Recently, Voso et al1 reported the frequent hypermethylation of the DAP-kinase gene in myelodysplastic syndrome (in 16 of 34 samples).
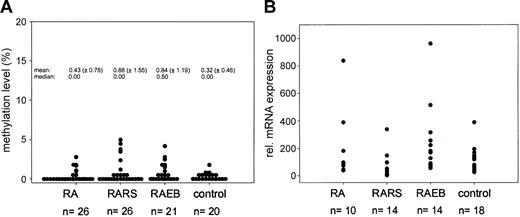
Since we are interested in the epigenetic profile of myelodysplastic syndrome (MDS), we assessed the methylation status of the DAP-kinase gene in a larger series of 73 bone marrow biopsies (26 refractory anemia [RA], 26 RA with ringed sideroblasts [RARS], and 21 RA with blast excess [RAEB]) using exactly the same primer set mentioned by Voso et al (from Katzenellenbogen et al2 ) and also our published quantitative real-time polymerase chain reaction (PCR)–based methylation assay.3 In 43% of all MDS biopsies analyzed, a signal for methylated DNA could be detected. This frequency is very similar to the one reported by Voso et al (47%), but the quantitative evaluation of the methylation level in each sample revealed that DAP-kinase gene hypermethylation is a minor event (methylation level, < 5%; Figure 1A). Since low levels of DAP-kinase hypermethylation were found in control cases (9 of 20; methylation level, 0.5%-2%) and have also been reported in normal lymphocytes,5 the biologic significance of this finding in MDS samples remains unclear.
Methylation and expression analysis ofDAP-kinasein MDS. (A) Results of quantitative methylation analysis of the DAP-kinase gene in MDS patients and control cases. (B) Expression analysis of DAP-kinase gene in MDS. Measurement of DAP-kinase mRNA levels using quantitative real-time PCR methodology. The mean expression level of the control group was set to 100% and all individual expression levels were calculated to this mean using the ΔΔCT-method.4 The mean relative expression level of the MDS samples is 127% (P = .5, Mann-Whitney test). Transcript levels were normalized to β-glucuronidase (β-GUS).
Methylation and expression analysis ofDAP-kinasein MDS. (A) Results of quantitative methylation analysis of the DAP-kinase gene in MDS patients and control cases. (B) Expression analysis of DAP-kinase gene in MDS. Measurement of DAP-kinase mRNA levels using quantitative real-time PCR methodology. The mean expression level of the control group was set to 100% and all individual expression levels were calculated to this mean using the ΔΔCT-method.4 The mean relative expression level of the MDS samples is 127% (P = .5, Mann-Whitney test). Transcript levels were normalized to β-glucuronidase (β-GUS).
Using real-time quantitative reverse transcription–PCR, we also measured the DAP-kinase mRNA expression level in 18 of 20 control biopsies, 12 MDS samples without any methylation, and 26 of 31 MDS samples displaying methylation signals. No significant reduction in DAP-kinase mRNA level could be observed. On the contrary, we found a weak trend toward increased DAP-kinase mRNA expression in the MDS biopsies in comparison with the control group (Figure 1B), which fits well to the proapoptotic function of death-associated protein kinase (DAP)–kinase6 and the well-known increase in apoptosis in the bone marrow of MDS patients.7 Parker et al8 clearly showed in their study, which is cited by Voso et al as reference 21, that the rate of apoptosis is significantly increased in CD34+ cells in RA, RARS, and RAEB. Therefore, this study rather contradicts than supports the statement that “a common feature of MDSs is a decreased apoptosis rate in bone marrow progenitor cells.”1 (pp 699)
Since Voso et al provide no details of the reaction conditions and do not show any primary data, evaluation of the reported results is difficult. From the context of paragraph one in “Results and discussion,” we assume that Voso et al carried out their mRNA expression studies with a subset of acute myeloid leukemia (AML) samples and not with MDS samples, which could explain in part the discrepancy of the results.
The well-known differences in the apoptosis rate of RA, RARS, and RAEB versus RAEB in transformation (RAEB-t), secondary AML, and AML might explain differences in the epigenetic inactivation pattern of the DAP-kinase gene in these entities, which has to be addressed using quantitative methylation and expression assays. Long-term follow-up studies will be necessary to evaluate the significance of low-level methylation in MDS for the clonal evolution to AML.
Finally, we would like to mention that 4 years ago in Blood, Aggerholm et al9 raised the question of overestimating the proportion of DAP-kinase gene methylation in AML by using methylation-specific PCR.
DAP-kinase hypermethylation in MDS
In their letter, Brakensiek et al comment on our recently published paper on DAP-kinase hypermethylation in therapy-related acute myeloid leukemia (AML) and myelodysplastic syndromes (MDSs).1
They report that DAP-kinase was hypermethylated in a proportion of MDS similar to the one we reported (43% vs 47%). Using a real-time quantitative approach, they extend this observation, showing that the frequency of methylation is less than 5% and question the biologic significance of DAP-kinase hypermethylation in MDS. We think that this is an interesting finding, and, since the bone marrow cell distribution in MDS is heterogeneous, studies are warranted to evaluate the role of DAP-kinase methylation in this disease, by analyzing bone marrow subpopulations, including CD34+ cells, their progeny, and CD19+ B lymphocytes.
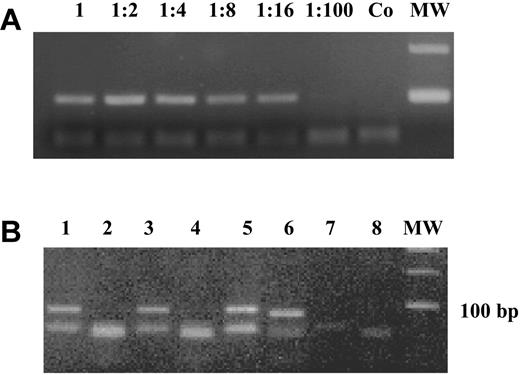
Low levels of DAP-kinase hypermethylation (0.003%-1.181%) have been reported in normal lymphocytes, especially in selected B cells (1%-6%), using a real-time methylation-specific polymerase chain reaction (MSP).2 We did not find any DAP-kinase hypermethylation in mononuclear cells isolated from 13 bone marrow and 15 peripheral blood samples from healthy individuals, of age similar to that of the patients, making it unlikely that the methylation we see in MDS could be due to contaminating B lymphocytes. Furthermore, the sensitivity of our polymerase chain reaction (PCR) technique might be too low to detect methylation at the low levels described by Reddy et al.2 We performed a dilution curve on 2 independent samples, in which we diluted a completely methylated sample into an unmethylated DNA: a distinct band was visible up to a dilution of 1:16 (6.75% positive; Figure 1A).
DAP-kinase methylation-specific PCR. (A) A completely methylated sample was diluted 1:2 to 1:100 into an unmethylated DNA and the “methylated” PCR reaction is shown. A distinct band was visible up to a dilution of 1:16 (6.75% positive). Co is the negative control and MW is the 100-bp molecular weight marker. (B) MSP for unmethylated and methylated DAP-kinase for 3 patients (patient a: lanes 1-2; patient b: lanes 3-4; and patient c: lanes 5-6). Adistinct band for the unmethylated reaction is visible at 106 bp for all patients, whereas only the DNAof patient c is methylated (98-bp band). Lanes 7 and 8 are negative controls for unmethylated and methylated DAP-kinase.
DAP-kinase methylation-specific PCR. (A) A completely methylated sample was diluted 1:2 to 1:100 into an unmethylated DNA and the “methylated” PCR reaction is shown. A distinct band was visible up to a dilution of 1:16 (6.75% positive). Co is the negative control and MW is the 100-bp molecular weight marker. (B) MSP for unmethylated and methylated DAP-kinase for 3 patients (patient a: lanes 1-2; patient b: lanes 3-4; and patient c: lanes 5-6). Adistinct band for the unmethylated reaction is visible at 106 bp for all patients, whereas only the DNAof patient c is methylated (98-bp band). Lanes 7 and 8 are negative controls for unmethylated and methylated DAP-kinase.
Apoptosis is truly an issue in MDS. Parker et al3 show that in MDS early disease is associated with excessive apoptosis and elevated ratio of apoptosis to proliferation. Increased proliferative rates are observed in RAEB, whereas leukemic transformation arises through inhibition of apoptosis. We agree that it would have been more correct to say “a common feature of high-risk MDSs is a decreased apoptosis rate in bone marrow progenitor cells.”
We used the methylation-specific PCR conditions reported by Katzenellenbogen et al,4 and data were not shown due to the limited space available for “Brief reports.” Figure 1B shows an example of MSP for unmethylated and methylated DAP-kinase.
Since bone marrow of patients with MDS is heterogeneous for cell content, we used for our expression analysis mRNA extracted from 37 bone marrow mononuclear cells, which usually contained more than 50% blasts, of patients with AML at the time of initial diagnosis. We showed that hypermethylation correlated to loss of expression. Aggerholm et al5 raised the question that MSP may overestimate the proportion of DAP-kinase gene methylation in AML, as they found methylation in 19 of 45 AML cases using MSP—confirming this finding by sequencing the PCR products— but only in 1 of 49 cases using a different technique (bisulfite-denaturing gradient gel electrophoresis [DGGE]). We (and many other authors) could confirm the functional role of DAP-kinase methylation, studied by MSP, by the corresponding lack of expression by reverse transcription (RT)–PCR. We agree that longitudinal studies from early disease to leukemic transformation, also using quantitative methylation assays, will help to clarify the contribution of DAP-kinase methylation to the pathogenesis and evolution of MDS and AML.
Correspondence: Maria Teresa Voso, Istituto di Ematologia, Universita' Cattolica S. Cuore, L.go A. Gemelli, 1 00168 Roma, Italy; e-mail: mtvoso@rm.unicatt.it

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal