Abstract
The functional analysis of the common prothrombin 20210 G>A(F2 20210*A) mutation has recently revealed gain of function of 3′end processing as a novel genetic mechanism predisposing to human disease. We now show that the physiologic G at the cleavage site at position 20210 is the functionally least efficient nucleotide to support 3′end processing but has evolved to be physiologically optimal. Furthermore, the F2 3′end processing signal is characterized by a weak downstream cleavage stimulating factor (CstF) binding site with a low uridine density, and the functional efficiency of F2 3′end processing can be enhanced by the introduction of additional uridine residues. The recently identified thrombosis-related mutation (F2 20221*T) within the CstF binding site up-regulates F2 3′end processing and prothrombin biosynthesis in vivo. F2 20221*T thus represents the first example of a likely pathologically relevant mutation of the putative CstF binding site in the 3′flanking sequence of a human gene. Finally, we show that the low-efficiency F2 cleavage and CstF binding sites are balanced by a stimulatory upstream uridine-rich element in the 3′UTR. The architecture of the F2 3′end processing signal is thus characterized by a delicate balance of positive and negative signals. This balance appears to be highly susceptible to being disturbed by clinically relevant gain-of-function mutations. (Blood. 2004;104:428-435)
Introduction
Blood coagulation is a tightly controlled process that, under physiologic conditions, maintains hemostasis and prevents intravascular thrombosis and embolism. Secondary hemostasis is activated and regulated by a network of endothelium, plasma, and platelet-associated coagulation enzymes that finally convert a small proportion of plasma fibrinogen to fibrin. Thrombin plays a key role in the regulation of the blood coagulation cascade because it acts as a procoagulant enzyme by activating factors XIII, XI, VIII, and V and the thrombin-activatable fibrinolysis inhibitor (TAFI) and by cleaving fibrinogen to release fibrin. Of equal importance, thrombin also acts as an anticoagulant enzyme by activating protein C.1
The pivotal role of thrombin in the regulation of hemostasis is illustrated by the identification of the common F2 20210*A allele. This mutation causes raised prothrombin plasma concentrations and predisposes carriers to develop thromboses.2-5 The F2 20210*A mutation is located in the 3′untranslated region (3′UTR) of the prothrombin mRNA at the most 3′ position where the premRNA is endonucleolytically cleaved and polyadenylated. The molecular mechanism of this mutation has previously been identified to represent a gain of function of 3′end formation6,7 and may also affect translation efficiency.8 It increases cleavage site (CS) recognition and accumulation of correctly 3′end processed mRNA accumulation in the cytoplasm, promoting protein synthesis. Enhanced mRNA 3′end formation efficiency thus emerged as a novel molecular principle causing pathologic gene expression and explains the role of F2 20210*A in the pathogenesis of thrombophilia.
The specificity and efficiency of 3′end processing is determined by the binding of a multiprotein complex to the 3′end processing signal.9,10 Most pre-mRNAs contain 2 core sequence elements. The canonical polyadenylation signal AAUAAA upstream of the CS is recognized by the cleavage/polyadenylation specificity factor (CPSF), which determines the site of cleavage 15 to 20 nucleotides (nt's) downstream. The second canonical sequence element is characterized by a high density of uridine residues and is located 10 to 30 nt's downstream of the CS (downstream U-rich sequence element). This sequence element is bound by the 64-kDa subunit of the heterotrimeric cleavage stimulating factor (CstF) that promotes the efficiency of 3′end processing.11 The CS itself is preferentially located immediately 3′ of a CA dinucleotide, although this is not stringently conserved.12 Mutations of the poly(A) signal commonly cause loss of function,13-21 whereas F2 20210*A represents the only known CS mutation and results in increased 3′end processing. Recently, a novel C>T mutation 11 nt's 3′ of the CS (F2 20221*T) has been identified in a child with an acute vascular rejection and intrarenal segmental arterial thrombosis of an allogeneic kidney transplant22 and a 28-year-old man with Budd-Chiari syndrome.23 The underlying molecular pathology has remained uncertain, although the patients' phenotypes suggest a gain of function. A systematic functional analysis of the entire F2 3′end processing signal has revealed an unusual architecture of noncanonical sequence elements, which appears to be susceptible to gain-of-function mutations.
Materials and methods
Constructs
The F2 wild-type (WT) hybrid gene construct has been described previously.6 In this construct, the 3′UTR and 62 nt's of the 3′flanking sequence (3′FS) of the β-globin gene (HBB) have been replaced by the 3′UTR and 140 nt's of the 3′FS of the human genomic F2 (schematically shown in Figures 1, 2, 3, 4, 5). Constructs F2 20210*A, F2 20210*C, F2 20210*T, F2 20221*A, F2 20221*G, and F2 20221*T were generated from the F2 WT hybrid construct by site-directed mutagenesis (GeneTailor site directed mutagenesis system; Invitrogen, Carlsbad, CA). In the hybrid gene construct F2-HBB, 140 nt's 3′ of the CS of F2 WT were replaced by the respective 62 nt's of the 3′FS of HBB by overlap polymerase chain reaction (PCR). Similarly, in HBB-F2 the 94-nt 3′UTR including the poly(A) signal (AATAAA) and the CS of F2 WT were replaced by the respective 133-nt 3′UTR sequences of HBB. The tandem CS constructs (F2/F2, 20210*A/F2, F2 20210*C/F2, F2 20210*T/F2, F2 20221*T/F2, F2-F2/F2, HBB-HBB/HBB, F2-F2/HBB, F2-HBB/F2, HBB-F2/F2) were generated by the downstream insertion of PCR-amplified fragments including either the F2 or the HBB 3′end formation signals as previously described.6
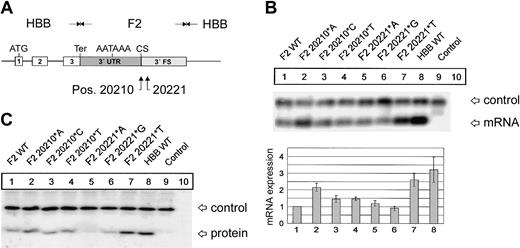
F2 mutations at positions 20210 and 20221 cause an increased abundance of F2 mRNA and protein. (A) In the hybrid genes, the 3′UTR including the poly(A) signal (AATAAA), the CS, the U-rich region, and 62 nt's of the 3′FS of the HBB gene were replaced by the respective sequences of F2 with either the normal F2 WT or the F2 20210*A, F2 20210*C, or F2 20210*T mutations at the CS (pos. 20210) or the F2 20221*A, F2 20221*G, or F2 20221*T mutations 11 nt's 3′ of the CS (pos. 20221). (B) Northern blot with an HBB-specific cRNA probe of cytoplasmic RNA preparations from cells cotransfected with the indicated hybrid constructs and the WT+300 E3 control. The RNA loaded in the control lanes 9 and 10 originate from cells that were either transfected with the WT+300 E3 control plasmid only or were not transfected. The bar diagram shows the fold difference relative to the F2 WT mRNA expression levels ± SD after normalization for transfection efficiency. Bars 1 to 8 represent quantification of lanes 1 to 8. (C) Immunoblot of protein lysates from cells transfected with constructs as shown in panel B. The blot was probed with an HBB-specific antibody.
F2 mutations at positions 20210 and 20221 cause an increased abundance of F2 mRNA and protein. (A) In the hybrid genes, the 3′UTR including the poly(A) signal (AATAAA), the CS, the U-rich region, and 62 nt's of the 3′FS of the HBB gene were replaced by the respective sequences of F2 with either the normal F2 WT or the F2 20210*A, F2 20210*C, or F2 20210*T mutations at the CS (pos. 20210) or the F2 20221*A, F2 20221*G, or F2 20221*T mutations 11 nt's 3′ of the CS (pos. 20221). (B) Northern blot with an HBB-specific cRNA probe of cytoplasmic RNA preparations from cells cotransfected with the indicated hybrid constructs and the WT+300 E3 control. The RNA loaded in the control lanes 9 and 10 originate from cells that were either transfected with the WT+300 E3 control plasmid only or were not transfected. The bar diagram shows the fold difference relative to the F2 WT mRNA expression levels ± SD after normalization for transfection efficiency. Bars 1 to 8 represent quantification of lanes 1 to 8. (C) Immunoblot of protein lysates from cells transfected with constructs as shown in panel B. The blot was probed with an HBB-specific antibody.
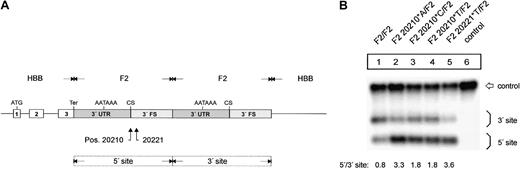
The F2 20210*A, F2 20210*C, F2 20210*T, and F2 20221*T mutations enhance F2 mRNA 3′end processing efficiency. (A) Schematic drawing of the constructs with tandem 3′end formation signals. (B) Northern blot of cells cotransfected with the tandem constructs and the WT+300 E3 control plasmid. The lower bands (5′ site) on the autoradiograph represent mRNAs that are cleaved and polyadenylated at the mutated 5′ site, whereas the upper bands (3′ site) correspond to transcripts that are processed at the reference 3′ site. Lane 6 shows the mRNA of cells transfected with the WT+300 E3 control only. The numbers shown below the panel represent the ratio of mRNA processed at the 5′ site relative to that processed at the 3′ site. The signals were quantified by phosphoimaging.
The F2 20210*A, F2 20210*C, F2 20210*T, and F2 20221*T mutations enhance F2 mRNA 3′end processing efficiency. (A) Schematic drawing of the constructs with tandem 3′end formation signals. (B) Northern blot of cells cotransfected with the tandem constructs and the WT+300 E3 control plasmid. The lower bands (5′ site) on the autoradiograph represent mRNAs that are cleaved and polyadenylated at the mutated 5′ site, whereas the upper bands (3′ site) correspond to transcripts that are processed at the reference 3′ site. Lane 6 shows the mRNA of cells transfected with the WT+300 E3 control only. The numbers shown below the panel represent the ratio of mRNA processed at the 5′ site relative to that processed at the 3′ site. The signals were quantified by phosphoimaging.
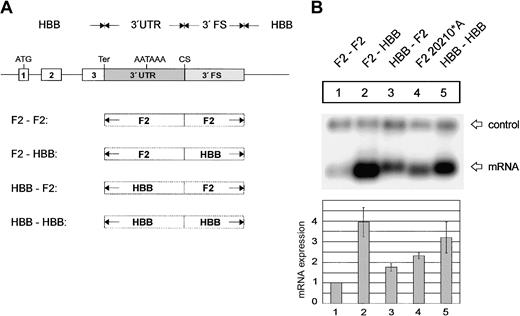
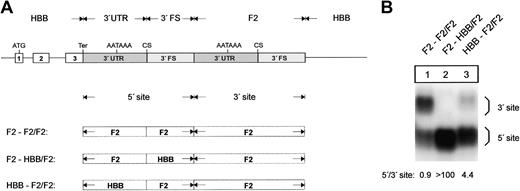
The F2 3′end formation signal is less efficient than the HBB signal. (A) Arrangement of the tandem constructs. (B) Northern blot of cytoplasmic RNA of cells cotransfected with the indicated tandem constructs and the WT+300 E3 control. The 3′FS of the HBB gene of the 5′ site is shorter than the 3′FS of the F2 gene. The HBB-HBB/HBB RNA (lane 2) processed at the 5′ site thus migrates faster than the respective F2-F2/F2 and F2-F2/HBB RNAs. The quantification of the signals (lanes 1-3) after normalization for the transfection efficiency control is shown in the bar diagram. The y-axis shows mRNA expression relative to F2-F2/F2, lane 1.
The F2 3′end formation signal is less efficient than the HBB signal. (A) Arrangement of the tandem constructs. (B) Northern blot of cytoplasmic RNA of cells cotransfected with the indicated tandem constructs and the WT+300 E3 control. The 3′FS of the HBB gene of the 5′ site is shorter than the 3′FS of the F2 gene. The HBB-HBB/HBB RNA (lane 2) processed at the 5′ site thus migrates faster than the respective F2-F2/F2 and F2-F2/HBB RNAs. The quantification of the signals (lanes 1-3) after normalization for the transfection efficiency control is shown in the bar diagram. The y-axis shows mRNA expression relative to F2-F2/F2, lane 1.
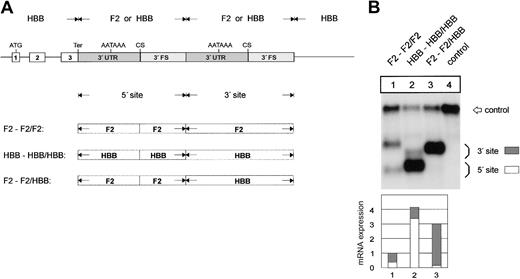
The F2 3′flanking sequence limits mRNA expression. (A) Arrangement of the hybrid constructs. (B) Northern blot of cytoplasmic RNAs of cells cotransfected with the indicated hybrid constructs and the WT+300 E3 control. The bar diagram shows the fold difference relative to the F2 WT mRNA expression levels ± SD after normalization for transfection efficiency. Bars 1 to 5 represent the quantification of lanes 1 to 5.
The F2 3′flanking sequence limits mRNA expression. (A) Arrangement of the hybrid constructs. (B) Northern blot of cytoplasmic RNAs of cells cotransfected with the indicated hybrid constructs and the WT+300 E3 control. The bar diagram shows the fold difference relative to the F2 WT mRNA expression levels ± SD after normalization for transfection efficiency. Bars 1 to 5 represent the quantification of lanes 1 to 5.
The 3′flanking sequence of the human prothrombin gene determines low efficiency of 3′end formation. (A) Arrangement of the tandem constructs. (B) Northern blot of cytoplasmic RNA of cells transfected with constructs shown in Figure 5A. The numbers shown below the panel represent the ratio of mRNA processed at the 5′ site relative to that processed at the 3′ site. The signals were quantified by phosphoimaging.
The 3′flanking sequence of the human prothrombin gene determines low efficiency of 3′end formation. (A) Arrangement of the tandem constructs. (B) Northern blot of cytoplasmic RNA of cells transfected with constructs shown in Figure 5A. The numbers shown below the panel represent the ratio of mRNA processed at the 5′ site relative to that processed at the 3′ site. The signals were quantified by phosphoimaging.
In the tandem constructs F2 ds U-rich(+1)/F2 and F2 ds U-rich(+2)/F2, one additional thymidine residue was inserted at the indicated positions 1 or 2 of the 5′ site of construct F2/F2, respectively (Figure 6A). In construct F2 ds U-rich(Δ)/F2, the sequence between positions 8 and 32 downstream of the 5′-located CS of F2/F2 was deleted. In construct F2 ds U-rich(HBB)/F2 these 24 nt's were replaced by the respective sequences of HBB. In constructs F2 USE(Δ)/F2 and F2 USE(N)/F2, a 15-nt U-rich sequence located between positions -33 and -17 relative to the 5′-located polyadenylation signal of F2/F2 was deleted or replaced by an unrelated sequence (5′-ACGAGACGAGCGCGC-3′). In construct HBB-F2/F2 + USE, a 15-nt sequence from position -33 to -17 relative to the 5′-located polyadenylation signal of HBB6 was replaced by the respective 15-nt sequence of the F2 gene. All modifications were confirmed by DNA sequencing.
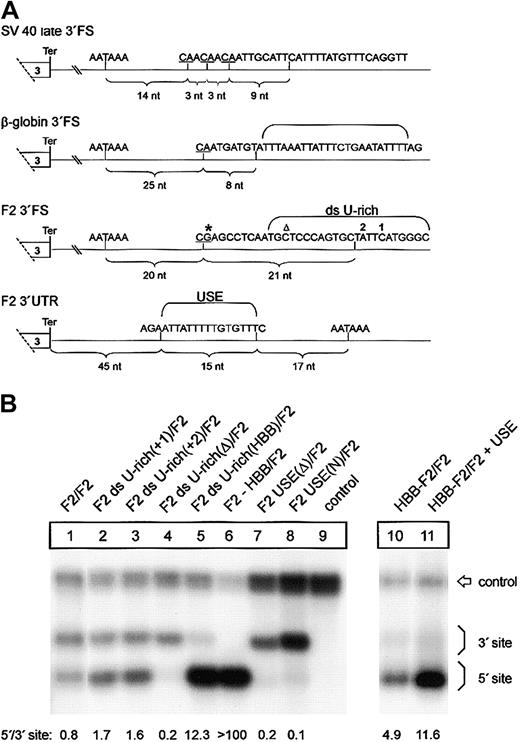
A low-efficiency downstream uridine-rich element and a noncanonical USE balance 3′end formation of the human prothrombin gene. (A) Comparison of the DNA encoding the 3′end processing signals of SV40 late, HBB, and F2. The CS in the pre-RNAs are underlined, and the respective distance to the polyA signals is indicated by brackets. The thymidine residues of the downstream U-rich element (ds U-rich) or the upstream U-rich element (USE) are highlighted. Positions 20210 and 20221 are indicated by * and ▵, respectively. (B) Northern blot of cytoplasmic RNA of cells transfected with tandem constructs with modifications of the ds U-rich element (lanes 2-5) or the USE (lanes 7-8). In construct HBB-F2/F2 + USE (lane 11), the F2 USE was inserted in the HBB 3′UTR of construct HBB-F2/F2 (lane 10). The numbers shown below the panel represent the ratio of mRNA processed at the 5′site relative to that processed at the 3′ site. The signals were quantified by phosphoimaging.
A low-efficiency downstream uridine-rich element and a noncanonical USE balance 3′end formation of the human prothrombin gene. (A) Comparison of the DNA encoding the 3′end processing signals of SV40 late, HBB, and F2. The CS in the pre-RNAs are underlined, and the respective distance to the polyA signals is indicated by brackets. The thymidine residues of the downstream U-rich element (ds U-rich) or the upstream U-rich element (USE) are highlighted. Positions 20210 and 20221 are indicated by * and ▵, respectively. (B) Northern blot of cytoplasmic RNA of cells transfected with tandem constructs with modifications of the ds U-rich element (lanes 2-5) or the USE (lanes 7-8). In construct HBB-F2/F2 + USE (lane 11), the F2 USE was inserted in the HBB 3′UTR of construct HBB-F2/F2 (lane 10). The numbers shown below the panel represent the ratio of mRNA processed at the 5′site relative to that processed at the 3′ site. The signals were quantified by phosphoimaging.
Cell culture and transfections
HeLa cells were grown in Dulbecco modified Eagle medium under standard conditions and were transiently transfected by calcium phosphate precipitation. For Northern blot analysis, 30 μg of the test plasmids were cotransfected with 3 to 5 μg WT+300 E3,24 which served as a control for transfection efficiency and gel loading. The cells were washed after 20 hours and harvested 24 hours after washing.
HepG2 and HUH-7 cells were grown in Dulbecco modified Eagle medium under standard conditions. Transfection of HUH-7 cells was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions with 16 μg test plasmid in 10 mL cell culture medium (10-cm-diameter cell culture dish) and 40 μL Lipofectamine 2000 reagent. The cells were harvested after 24 hours after transfection.
RNA analysis
Northern blot analysis was performed as previously described with 5 μg total cytoplasmic RNA.25 For the high-resolution Northern blot analysis of mRNA in the in vivo competition assay, 7.5 μg total cytoplasmic RNA was deadenylated by hybridization of oligo(dT) and subsequent treatment with RNAse H as previously described.26 The gel electrophoresis was then carried out on a 1.7% agarose gel.
Ribonuclease protection analysis (RPA) was performed with 10 μg total cytoplasmic RNA using a probe specific for the F2 3′UTR and 140 nt's of the 3′flanking sequence as previously described.27
Autoradiographic signals were quantified by imaging in a phosphoimager FLA-3000 (Fujifilm, Stamford, CT).
For the poly(A) test (PAT) assay 1.5 μg cytoplasmic HUH-7 mRNA was first reverse transcribed. To generate highly resolvable polyadenylation site-specific cDNAs we have developed a modified reverse transcription technique using equimolar amounts of 3 different anchor-oligo(dT)16X primers (5′-TGCTGGTCGATGCACGTGACGC(T)16A-3′, 5′-TGCTGGTC GATGCACGTGACGC(T)16C-3′, and 5′-TGCTGGTCGATGCACGTGACGC(T)16G-3′) using SuperScript (Invitrogen) according to the manufacturers' instruction manual. Thus, a homogeneous (“focused”) pool of 2 different cDNA species primed at the most 5′ end of each poly(A) tail was synthesized after reverse transcription. This technique, followed by a PCR reaction, allowed a reliable quantification even of trace amounts of cDNAs representing the mRNAs processed at either the 5′ or the 3′ polyadenylation site. The subsequent PCR amplification of the cDNAs representing the alternatively polyadenylated mRNA species was carried out with a β-globin-specific exon 2 sense primer (5′-TGGTCTACCCTTGGACCCAGAGGTTCTTT-3′) and an anchor-specific antisense primer (5′-TGCTGGTCGATGCACGTGACGC-3′) as previously described.27 The result was visualized on a 2% agarose gel stained with ethidium bromide, and the band intensities were quantified by using the Quantity One software (Bio-Rad, Hercules, CA).
Immunoblotting was performed as previously described.25
Results
Prothrombin (F2)3′end formation is enhanced by mutations at positions 20210 and 20221
As a first step to gain insight into the physiology and pathology of 3′end formation, we have systematically analyzed sequence elements that determine F2 mRNA 3′end formation efficiency. The first target of our analysis was position 20210, the site of the common G>A mutation. The physiologic G at position 20210 of the hybrid constructs (Figure 1A) was additionally changed to C and T. The resulting F2 20210*C and F2 20210*T constructs were transiently cotransfected with the WT+300 E3 control plasmid. Compared with the F2 WT (Figure 1B, lane 1), both the F2 20210*C (Figure 1B, lane 3) and F2 20210*T (Figure 1B, lane 4) mutations led to a 1.4- (± 0.2) and 1.5- (± 0.1) fold increased mRNA expression, whereas the disease-related F2 20210*A mutation (Figure 1B, lane 2) increased mRNA abundance by 2.15- (± 0.2) fold. This increased mRNA expression correlates with increased protein synthesis (Figure 1C). This result is consistent with an in vitro analysis of general 3′end processing signals, which showed the highest 3′end formation efficiency with an A and the lowest with a G at the CS, while C and T were intermediate.12 Interestingly, therefore, the least efficient nucleotide has evolved as the physiologic and presumably optimal F2 CS. This explains the unusual susceptibility of the F2 CS to gain-of-function mutations.
Recently, intrarenal segmental arterial thromboses of an allogeneic kidney transplant have been reported in a 9-year-old patient with a novel F2 C>T mutation at position 20221.22 Recently, the same mutation has also been identified in a 28-year-old man with Budd-Chiari syndrome.23 This mutation is of particular interest because it is located 11 nt's 3′ of the CS and is not included in the mature mRNA. The phenotype of these patients is consistent with a gain of function of this mutation and suggests that the F2 3′end formation signal contains another low-efficiency wild-type signal. We have therefore systematically analyzed the effect of mutations at position 20221 on F2 mRNA expression (Figure 1B, lanes 5-7). The F2 20221*A and F2 20221*G mutation (Figure 1B, lanes 5-6) show similar mRNA levels compared with F2 WT (Figure 1B, lane 1), whereas the disease-related F2 20221*T mutation (Figure 1B, lane 7) increases the mRNA expression levels by 2.6- (± 0.4) fold (Figure 1B, lane 7), which is also reflected at the protein level (Figure 1C). This is similar to the expression levels of a β-globin mRNA (Figure 1B, HBB WT, lane 8) that is known to be efficiently 3′end processed.6 Furthermore, this result indicates that the F2 3′flanking sequence (FS) is an important component of the 3′end processing machinery and is sensitive to gain-of-function mutations.
We next studied the mechanism of the increased mRNA expression by an analysis of the mutations that we refer to as an in vivo 3′end processing competition assay6 to differentiate this experimental system from cell-free analyses. We generated constructs that contain a tandem array of 3′end formation signals with mutations at positions 20210 or 20221 within the 5′ site (Figure 2A). In contrast, the unmodified 3′ site consists of sequences originating from the wild-type F2 3′UTR and 3′FS. The smaller mRNA species in the Northern blot analysis is cleaved and polyadenylated at the 5′ site, whereas the longer mRNAs are processed at the 3′ site. Thus, this experimental setting enabled us to directly compare the processing efficiency of the (mutated) 5′ site in relation to the 3′ site. Importantly, this setting also provides an internal control for other mechanisms, such as transcription, splicing efficiency, or mRNA stability,28 which may potentially influence the abundance of the mRNA encoded by the transfected constructs.
The in vivo competition assay (Figure 2B) shows that the F2 20210*C (Figure 2B1, F2 20210*C/F2, lane 3) and F2 20210*T (Figure 2B, F2 20210*T/F2, lane 4) mutations enhance 3′end formation efficiency when compared with the F2 20210*G (Figure 2B, F2 WT, lane 1) although this effect is not as strong as it is for F2 20210*A (Figure 2B, lane 2). This result is consistent with the different levels of mRNA expression observed with the constructs containing only one 3′end processing site (Figure 1B, lanes 2-4). Furthermore, the in vivo competition assay revealed that the F2 20221*T mutation enhances 3′end formation to a similar extent as does F2 20210*A, which demonstrates that this point mutation in the 3′FS that is not included in the mature RNA can up-regulate F2 3′end formation. F2 20221*T thus represents a clinically relevant bona fide gain-of-function mutation that is located outside the mature mRNA that is itself not altered in its sequence.
The 3′flanking sequence is a second major determinant of the low F2 3′end formation efficiency
Next, we analyzed which region of the 3′end formation signal contains the sequence elements that determine 3′end formation efficiency. The constructs used for this analysis contain a tandem array of F2 3′end formation signals originating from the F2 or the HBB genes (Figure 3A). The hybrid mRNA with an array of 2 F2 3′end formation signals is preferentially processed at the 3′ site (Figure 3B, lane 1), whereas the RNA with a set of 2 HBB sites is preferentially cleaved at the 5′ site (Figure 3B, lane 2). Furthermore, the overall expression level of the mRNA with the HBB sites is approximately 4 times that of the mRNA with the F2 sites (Figure 3B, compare lanes 1 and 2). When the 3′ F2 signal is replaced by HBB sequences, processing is almost completely shifted to the 3′ site (Figure 3B, lane 3), while the overall expression level is only slightly reduced in comparison with the construct containing 2 HBB signals. The HBB signal thus confers high-level overall mRNA expression and competes efficiently with the weaker F2 3′end processing signal.
We further determined whether the sequences conferring these differences in 3′end processing efficiency are contained in the 3′UTR or in the 3′FS. This was tested with constructs containing single low-efficiency, single high-efficiency, or hybrid 3′end formation signals (Figure 4A). The replacement of the F2 3′FS in construct F2-F2 by the HBB 3′FS (construct F2-HBB) results in a 3.9- (± 0.7) fold increase of mRNA expression (Figure 4B, lane 2), which is similar to the expression of the construct with a complete HBB 3′end formation signal (Figure 4B, HBB-HBB, lane 5). By contrast, when the F2 3′UTR is replaced by the HBB 3′UTR (construct HBB-F2) there is only a 1.7- (± 0.2) fold increase of mRNA expression (Figure 4B, lane 3). This increase is similar to that caused by the F2 20210*A mutation (Figure 4B, lane 4) and can thus likely be explained by the presence of the CA dinucleotide at the CS of construct HBB-F2. The slight but reproducible difference between the expression of constructs HBB-F2 (Figure 4B, lane 3) and F2 20210*A (Figure 4B, lane 4) can probably be accounted for by the lack of the upstream uridine-rich element in the HBB 3′UTR (see the next section). These data indicate that in addition to the G residue at the CS the major determinant of the weak F2 3′end processing signal resides in the 3′FS.
Next, we analyzed the specificinfluence of the 3′FS and 3′UTR determinants on 3′end processing efficiency in tandem array constructs (Figure 5A). When the F2 3′FS was replaced by the HBB 3′FS at the 5′ site, processing was almost completely shifted to the 5′ site (Figure 5B, lanes 1-2). Replacement of the F2 3′UTR by the HBB 3′UTR also resulted in a shift of 3′end processing although this was not as complete as the shift affected by the 3′FS (Figure 5B, lane 3). The F2 3′end processing signal thus contains another low-efficiency sequence element.
A noncanonical low-efficiency downstream U-rich region and an unusual stimulatory upstream U-rich region balance F2 3′end formation
We next sought to determine the sequence elements in the 3′FS that confer the differential behavior of the low-efficiency type F2 and the high-efficiency type HBB 3′end processing signals. A sequence comparison with high-efficiency 3′end processing signals such as those derived from SV40 late or HBB shows that the F2 3′FS between positions +9 and +30 contains a low number of uridine residues (Figure 6A). This area, referred to as the downstream U-rich sequence element, is expected to recruit the 64-kDa CstF subunit that plays a key role in 3′end processing.9,10 Therefore, we tested the hypothesis that the low uridine content of the F2 3′FS causes weak 3′end processing. We introduced one additional uridine residue into the 3′FS at the 5′ site of constructs with a tandem array of F2 3′end formation signals at 2 different positions, 1 or 2 (Figure 6A), which enhances processing at this site (Figure 6B, F2 ds U-rich(+1)/F2, F2 ds U-rich(+2)/F2, lanes 2 and 3, respectively). The degree of up-modulation caused by these mutations appears to be less than for the F2 20221*T mutation, which may be related to the longer distance between positions 1 and 2 than position 20221 from the CS. We further increased the number of uridines in the F2 3′FS between positions +8 and +32 downstream of the 5′-located CS from 7 to 14 by replacing the entire downstream U-rich sequence by the corresponding region of the HBB gene (Figure 6B, lane 5, F2 ds U-rich(HBB)/F2). This increase of uridine number further enhanced 3′end processing at the mutated site (Figure 6B, lane 5). By contrast, deletion of the downstream U-rich region at the 5′ site (Figure 6B, lane 4, F2 ds U-rich(Δ)/F2) abolishes processing at this site almost completely, whereas processing at the 3′ site remains essentially unaltered (Figure 6B, lane 4). This result confirms the critical role of the downstream U-rich region in 3′end processing and demonstrates that the weak F2 3′end formation efficiency can be attributed to the low number of uridines within the downstream element in addition to the G residue at the CS. Furthermore, this finding is also consistent with the observation in an in vitro analysis that increasing the number of uridines within a downstream U-rich element promotes CstF binding and leads to enhanced 3′end maturation and increased mRNA expression.29 Thus, the novel F2 20221*T mutation increases the physiologically low number of uridines within the downstream U-rich element and illustrates that this noncanonical sequence is prone to clinically relevant gain-of-function mutations.
In light of the unusually inefficient CS and downstream U-rich element, the question arises as to how F2 3′end processing is achieved. In viral genes, 3′end formation efficiency can be promoted by upstream sequence elements (USEs) that are typically located within the 3′UTRs.30 More recently, human genes with noncanonical polyadenylation signals have been identified to require auxiliary upstream U-rich elements to promote efficient 3′end formation.31-33 Interestingly, the F2 3′UTR contains a 15-nt U-rich region (Figure 6A) that is absent in HBB and is located at a position where a uridine-rich stretch promotes 3′end processing in other genes with noncanonical polyadenylation signals as shown for the C2 complement gene,31 the lamin B gene,32 or human collagen genes.33 We tested the putative stimulatory role of this region and either deleted or replaced this 15 nt region by an unrelated sequence at the 5′ site of constructs with a tandem array of F2 3′end formation signals (Figure 6B, constructs F2 USE(Δ)/F2 and F2 USE(N)/F2, lanes 7 and 8, respectively). When the USE was either deleted or replaced by an unrelated sequence, processing at the 5′ site was almost completely abolished whereas processing at the 3′ site remained essentially unaltered (Figure 6B, lanes 1 and 7-8; compare 3′ signal intensities relative to the control). By contrast, introduction of the 15-nt USE into the 3′UTR of a heterologous HBB context in construct HBB-F2/F2 + USE (Figure 6B, lane 11) further promotes 3′end processing of transcripts generated at this site (Figure 6B, compare lanes 10 and 11).
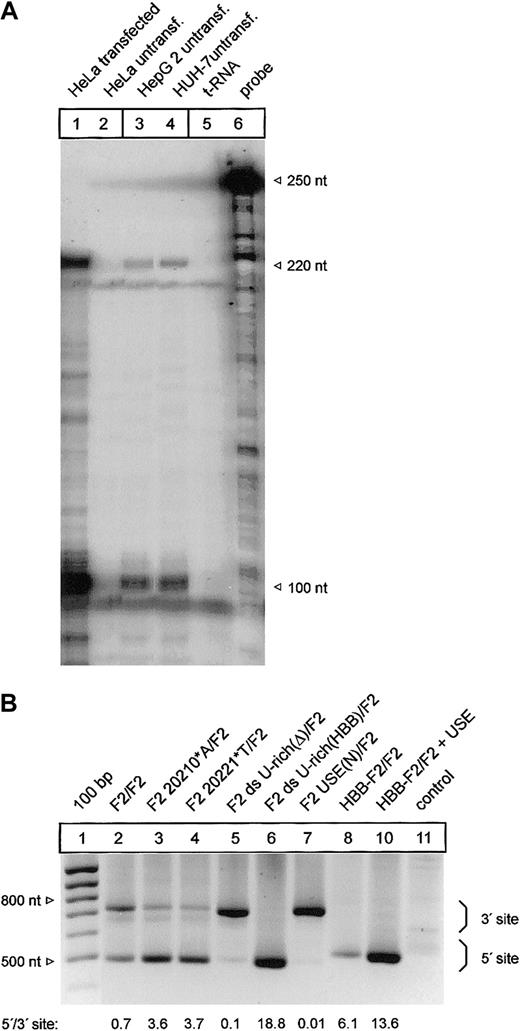
It has been shown that mRNA processing can occur in a highly tissue-dependent manner.34 We have therefore analyzed the F2 3′end processing signal in HepG2 and HUH-7 liver cell lines that constitutively express prothrombin (Figure 7A, lanes 3-4). Importantly, this ribonuclease protection analysis also revealed that transfected HeLa cells that have been used for all previous analyses express F2 mRNA whose 3′end structure is identical to the endogenous F2 mRNA expressed in HepG2 and HUH-7 cells (Figure 7A, compare lanes 1, 3, and 4).
Mutations of the cleavage site and the U-rich element in the 3′flanking sequence affect 3′end processing in a cell line expressing the endogenous F2 gene. (A) Comparison of the F2 3′end mRNA structure by ribonuclease protection analysis of transfected HeLa cells (lane 1) and HepG 2 and HUH-7 cells (lanes 3-4). (B) Modified PAT analysis of cytoplasmic RNA of HUH-7 cells transfected with tandem constructs shown in Figures 2 and 6B. The numbers shown below the panel reflect semiquantitative estimates of the signals generated by limited PCR.
Mutations of the cleavage site and the U-rich element in the 3′flanking sequence affect 3′end processing in a cell line expressing the endogenous F2 gene. (A) Comparison of the F2 3′end mRNA structure by ribonuclease protection analysis of transfected HeLa cells (lane 1) and HepG 2 and HUH-7 cells (lanes 3-4). (B) Modified PAT analysis of cytoplasmic RNA of HUH-7 cells transfected with tandem constructs shown in Figures 2 and 6B. The numbers shown below the panel reflect semiquantitative estimates of the signals generated by limited PCR.
Finally, we used a modified and highly sensitive PCR-based poly(A) test (PAT) assay to specifically analyze the F2 3′end processing efficiency of mRNA derived from transfected HUH-7 cells with constructs with a tandem array of F2 3′end formation signals (Figure 7B). The PAT analysis confirmed that both the G>A mutation at position 20210 (Figure 7B, F2 20210*A/F2, lane 3) and the C>T mutation at position 20221 (Figure 7B, F2 20221*T/F2, lane 4) increase 3′end processing efficiency at the mutated 5′ site when compared with the reference 3′ site. In this semiquantitative analysis, the degree of up-modulation by the F2 20210*A and the 20221*T mutations appeared to be comparable to the data generated in transfected HeLa (Figure 2, lanes 2 and 5). Furthermore, the deletion of the ds U-rich element (Figure 7B, F2 ds U-rich(Δ)/F2, lane 5), the replacement of the USE (Figure 7B, F2 USE(N)/F2, lane 7), or the insertion of the HBB-specific ds U-rich element (Figure 7B, F2 ds U-rich(HBB)/F2, lane 6) and the introduction of the F2 USE in a heterologous HBB context (Figure 7B, HBB-F2/F2 + USE, lane 10) resulted in similar changes of the F2 3′end processing efficiency in HUH-7 cells as were observed in HeLa cells (Figure 6B).
These data demonstrate that the USE within the F2 3′UTR balances the low efficiency of the CS and the putative CstF-64 binding site and indispensably maintains F2 mRNA expression by stimulating 3′end processing both in an epithelial cell line that does not express endogenous F2 and in a liver cell line that does express endogenous F2. Moreover, this unusual architecture of the 3′end processing signal appears to be highly susceptible to clinically relevant gain-of-function mutations.
Discussion
The complexity of the human genome is determined by both the number of genes and by alternative mRNA processing such as alternative splicing,35 pre-mRNA editing,36 or alternative polyadenylation.30,37 Consequently, it is essential that the functional diversity of gene expression that results from alternative and regulated mRNA processing is monitored by proofreading mechanisms.38-42 The medical relevance of RNA processing is highlighted by a large number of disease-related mutations that affect mRNA metabolism and its proofreading.43 This is exemplified by mutations of the highly conserved polyadenylation signal (AAUAAA), which invariably inactivate gene expression,13-21 and the loss-of-function mutation of the poly(A) binding protein 2 (PABP2) in patients with oculopharyngeal muscular dystrophy.44 In contrast, the F2 RNA contains a 3′end processing signal that is sensitive for a novel type of mutation that results in gain of function of RNA maturation.
Our systematic analysis of the F2 3′end processing signal reveals an unusual architecture of noncanonical sequence elements that appears to balance F2 gene expression. The F2 cleavage site and uridine-rich element in the 3′ flanking sequence are functionally less efficient than those of β-globin. In contrast, the F2 3′UTR contains a USE that promotes 3′end processing and is absent in β-globin. We now demonstrate that both the F2 CS and the F2 uridine-rich element in the 3′FS predispose to gain-of-function mutations. This is exemplified by the identification of the F2 20221*T allele that to our knowledge represents the first disease-related mutation of the downstream U-rich element to be of documented functional and clinical significance. In this context it is important to note that this posttranscriptionally relevant mutation escapes detection on the basis of reverse transcriptase (RT)-PCR sequencing, and diagnosis must be ascertained by genomic DNA analysis. Moreover, it is interesting that the effect of the F2 20210*A mutation may be further modulated by the general architecture of the 3′FS and by variations of intronic sequences that have been reported to affect splicing efficiency.45
The USE in the F2 3′UTR is of particular interest. This previously unrecognized sequence element is located between positions -33 and -17 5′ of the polyadenylation signal and promotes 3′end processing in the context of the poor CS and 3′FS of F2 and in the context of the high-efficiency CS and 3′FS of β-globin. Similar sequence elements have previously been identified in viral genes and have been referred to as upstream sequence elements (USEs). These noncanonical sequence elements are often U-rich and thought to serve as recognition sites for factors that stabilize the polyadenylation complex.30 In contrast, very little is known about USEs in mammalian genes.31-33 It is largely unknown how USEs promote 3′end formation, although USEs in mammalian genes appear to be capable of directly stabilizing the binding of CPSF to the poly(A) signal.32 More recently, human Fip1 has been identified as a subunit of CPSF that binds to U-rich RNA elements and stimulates the poly(A) polymerase.46
An interesting question that has not been directly addressed here relates to the physiological significance of the unique architecture of the F2 3′end processing signal. 3′end processing can be modified during different phases of the cell cycle,47 at different growth conditions,48 in tissue-specific gene expression,49 and at different developmental states.30,37,50 Further analyses will address the question of whether the overall low efficiency of F2 3′end processing and the synthesis of the procoagulatory prothrombin may be up-regulated physiologically as a response to exogenous, possibly stress-related, stimuli.
Prepublished online as Blood First Edition Paper, April 1, 2004; DOI 10.1182/blood-2003-08-2894
Supported by the Deutsche Forschungsgesellschaft (DFG) and the DFG Forschergruppe (FOR 426): “complex RNA-protein interactions in the maturation and function of eukaryotic mRNA.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal