for α-tubulin demonstrates equivalent loading of total protein. Results are representative of 3 separate experiments, and quantitation of the 3 experiments was performed by densitometry with VE-cadherin levels normalized to α-tubulin (B). *P < .001 for sample means compared with the empty vector control.
Constitutively-active RBP-Jκ inhibits endothelial sprouting
Given that the Notch4 deletion mutants that were able to activate RBP-Jκ-dependent genes were also able to inhibit endothelial sprouting, we attempted to determine whether Notch-independent activation of RBP-Jκ-dependent gene expression could inhibit endothelial sprouting. Attempts were made to determine to what degree Notch4-induced inhibition of sprouting was mediated by RBP-Jκ. To this end, a dominant-negative human RBP-Jκ was constructed on the basis of RBP-Jκ R218H, an established dominant-negative mouse RBP-Jκ that cannot bind DNA.61 Cotransduction of dominant-negative human RBP-Jκ and N4IC into HMECs only partially blocked RBP-Jκ-dependent gene activation, as seen by RBP-Jκ-dependent promoter-reporter assays and RTPCR (data not shown). This is due to at least 2 reasons: The first is that even with retrovirally mediated gene transfer, in our hands, the proportion of selected cells expressing a given transgene can vary from 50% to 80%. Secondly, the vast excess of dominant-negative RBP-Jκ protein expression required to inhibit Notch function is likely not achievable using this strategy. Transient transfections in which the dominant-negative can be added in large excess are not practical because of the low efficiency of these methods in endothelial cells.
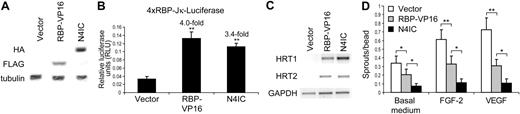
Thus, an alternative approach to determine the role of RBP-Jκ in Notch-mediated inhibition of endothelial sprouting was attempted. A constitutively active RBP-Jκ was constructed on the basis of an established mutant that fuses the transactivation domain of the herpes simplex virus transcription factor VP16 to the C-terminus of RBP-Jκ (RBP-VP16).48 FLAG-tagged RBP-VP16 or HA-tagged N4IC were transduced into HMECs, and their expression was confirmed by immunoblotting (Figure 6A). Activation of the 4xRBP-Jκ luciferase reporter and up-regulation of endogenous transcripts confirmed RBP-VP16 activity. RBP-VP16 (4.0-fold up-regulation) and N4IC (3.4-fold up-regulation) induced similar levels of reporter activity (Figure 6B). RBP-VP16 and N4IC also induced similar levels of HRT1 and HRT2 expression (Figure 6C).
Constitutively active RBP-Jκ inhibits endothelial sprouting. (A) Expression of RBP-VP16 and N4IC in HMECs was detected by immunoblotting of total cellular lysates with a monoclonal anti-FLAG antibody and a monoclonal anti-HA antibody, respectively. Immunoblotting for tubulin demonstrates equivalent loading of total protein. (B) 4xRBP-Jκ luciferase reporter activity in HMECs transduced with RBP-VP16, N4IC, or empty vector control. Data are means + SD for a single experiment done in triplicate. Fold increases are reported for RBP-VP16 and N4IC cell lines as compared with the empty vector control. **P < .001 for sample means compared with the empty vector control. The relative RLU patterns are representative of at least 3 separate experiments. (C) RT-PCR was performed by using single-stranded cDNA reverse-transcribed from total RNA isolated from HMECs transduced with RBP-VP16, N4IC, or empty vector control. PCR amplifications were done with primers specific for fragments of the HRT1-3 and GAPDH cDNA sequences. Amplification of the GAPDH fragment demonstrates equivalent levels of cDNA input. The relative patterns of mRNA expression are representative of at least 3 separate experiments. (D) Endothelial sprouting assay for HMECs transduced with RBP-VP16, N4IC, or empty vector control. Assays were quantitated and graphed as means + SE for an average of 4 experiments, each done in triplicate. *P < .05 and **P < .001 for the indicated comparisons.
Constitutively active RBP-Jκ inhibits endothelial sprouting. (A) Expression of RBP-VP16 and N4IC in HMECs was detected by immunoblotting of total cellular lysates with a monoclonal anti-FLAG antibody and a monoclonal anti-HA antibody, respectively. Immunoblotting for tubulin demonstrates equivalent loading of total protein. (B) 4xRBP-Jκ luciferase reporter activity in HMECs transduced with RBP-VP16, N4IC, or empty vector control. Data are means + SD for a single experiment done in triplicate. Fold increases are reported for RBP-VP16 and N4IC cell lines as compared with the empty vector control. **P < .001 for sample means compared with the empty vector control. The relative RLU patterns are representative of at least 3 separate experiments. (C) RT-PCR was performed by using single-stranded cDNA reverse-transcribed from total RNA isolated from HMECs transduced with RBP-VP16, N4IC, or empty vector control. PCR amplifications were done with primers specific for fragments of the HRT1-3 and GAPDH cDNA sequences. Amplification of the GAPDH fragment demonstrates equivalent levels of cDNA input. The relative patterns of mRNA expression are representative of at least 3 separate experiments. (D) Endothelial sprouting assay for HMECs transduced with RBP-VP16, N4IC, or empty vector control. Assays were quantitated and graphed as means + SE for an average of 4 experiments, each done in triplicate. *P < .05 and **P < .001 for the indicated comparisons.
In keeping with activation of the RBP-Jκ-dependent promoters, RBP-VP16 inhibited endothelial sprouting in response to basal medium, as well as medium supplemented with FGF-2 and VEGF (Figure 6D). However, inhibition of sprouting by RBP-VP16 was significantly less than that induced by activated Notch4 for all stimuli (P < .05). These findings suggest that constitutively active RBP-Jκ partially mimics the Notch4-mediated inhibition of endothelial sprouting and imply that Notch4 may block morphogenesis through both RBP-Jκ-dependent and RBP-Jκ-independent pathways.
Inhibition of endothelial migration partially explains the antisprouting effect of Notch4
We have previously shown that Notch4 does not affect HMEC proliferation.42 Similarly, RBP-VP16 did not decrease HMEC proliferation in response to VEGF (30 ng/mL) or FGF-2 (15 ng/mL). In response to VEGF, RBP-VP16 HMECs showed a 1.88 ± 0.27-fold increase in cell number over 72 hours compared with 1.70 ± 0.10-fold increase in vector-transduced cells. FGF-2-stimulated HMECs showed a 2.00 ± 0.25-fold and 1.97 ± 0.14-fold increase in RBP-VP16 and vector-transduced HMECs, respectively.
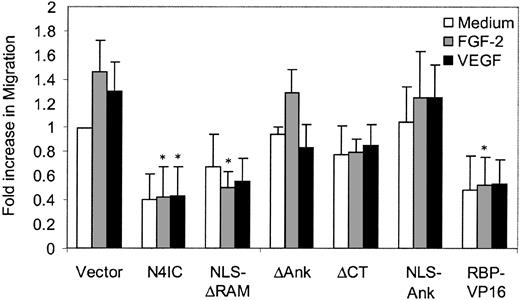
The ability of Notch4 to inhibit endothelial sprouting is in part related to the inhibition of HMEC migration across collagen but not fibrinogen.42 To investigate the effect of the various Notch4 mutants and RBP-VP16 on HMEC migration, a Transwell filter assay was carried out. Although Notch4IC inhibited migration, as expected, the NLS-Ank mutant had no effect on migration (Figure 7), despite a significant effect on sprouting (Figure 2C). In contrast, RBP-VP16 had a significant inhibitory effect on HMEC migration across collagen even though the antisprouting effect was not as potent as that of Notch4IC. These studies highlight the multiple steps required for an endothelial sprout to form and provide further evidence that Notch must act at several steps to block angiogenesis.
Constitutively active RBP-Jκ inhibits endothelial migration. HMEC migration was assayed by a Transwell filter assay in response to basal medium, FGF-2 (15 ng/mL), or VEGF (30 ng/mL). Cells migrating to the underside of the filter were quantitated and graphed as means + SE for the average of 3 experiments, each done in duplicate. *P < .05 compared with the vector control.
Constitutively active RBP-Jκ inhibits endothelial migration. HMEC migration was assayed by a Transwell filter assay in response to basal medium, FGF-2 (15 ng/mL), or VEGF (30 ng/mL). Cells migrating to the underside of the filter were quantitated and graphed as means + SE for the average of 3 experiments, each done in duplicate. *P < .05 compared with the vector control.
Discussion
Angiogenesis, the development of new blood vessels from the existing microvasculature, contributes to the pathogenesis of many human diseases, including cancer and cardiovascular disease.62 Notch4 functions in endothelial cells to regulate angiogenic remodeling of the vasculature. Notch4 is predominantly expressed in the vascular endothelium and loss of Notch4 function in mice enhances the vascular remodeling defects caused by loss of Notch1 function.34 Intriguingly, a similar failure in mouse vascular development occurs when constitutively active Notch4 is targeted to the endothelium under control of the regulatory elements of the VEGFR-2 gene.36 Enforced expression of activated Notch4 also blocks angiogenesis in vivo in the chick chorioallantoic membrane and inhibits endothelial sprouting in vitro, pointing to a requirement for fine regulation of Notch signaling in vascular homeostasis.42
The elucidation of the functional domains required for Notch4-mediated inhibition of angiogenesis in this study identifies the ankyrin repeats as the crucial motif for Notch4 function. The necessity of the ankyrin repeats for Notch4 function in HMECs is not entirely surprising, given the established significance of this domain for the activity of Notch proteins across species.7,63-66 In particular, deletions or loss-of-function mutations of this motif invariably abolish transactivation of RBP-Jκ-dependent genes.29,65-68 The requirement of the ankyrin repeats for RBP-Jκ-dependent signaling probably owes to the interaction with multiple factors in the coactivator complex, including RBP-Jκ, SKIP (Ski-interacting protein), MAML (Mastermind-Like-1), and the histone acetyltransferases, PCAF (p300/CBP-associated factor) and GCN5 (general control of amino-acid synthesis 5).9,13,29,31
Although the ankyrin repeats are necessary for Notch4 function in HMECs, they only provide partial activity on their own. Numerous studies have shown that the Notch1 ankyrin repeats are insufficient for transactivation of RBP-Jκ-dependent reporter constructs.19,20,69,70 Our results with Notch4 using RBP-Jκ-dependent promoter-reporter assays are consistent with this finding. However, the ankyrin repeats do elevate expression of certain endogenous RBP-Jκ-dependent genes. The ankyrin repeats have been reported to be sufficient for Notch-induced inhibition of C2C12 myoblast differentiation and Notch-induced neoplastic transformation of RK3E rat kidney cells.20,69 In both instances, the ankyrin domain-mediated effects were proposed to be RBP-Jκ independent, based largely on the fact that this motif could not activate luciferase reporters driven by promoters containing multimerized RBP-Jκ-binding elements. As shown here, it cannot be assumed that a given Notch mutant is unable to regulate all endogenous RBP-Jκ-dependent genes based solely on the fact that the mutant does not activate a reporter construct with RBP-Jκ-binding sites in its promoter.
The varying ability of the ankyrin domain alone to transactivate distinct RBP-Jκ-dependent genes suggests that different targets require varying degrees or aspects of Notch function for their transcription. Genetic studies in Drosophila support the theory that the transcription of some genes only requires Notch to relieve the repression mediated by RBP-Jκ, whereas other targets require both depression and the recruitment of coactivators for transcription.71 Alleviation of RBP-Jκ-mediated repression may be sufficient for transcriptional activation when Notch-independent transcriptional activators are available or already in place, but are inhibited by the RBP-Jκ corepressor complex. It is possible that in such instances the Notch ankyrin domain alone may be sufficient to disrupt the corepressor complex and allow transcription. However, the ankyrin repeats may not be sufficient to activate regulatory sites that require Notch-mediated derepression and coactivation. Regardless of the specific requirements of individual promoters, the finding that the ankyrin repeats alone were able to significantly inhibit endothelial sprouting, despite having a minimal effect on migration, suggests that multiple distinct events play roles in Notch4-mediated angiogenesis inhibition. That multiple events are required for Notch4-directed antiangiogenic activity is bolstered by the fact that constitutive activation of RBP-Jκ, albeit having a significant effect, is also not sufficient for complete inhibition of endothelial sprouting.
In this regard, the ability of Notch4 to block both FGF-2 and VEGF-induced sprouting is significant because these growth factors are able to induce angiogenesis through distinct pathways. For example, Src-family kinases were shown to be required for angiogenesis induced by VEGF, but not FGF-2, in chick embryo and mouse in vivo models.72 Although Notch activation has been shown to down-regulate VEGFR-2 expression, this in itself is not sufficient to explain inhibition of VEGF-induced sprouting, because the ΔCT mutants inhibit sprouting to the same degree without affecting VEGFR-2 levels. Moreover, FGFR-1 mRNA expression is not affected by Notch4 activation, thus precluding receptor down-regulation as the principal mode of Notch-mediated antiangiogenic function. Finally, the demonstration that mutants that do not down-regulate VE-cadherin still inhibit endothelial sprouting provides additional evidence for Notch inhibiting endothelial sprouting through multiple pathways.
The RAM and CT domains are not required for Notch4-induced inhibition of endothelial sprouting and are largely dispensable for up-regulation of RBP-Jκ-dependent genes. The primary function of the RAM domain is to bind RBP-Jκ, although this activity is not strictly required for interaction between Notch and RBP-Jκ.30,31 Presumably, multiple contacts between the other Notch domains, in particular the ankyrin repeats, and the RBP-Jκ-coactivator complex can compensate for loss of the RAM. Nevertheless, the RAM domain does provide specific functional activity in endothelial cells. For instance, we have seen in other studies, that deletion of the RAM domain reduces the antiapoptotic activity provided by Notch4.73 The CT domain of Notch1, but not Notch4, has intrinsic transcriptional activation capability when analyzed in COS7, NIH3T3, and C2C12 cells.65 The CT is the least conserved region among human Notch proteins and this divergence may explain the differential responses evolved by Notch1 and Notch4. The RAM and CT domains are not completely superfluous because the addition of either motif to the ankyrin repeats greatly enhances the inhibition of endothelial sprouting and the activation of RBP-Jκ (Figures 2C and 3). The RAM and CT motifs may directly potentiate the activity of the ankyrin repeats or, alternatively, the ankyrin repeats may only require one of the RAM or CT for proper protein folding following translation.
In conclusion, Notch4-induced inhibition of endothelial sprouting requires the ankyrin repeats and appears to involve RBP-Jκ-dependent and -independent signaling. Of interest, it appears that down-regulation of VEGFR-2 and VE-cadherin, and functionally inhibition of migration, by Notch4 is not sufficient to explain the inhibitory effects of Notch4 on endothelial sprouting and indicate that Notch uses multiple pathways to restrict vascular morphogenesis and angiogenesis.
Prepublished online as Blood First Edition Paper, June 8, 2004; DOI 10.1182/blood-2003-12-4244.
Supported by grants from the Heart and Stroke Foundation of British Columbia and the Yukon (A.K.), the National Cancer Institute of Canada with funds from the Canadian Cancer Society (A.K.), and the Canadian Institutes of Health Research (A.K.). P.D. was supported by a Postdoctoral Fellowship Award from the Heart and Stroke Foundation of Canada. B.L. was supported by a Doctoral Research Award from the Heart and Stroke Foundation of Canada. A.K. is a Clinician-Scientist of the Canadian Institutes of Health Research and a Scholar of the Michael Smith Foundation for Health Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank E. Manet, R. Schmid, S.D. Hayward, and E.N. Olson for providing plasmids, and D. Walker for assistance with electron microscopy.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal