Imatinib is a tyrosine kinase inhibitor that is effective in the treatment of chronic myeloid leukemia (CML). Not all patients achieve cytogenetic response. Some patients even lose the initial cytogenetic response. In this study, we investigated the active cellular transport of imatinib to gain a better understanding of the possible mechanisms of imatinib resistance. We used the leukemic cell line CCRFCEM and its drug-resistant subline VBL100 to measure the uptake of carbon 14 (14C)-labeled imatinib. Imatinib uptake was temperature dependent, indicative of an active uptake process. Additionally, incubations with transport inhibitors showed that verapamil, amantadine, and procainamide, inhibitors of the human organic cation transporter 1 (hOCT1), significantly decreased imatinib uptake into CEM cells, whereas the inhibition of hOCT2 or hOCT3 had no effect, indicating that influx into the cells is an active process likely to be mediated by hOCT1. Studies using transfected MDCK cell lines revealed an active efflux component attributable to MDR1 (ABCB1). Both hOCT1 and MDR1 were expressed in CML primary cells and cell lines. The results indicate that active transport processes mediate the influx and efflux of imatinib. Differential expression of influx (hOCT1) and efflux (MDR1) transporters may be a critical determinant of intracellular drug levels and, hence, resistance to imatinib.
Introduction
Approximately 95% of patients with chronic myeloid leukemia (CML) are positive for the Philadelphia chromosome, 22q-, which results from a reciprocal translocation between chromosomes 9 and 22.1 A result of this is production of the fusion protein, BCR-ABL, a constitutively active tyrosine kinase thought to be responsible for cell transformation through the activation of downstream signaling pathways.2 Screening of a group of compounds, 2-phenylaminopyrimidines, resulted in the discovery of imatinib mesylate, a potent and selective inhibitor of BCR-ABL.3 Imatinib has since been shown in clinical trials to be highly effective in CML.4 In a phase 2 study of patients in chronic phase for whom interferon-α (IFN-α) was failing, 95% of patients achieved complete hematologic response (CHR), and 60% achieved major cytogenetic response (MCR).5 In a recent phase 3 trial in newly diagnosed disease, imatinib had superior CHR and MCR compared with IFN.6 However, as clinical experience with imatinib grows, it is clear that some patients are resistant to imatinib, and cytogenetic response does not occur. Other patients experience relapse after initial success with the drug.7
Resistance to drug therapy is a common problem in many diseases, particularly in malignant conditions. Some of the mechanisms leading to imatinib resistance have already been elucidated. For example, amplification of the BCR-ABL gene and overexpression of the protein have been shown to occur in vitro and in vivo,8-10 and consequently a higher drug concentration is required to drive the cell to apoptosis.11 Additionally, mutations in the BCR-ABL protein can confer imatinib resistance, either by altering imatinib binding characteristics or through indirect modulation of kinase function.12-15
Little, however, is known about how imatinib enters and leaves a cell and about whether those actions are mediated by active transport and may be contributory mechanisms of resistance. Recent papers have shown that imatinib is a substrate for P-glycoprotein (MDR1, ABCB1), a drug transporter commonly involved in multidrug resistance.16-18 However, ABCB1 is only one of many different transport proteins, many of which have also been implicated in drug resistance.19,20
In addition to the transporters involved in drug efflux, a number of transporters are responsible for drug uptake. These are important in drug absorption, distribution, and elimination in vivo and have been particularly well studied in organs such as the kidneys and the gastrointestinal tract.21 This group of transporters includes such families as the organic cation transporters (OCTs), and organic anion transporters (OATs). Whether imatinib is a substrate for other transporters, including those responsible for uptake, has not been investigated. The aim of this study, therefore, was to assess the contribution of other drug transporters to the cellular transport of imatinib to provide insight into other possible mechanisms by which patients with CML are resistant to imatinib.
Patients, materials, and methods
Materials
CEM and VBL100 cells were kind gifts from Dr R. Davey (Sydney, Australia), and MDCK and sublines transfected with ABCB1 were kind gifts from Prof P. Borst (Netherlands Cancer Institute, Amsterdam, the Netherlands). Carbon 14 (14C)-labeled imatinib was kindly provided by Novartis (Basel, Switzerland) (specific activity, 3.386 MBq/mg). Ultima Gold scintillation cocktail was purchased from Canberra Packard (Didcot, Oxfordshire, United Kingdom). All other drugs and chemicals, unless otherwise stated, were purchased from Sigma (Poole, Dorset, United Kingdom).
Cell culture
CCRF-CEM cells are a human T-lymphoblastoid cell line derived from a patient with acute lymphoblastic leukemia. VBL100 cells constitute a multidrug-resistant subline that overexpresses ABCB1 through exposure to vinblastine. CCRF-CEM and VBL100 cells were maintained in RPMI 1640 supplemented with 2 mM l-glutamine and 10% fetal calf serum (FCS). VBL100 cells were treated every 6 weeks with vinblastine (100 ng/mL) to maintain resistance. MDCK II cells are a canine kidney cell line shown to be useful in the study of permeability and drug transport by efflux transporters such as MDR1.22 They can be grown on filters to create a polarized epithelial cell monolayer, across which the movement of chemical compounds can be measured. MDCK II cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FCS, 2 mM l-glutamine, and penicillin and streptomycin. They were passaged every 3 to 4 days using trypsin/EDTA (ethylenediaminetetraacetic acid) to detach the cells from the flasks. Transport media consisted of Hanks balanced salt solution (HBSS) with 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) at pH 7.4.
CML cell lines and patients
The CML lines KYO1, LAMA84, KCL22, and K562 (donated by the LRF Leukaemia Unit, Hammersmith Hospital, London, United Kingdom) were maintained in RPMI 1640 medium supplemented with l-glutamine and 10% FCS. Peripheral blood leukocytes were collected at the start of imatinib treatment for 4 patients with CML in first chronic phase (CP) and 2 patients in accelerated phase (AP). Ethical approval was obtained from the Liverpool Local Research Ethics Committee. Informed consent was provided according to the Declaration of Helsinki.
Radiolabeled drug uptake
14C-labeled imatinib was used in all experiments at a concentration of 1 μg/mL, and nonradiolabeled imatinib was added to give final concentrations of 5 μM for the time-course and uptake experiments or variable concentrations, as appropriate.
Cells were suspended in transport media, and 2 million cells were added to imatinib-containing incubation media with or without transport inhibitors. Verapamil (20 μM) was used as an inhibitor initially of ABCB1 and subsequently of human organic cation transporter 1 (hOCT1). PSC-833 is a more specific inhibitor of ABCB1 and was used at a concentration of 10 μM to inhibit the transporter. A transport inhibitor that affects imatinib efflux through ABCB1 would be expected to cause an increase in imatinib accumulation in VBL100 cells only, not in CEM cells, because these show only low expression of ABCB1. Inhibitors affecting imatinib uptake, however, cause a decrease in imatinib uptake in CEM cells and should also affect accumulation in VBL100 cells. Cells and media were gently mixed and incubated at 37°C for the appropriate length of time—60 minutes for uptake experiments and up to 2 hours for time-course experiments. After incubation, ice-cold phosphate-buffered saline (PBS) was added to the cells and was centrifuged at 700g, 4°C, for 3 minutes. The resultant cell pellet was washed twice with ice-cold PBS and again centrifuged. One hundred microliters distilled water was added to the cells and vortexed to break up the pellet. Cells were solubilized by incubation with 200 μL 0.2 M NaOH and 1% sodium dodecyl sulfate (SDS) for 1 hour. The resultant solution was mixed with 3.5 mL scintillation fluid, and radioactivity was counted using a β-counter (1500 Tri Carb LS Counter; Packard).
Measurement of transepithelial drug transport
MDCK II cells, transfected with MDR1 or nontransfected controls, were seeded at a density of 2 × 106 cells per well onto a 23.1-mm diameter, 3-μm pore size, presoaked semipermeable membrane (FalconCell Culture Inserts; Fred Baker Scientific, Runcorn, Cheshire, United Kingdom). Cells were cultured for 10 days, and media were changed in apical and basal compartments every day until they formed a confluent monolayer. On the day of the assay, the cells were allowed to equilibrate in the transport media at 37°C. Transepithelial resistance for each of the monolayers was measured to check the integrity of the monolayer. Monolayer transepithelial electrical resistance (TEER) was measured in Ω/cm2 using a Millicell-ERS Voltohmmeter (Millipore, Watford, United Kingdom), and a corrected value was obtained by subtracting the TEER value of a blank filter from the value obtained from the monolayer. Monolayers with TEERs less than 90 Ω/cm2 were not used because this has previously been determined to be the lower limit for MDCK monolayers.22 A tracer amount of 14C-labeled imatinib was added to the donor well, and enough nonradiolabeled drug was added to give a final concentration of 5 μM. Monolayers were incubated with the drug for a total of 3 hours, and at various time points 50 μL solution was sampled from the acceptor compartment and was replaced with 50 μL warmed transport media. This was then added to 3.5 mL scintillation cocktail and was counted in a beta scintillation counter.
Manual and real-time quantitative reverse transcription-polymerase chain reaction
To detect the expression of MDR1 and hOCT1 in CML cell lines and primary cells from patients with CML, RNA was extracted and cDNA was synthesized using standard methodologies. cDNA was amplified through polymerase chain reaction (PCR) on a LightCycler (Roche Diagnostics, East Sussex, United Kingdom) using LightCycler FastStart DNA MasterPlus SYBR Green I (Roche Diagnostics) with 0.5 μM each primer (MWG Biotech, Milton Keyes, United Kingdom). PCR was performed at 95°C for 10 minutes followed by 40 cycles of denaturation at 95°C for 2 seconds, annealing for 10 seconds, extension at 72°C for 20 seconds, and single signal acquisition for 10 seconds. Annealing temperature, signal acquisition temperature, and primer sequences for each gene are listed in Table 1. For the hOCT1 gene, primers were designed to amplify cDNA of the full-length hOCT1 isoform only. GAPDH gene expression was used as an internal control. To design primers specific for GAPDH gene amplification, GAPDH pseudogene sequences were searched from the National Center for Biotechnology Information (NCBI) GenBank database. Homology of the genuine GAPDH sequence (GenBank accession number NM_002046) to the sequences of each of the 17 known GAPDH pseudogenes was assessed using the ClustalW multiple sequence alignment tool provided by European Bioinformatics Institute (EBI). Using this strategy, GAPDH reverse transcription (RT)-PCR primers were designed to bind to regions that have low sequence homology to the GAPDH pseudogenes. RNA that had not been reverse transcribed was also amplified for all 3 genes to ensure there was no amplification of any contaminating genomic DNA. Primers were designed, and PCR conditions were optimized to obtain a PCR product with a single peak on melting curve analysis on the LightCycler. Standard curves for each gene analyzed were constructed from serial dilutions of VBL cell cDNA to obtain the linear range for quantification of gene expression. PCR product was also run on a 1.0% agarose gel and stained with ethidium bromide to obtain a single band of the expected size. PCR products from all 3 genes were gel-purified by using GenElute Agarose Spin Column (Sigma, Poole, Dorset, United Kingdom) and were directly sequenced using forward and reverse PCR primers, respectively (Lark Technologies, Essex, United Kingdom).
PCR primer sequences and conditions
Genes . | Forward and reverse primers . | Annealing temperature, °C . | Acquisition temperature, °C . | Amplicon size, bp . |
|---|---|---|---|---|
| hOCT1 | OCT3F: 5′-GGGCAGCCTGCCTCGTCATG | 68 | 82 | 229 |
| OCT1503R: 5′-ACCTCCCTCAGCCTGAAGAC | ||||
| MDR1 | MDR744F: 5′-TGGAGGAAGACATGACCAGG | 65 | 80 | 455 |
| MDR1178R: 5′-CAAGACCTCTTCAGCTACTGC | ||||
| GAPDH | G12: 5′-AGCCACATCGCTCAGACAC | 65 | 82 | 472 |
| G471: 5′-GAGGCATTGCTGATGATCTTG |
Genes . | Forward and reverse primers . | Annealing temperature, °C . | Acquisition temperature, °C . | Amplicon size, bp . |
|---|---|---|---|---|
| hOCT1 | OCT3F: 5′-GGGCAGCCTGCCTCGTCATG | 68 | 82 | 229 |
| OCT1503R: 5′-ACCTCCCTCAGCCTGAAGAC | ||||
| MDR1 | MDR744F: 5′-TGGAGGAAGACATGACCAGG | 65 | 80 | 455 |
| MDR1178R: 5′-CAAGACCTCTTCAGCTACTGC | ||||
| GAPDH | G12: 5′-AGCCACATCGCTCAGACAC | 65 | 82 | 472 |
| G471: 5′-GAGGCATTGCTGATGATCTTG |
Raw data collected from the LightCycler were analyzed using LightCycler Software Version 3.5 (Roche Diagnostics). hOCT1 and MDR1 gene expression levels were normalized with GAPDH gene expression levels in the same sample. hOCT1 and MDR1 gene expression was compared between samples by relating them to the respective levels in VBL cells, which were arbitrarily scored as 1.0.
Statistics
Time-course experiments for drug uptake were analyzed using analysis of variance (ANOVA) with repeated measures, after testing whether the data were normally distributed. For uptake in the presence of inhibitors, an unpaired t test was used. Data are all presented as mean ± SEM. For the transepithelial transport assays, results are presented as drug transport per square centimeter. Data are shown as mean ± SEM from observations of 4 to 6 separate monolayers.
Apparent permeability (Papp, expressed in cm/s) of the MDCK monolayers was calculated from the following formula:
where dC/dt is a change in drug concentration in the acceptor compartment per unit time, V is the volume (mL) of the acceptor compartment, A is the area (cm2) of the membrane, and Co is the initial concentration (mg/mL) of the drug in the donor compartment.22 These data were analyzed using one-way ANOVA with Tukey HSD. From this the efflux ratio of the monolayers was calculated using the following formula:
Results
Imatinib uptake into CEM and VBL100 cells
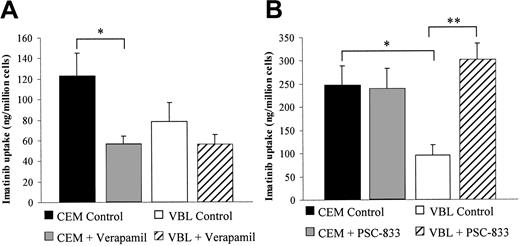
Uptake of imatinib into CEM and VBL100 (the ABCB1-overexpressing subline of CEM) cells was compared with and without the use of transport inhibitors. The P-glycoprotein (ABCB1) inhibitors verapamil (20 μM) and PSC-833 (10 μM) were used initially to determine whether ABCB1 contributed to cellular efflux transport. Data given in Table 2 and Figure 1 show that PSC-833, a specific ABCB1 inhibitor,23 had little effect on CEM cells but approximately doubled imatinib accumulation in VBL cells. Verapamil, a less specific ABCB1 inhibitor,24 actually decreased imatinib uptake in CEM cells.
Comparison of imatinib uptake by CEM and VBL100 cells in the presence or absence of ABCB1 inhibitors
. | Cell line . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Drug . | CEM* . | . | VBL100† . | . | |||
| Imatinib control | 123.1 (22.2) | P < .05 | 78.3 (18.4) | N/S | |||
| Imatinib + verapamil | 56.9 (7.4) | 56.3 (9.1) | |||||
| Imatinib control | 247.3 (42.0) | N/S | 95.5 (23.2) | P < .001 | |||
| Imatinib + PSC-833 | 239.7 (44.1) | - | 301.7 (35.8) | - | |||
. | Cell line . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Drug . | CEM* . | . | VBL100† . | . | |||
| Imatinib control | 123.1 (22.2) | P < .05 | 78.3 (18.4) | N/S | |||
| Imatinib + verapamil | 56.9 (7.4) | 56.3 (9.1) | |||||
| Imatinib control | 247.3 (42.0) | N/S | 95.5 (23.2) | P < .001 | |||
| Imatinib + PSC-833 | 239.7 (44.1) | - | 301.7 (35.8) | - | |||
Data are presented as mean (SEM) of 4 observations.
In the CEM cell line, P < .05 when imatinib alone was compared with imatinib plus verapamil; the P value was not significant when imatinib alone was compared with imatinib plus PSC-833
In the VBL 100 cell line, the P value was not significant when imatinib alone was compared with imatinib plus verapamil; P < .001 when imatinib alone was compared with imatinib plus PSC-833
Uptake of imatinib into CEM and VBL100 cells in the presence of ABCB1 inhibitors. Cells were incubated for 60 minutes in transport media with 14C-labeled imatinib and (A) 20 μM verapamil or (B) 10 μM PSC-833. Data represent mean ± SEM from 4 experiments. *Significantly different from CEM control (P < .05). **Significantly different from VBL control (P < .001).
Uptake of imatinib into CEM and VBL100 cells in the presence of ABCB1 inhibitors. Cells were incubated for 60 minutes in transport media with 14C-labeled imatinib and (A) 20 μM verapamil or (B) 10 μM PSC-833. Data represent mean ± SEM from 4 experiments. *Significantly different from CEM control (P < .05). **Significantly different from VBL control (P < .001).
Effect of temperature on imatinib transport
Because verapamil is known to interact with transport mechanisms other than ABCB1,25 the effect on imatinib transport of lowering the incubation temperature to 4°C was investigated. Several concentrations of imatinib were used in the uptake experiment, ranging from 0.5 to 100 μM. There was a significant difference in the uptake at 4°C when compared with 37°C for all concentrations of imatinib tested, except 0.5 μM (Table 3). These data demonstrate that imatinib uptake is an active process.
Comparison of imatinib uptake by CEM cells at 4°C and 37°C
. | Imatinib uptake, ng/million cells . | . | |
|---|---|---|---|
| Concentration, μM . | 4°C . | 37°C . | |
| 0.5 | 4.0 (11) | 10.5 (2.7) | |
| 1 | 10.7 (2.1) | 32.5 (6.4)* | |
| 5 | 40.4 (2.9) | 420.5 (70.2)* | |
| 10 | 51.9 (6.7) | 794.6 (159.5)* | |
| 50 | 249.9 (54.3) | 3156.1 (489.6)† | |
| 100 | 606.9 (108.8) | 4243.0 (417.5)† | |
. | Imatinib uptake, ng/million cells . | . | |
|---|---|---|---|
| Concentration, μM . | 4°C . | 37°C . | |
| 0.5 | 4.0 (11) | 10.5 (2.7) | |
| 1 | 10.7 (2.1) | 32.5 (6.4)* | |
| 5 | 40.4 (2.9) | 420.5 (70.2)* | |
| 10 | 51.9 (6.7) | 794.6 (159.5)* | |
| 50 | 249.9 (54.3) | 3156.1 (489.6)† | |
| 100 | 606.9 (108.8) | 4243.0 (417.5)† | |
Data are presented as mean (SEM) of 4 observations.
Significantly different from incubation at 4°C (P < .05)
Significantly different from incubation at 4°C (P < .005)
Inhibition of organic cation transporters
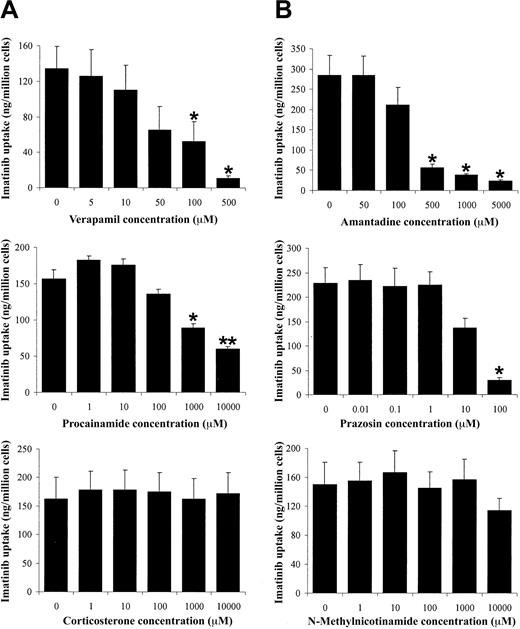
Verapamil has been shown to interact with the hOCT1.25 Therefore, to determine whether hOCT was involved in imatinib uptake, a number of other hOCT-inhibiting compounds were coincubated with imatinib (Figure 2; Table 4). A number of different transport inhibitors had to be used because of the overlapping substrate specificities of many of the organic cation transporters. In addition, hOCT, hOCT2, and hOCT3 were investigated because of the known interaction of verapamil with hOCT125 and the overlapping substrate specificities seen between these transporters.
Effect of OCT inhibitors on imatinib uptake by CEM cells. (A) Verapamil, a P-glycoprotein and hOCT1 inhibitor, decreased imatinib uptake. (B) Amantadine and (C) procainamide, both hOCT1 and hOCT2 inhibitors, also decreased imatinib uptake. (D) Prazosin, an hOCT1 and hOCT3 inhibitor, decreased imatinib uptake. (E) The hOCT3 inhibitor corticosterone had no effect on imatinib uptake. (F) N-methylnicotinamide, an hOCT2 inhibitor, had no effect on imatinib uptake. Data are presented as mean plus or minus SEM of 4 observations. *Significantly different from control (P < .05). **Significantly different from control (P < .01).
Effect of OCT inhibitors on imatinib uptake by CEM cells. (A) Verapamil, a P-glycoprotein and hOCT1 inhibitor, decreased imatinib uptake. (B) Amantadine and (C) procainamide, both hOCT1 and hOCT2 inhibitors, also decreased imatinib uptake. (D) Prazosin, an hOCT1 and hOCT3 inhibitor, decreased imatinib uptake. (E) The hOCT3 inhibitor corticosterone had no effect on imatinib uptake. (F) N-methylnicotinamide, an hOCT2 inhibitor, had no effect on imatinib uptake. Data are presented as mean plus or minus SEM of 4 observations. *Significantly different from control (P < .05). **Significantly different from control (P < .01).
Effect of OCT inhibitors on imatinib uptake by CEM cells
Inhibitor . | Inhibitor concentration, μM . | Imatinib uptake, ng/million cells . | Comments . |
|---|---|---|---|
| Verapamil | Control | 133.9 (25.3) | Known to inhibit hOCT125 |
| 5 | 125.9 (29.7) | ||
| 10 | 110.3 (27.9) | ||
| 50 | 65.5 (26.1) | ||
| 100 | 52.4 (22.5)* | ||
| 500 | 10.9 (2.7)* | ||
| Amantadine | Control | 284.2 (48.6) | Inhibits hOCT1 and hOCT226 |
| 50 | 284.8 (47.2) | ||
| 100 | 211.8 (42.8) | ||
| 500 | 56.6 (8.3)* | ||
| 1000 | 39.4 (1.9)* | ||
| 5000 | 24.5 (2.4)* | ||
| Procainamide | Control | 156.8 (12.6) | Inhibits hOCT1 and hOCT226 |
| 1 | 182.8 (8.8) | ||
| 10 | 175.6 (6.8) | ||
| 100 | 136.0 (5.6) | ||
| 1000 | 89.1 (3.5)* | ||
| 10 000 | 59.9 (2.2)† | ||
| Prazosin | Control | 228.6 (32.3) | Inhibits hOCT1 and hOCT327 |
| 0.01 | 235.1 (31.3) | ||
| 0.1 | 222.5 (36.2) | ||
| 1 | 224.8 (26.9) | ||
| 10 | 136.5 (20.2) | ||
| 100 | 29.4 (5.0)* | ||
| Corticosterone | Control | 162.7 (37.3) | Inhibits hOCT327 |
| 0.001 | 178.7 (31.9) | ||
| 0.01 | 178.1 (34.6) | ||
| 0.1 | 175.4 (33.4) | ||
| 1 | 162.8 (34.9) | ||
| 10 | 171.4 (37.0) | ||
| N-methylnicotinamide | Control | 150.1 (30.7) | Inhibits hOCT2 at nanomolar and micromolar concentrations28 |
| 1 | 154.7 (26.9) | ||
| 10 | 166.8 (30.2) | ||
| 100 | 145.0 (22.4) | ||
| 1000 | 156.8 (27.8) | ||
| 10 000 | 114.3 (16.6) |
Inhibitor . | Inhibitor concentration, μM . | Imatinib uptake, ng/million cells . | Comments . |
|---|---|---|---|
| Verapamil | Control | 133.9 (25.3) | Known to inhibit hOCT125 |
| 5 | 125.9 (29.7) | ||
| 10 | 110.3 (27.9) | ||
| 50 | 65.5 (26.1) | ||
| 100 | 52.4 (22.5)* | ||
| 500 | 10.9 (2.7)* | ||
| Amantadine | Control | 284.2 (48.6) | Inhibits hOCT1 and hOCT226 |
| 50 | 284.8 (47.2) | ||
| 100 | 211.8 (42.8) | ||
| 500 | 56.6 (8.3)* | ||
| 1000 | 39.4 (1.9)* | ||
| 5000 | 24.5 (2.4)* | ||
| Procainamide | Control | 156.8 (12.6) | Inhibits hOCT1 and hOCT226 |
| 1 | 182.8 (8.8) | ||
| 10 | 175.6 (6.8) | ||
| 100 | 136.0 (5.6) | ||
| 1000 | 89.1 (3.5)* | ||
| 10 000 | 59.9 (2.2)† | ||
| Prazosin | Control | 228.6 (32.3) | Inhibits hOCT1 and hOCT327 |
| 0.01 | 235.1 (31.3) | ||
| 0.1 | 222.5 (36.2) | ||
| 1 | 224.8 (26.9) | ||
| 10 | 136.5 (20.2) | ||
| 100 | 29.4 (5.0)* | ||
| Corticosterone | Control | 162.7 (37.3) | Inhibits hOCT327 |
| 0.001 | 178.7 (31.9) | ||
| 0.01 | 178.1 (34.6) | ||
| 0.1 | 175.4 (33.4) | ||
| 1 | 162.8 (34.9) | ||
| 10 | 171.4 (37.0) | ||
| N-methylnicotinamide | Control | 150.1 (30.7) | Inhibits hOCT2 at nanomolar and micromolar concentrations28 |
| 1 | 154.7 (26.9) | ||
| 10 | 166.8 (30.2) | ||
| 100 | 145.0 (22.4) | ||
| 1000 | 156.8 (27.8) | ||
| 10 000 | 114.3 (16.6) |
Data are presented as mean (SEM) of 4 observations.
Significantly different than control (P < .05)
Significantly different than control (P < .01)
Figure 2 shows that of the 6 inhibitors used, verapamil (an hOCT1 inhibitor25 ), amantadine and procainamide (both hOCT1 and hOCT2 inhibitors26 ), and prazosin (an hOCT1 and an hOCT3 inhibitor27 ) decreased imatinib uptake in a concentration-dependent manner. However, when the relatively specific hOCT3 inhibitor corticosterone27 was used, there was no change in imatinib uptake across the concentration range used, implying that in these cells hOCT3 does not contribute to imatinib uptake and that the effect of prazosin was the result of an interaction with hOCT1. N-methylnicotinamide, at nanomolar and micromolar concentrations, inhibits hOCT2.28 Adding this compound, however, had no significant effect on imatinib uptake, suggesting that hOCT2 is unlikely to transport imatinib. Taken together, our data suggest that imatinib uptake into the CEM and VBL cell lines was mediated by hOCT1.
Transport of imatinib in transfected MDCK monolayers
To further confirm the involvement of ABCB1 (MDR1) in imatinib transport, polarized MDCK cells were grown in monolayers on a semipermeable filter. The formation of confluent, functional MDCK monolayers was confirmed by the use of the TEER measurement. Mean TEER values for MDCK II and MDCKII-MDR1 monolayers used in the experiments were 183 and 205 Ω/cm2, respectively. These were all above the recommended lower limit for MDCK monolayers.22 The presence of the transfected proteins in these cells lines has previously been confirmed through Western blotting in our laboratory.29
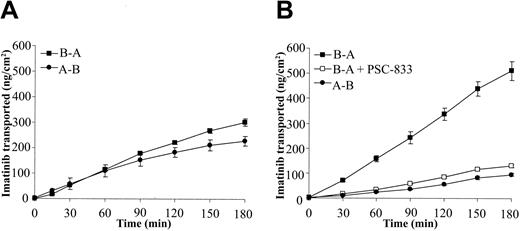
Figure 3 shows the transport from the basolateral to apical (B-A, secretory transport) sides of the monolayer and from the apical to basolateral sides (A-B, absorptive transport), in addition to transport in the presence of the ABCB1 inhibitor, PSC-833. In the nontransfected (Figure 3A) cells, there was no significant difference between B-A and A-B transport, indicating that there was minimal transport of imatinib in these cells. In the ABCB1-transfected cells (Figure 3B), however, there was a much greater rate of B-A transport of imatinib than of A-B transport, indicating the presence of an active secretory process on the apical side of the cell (Table 5). B-A transport was inhibited by the transport inhibitor PSC-833 (100 μM), a specific ABCB1 inhibitor.23 The apparent permeability of the monolayers for transport of imatinib in each direction was calculated (Table 5), and the efflux ratio was determined. Together, these data confirm the involvement of ABCB1 in imatinib transport.
Transport of imatinib across transfected MDCK monolayers. MDCK monolayers were incubated with transport media with or without imatinib and transport inhibitors. The transport of 14C-labeled imatinib was measured every 30 minutes for 3 hours in the apical-basolateral (A-B, •) and the basolateral-apical (B-A, ▪) directions. A transport inhibitor, PSC-833 (10 μM), was included (□) in the media of the (B) MDR1-transfected monolayers but not in the (A) nontransfected parental cell line. Data represent mean ± SEM from 4 to 6 separate monolayers.
Transport of imatinib across transfected MDCK monolayers. MDCK monolayers were incubated with transport media with or without imatinib and transport inhibitors. The transport of 14C-labeled imatinib was measured every 30 minutes for 3 hours in the apical-basolateral (A-B, •) and the basolateral-apical (B-A, ▪) directions. A transport inhibitor, PSC-833 (10 μM), was included (□) in the media of the (B) MDR1-transfected monolayers but not in the (A) nontransfected parental cell line. Data represent mean ± SEM from 4 to 6 separate monolayers.
Apparent permeability of imatinib across MDCK cell monolayers
. | MDCK II cm/s × 108 . | MDCK-MDR1 cm/s × 108 . |
|---|---|---|
| Apical-basal | 1.0 (0.22) | 0.64 (0.03) |
| Basal-apical | 2.1 (0.10) | 4.6 (0.37)* |
| Basal-apical + transport inhibitor | N/A | 1.08 (0.04)† |
| Efflux ratio | 2.1 | 7.2 |
. | MDCK II cm/s × 108 . | MDCK-MDR1 cm/s × 108 . |
|---|---|---|
| Apical-basal | 1.0 (0.22) | 0.64 (0.03) |
| Basal-apical | 2.1 (0.10) | 4.6 (0.37)* |
| Basal-apical + transport inhibitor | N/A | 1.08 (0.04)† |
| Efflux ratio | 2.1 | 7.2 |
Data are presented as mean (SEM).
Significantly different from apical-basal transport (P < .001)
Significantly different from basal-apical transport (P < .001)
Expression of hOCT1 and MDR1 in cell lines and primary CML cells
It has previously been demonstrated that CEM cells express several different types of OCT.30 To confirm that hOCT1 was expressed in our cell lines, RT-PCR was undertaken using specific primers. A 229-bp fragment was detected in the cDNA of CEM and VBL100 cells (Figure 4). Direct sequencing of the PCR product confirmed that this fragment was amplified from the full-length functional isoform of the hOCT1 gene. Similar direct sequencing experiments of the MDR1 gene PCR product confirmed the expected 455-bp sequence. The RT-PCR product (472 bp) for GAPDH was also directly sequenced and was found to be an exact match of the expected genuine GAPDH gene sequence, but it did not match any of the available GAPDH pseudogene sequences (data not shown). There was no specific amplification of any contaminating genomic DNA for any of the 3 genes (Figure 4).
hOCT1 and MDR1 expression in cell lines and primary CML cells. RT-PCR products were run on an agarose gel to show expression of the hOCT1 and MDR1 genes. The expected 229-bp product for hOCT1 and the 455-bp product for MDR1 were present in VBL, CEM cell lines, and CML cell lines LAMA84, KYO1, and KCL22. Both hOCT1 and MDR1 were expressed in 4 cases of CP (CP1 to CP4) and 2 cases of AP (AP1 and AP2). The 472-bp product for GAPDH gene expression was present in all cell lines and CML cells.
hOCT1 and MDR1 expression in cell lines and primary CML cells. RT-PCR products were run on an agarose gel to show expression of the hOCT1 and MDR1 genes. The expected 229-bp product for hOCT1 and the 455-bp product for MDR1 were present in VBL, CEM cell lines, and CML cell lines LAMA84, KYO1, and KCL22. Both hOCT1 and MDR1 were expressed in 4 cases of CP (CP1 to CP4) and 2 cases of AP (AP1 and AP2). The 472-bp product for GAPDH gene expression was present in all cell lines and CML cells.
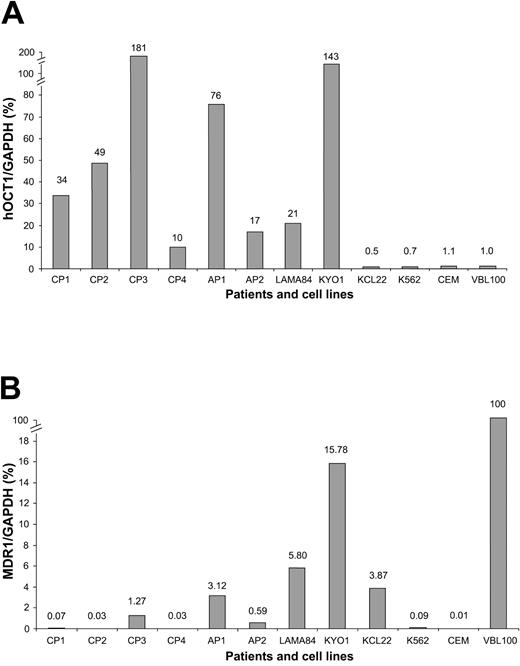
RT-PCR results also showed that hOCT1 was expressed in all 4 CML cell lines. KYO1 and LAMA84 cells expressed hOCT1 at higher levels than the other cell lines—approximately 21- and 143-fold higher than in VBL cells, respectively (Figure 5A). MDR1 gene transcripts were also detected in all 4 cell lines (Figures 4, 5), though expression was lower than 16% of VBL MDR1 expression (Figure 5B).
Quantification of hOCT and MDR1 gene expression in cell lines and primary CML cells by real time RT-PCR. (A) hOCT1 and (B) MDR1 levels are expressed relative to those in VBL cells (arbitrary level of 1.0).
Quantification of hOCT and MDR1 gene expression in cell lines and primary CML cells by real time RT-PCR. (A) hOCT1 and (B) MDR1 levels are expressed relative to those in VBL cells (arbitrary level of 1.0).
In the primary CML cells, hOCT1 and MDR1 were expressed in all 6 patients before imatinib treatment at varying levels (Figures 4, 5). All samples expressed greater levels of hOCT1 than VBL cells (range, 10- to 181-fold; Figure 5A). Conversely, all samples expressed lower levels of MDR1 than VBL cells (range, 0.03-3.1%; Figure 5B). GAPDH gene amplification (internal control) was present in all cell lines and primary CML cells.
Discussion
Imatinib has become a key player in the treatment of CML through its novel mechanism of action.4 However, some patients show de novo resistance, and others develop resistance after an initial response. Some of the mechanisms of resistance include gene and protein amplification,12 mutations in the BCR-ABL protein rendering the cells insensitive to imatinib, binding of imatinib to α1-acid glycoprotein in the plasma,31 and further genetic alterations that may confer an additional growth advantage on the cells.13 However, few of these studies have investigated the role of drug transporters, a well-known mechanism of resistance in a number of other malignant diseases.32 Drug transporters play a key role in drug absorption, distribution, and elimination in vivo21 and, at the cellular level, in the transport of drugs across cell membranes and, hence, in determining intracellular concentrations. Investigation of the role of these transporters in drug efflux and influx has largely taken place using the same cell systems used in this study.21,29
In this report, we show 3 important findings relevant to the intracellular concentrations of imatinib. First, uptake of imatinib into the cell is predominantly an active rather than a passive process, as demonstrated by the temperature dependency of the uptake process. Second, the influx of imatinib seems to be mediated by hOCT1. The OCT proteins are responsible for the cellular uptake and excretion of a number of exogenous and endogenous cationic and uncharged substances.33 Compared with the ABC transporter family,34 however, the role of OCTs in multidrug resistance has been less well studied. Our data using a number of inhibitors suggest that imatinib is a substrate for hOCT1, but not for hOCT2 or hOCT3. Because hOCT1 mediates drug uptake rather than efflux, its down-regulation may influence the response to chemotherapy. In accordance with this, hOCT1 expression was shown not only in the cell lines used in the study but also in primary cells taken from patients with CML at varying levels before imatinib treatment (Figure 5). Theoretically, therefore, fewer transporters would result in a lower intracellular concentration of imatinib (Figure 6). Whether this is indeed the case must be investigated in patients followed up longitudinally to determine whether hOCT1 correlates with primary or acquired resistance to imatinib.
Consequences of altered transporter expression on intracellular imatinib accumulation. (A) Under normal conditions (A), an adequate imatinib concentration is achieved inside the cell and the leukemia cell undergoes cell death. (B) With the underexpression of uptake transporters or the overexpression of efflux transporters, the intracellular imatinib concentration is too low to inhibit BCR-ABL to an extent that will cause cell death. Im indicates imatinib.
Consequences of altered transporter expression on intracellular imatinib accumulation. (A) Under normal conditions (A), an adequate imatinib concentration is achieved inside the cell and the leukemia cell undergoes cell death. (B) With the underexpression of uptake transporters or the overexpression of efflux transporters, the intracellular imatinib concentration is too low to inhibit BCR-ABL to an extent that will cause cell death. Im indicates imatinib.
Third, we show that imatinib is a substrate for ABCB1, which confirms the findings of Dai et al16 and Mahon et al.18 Additionally, Widmer et al35 and Illmer et al36 have recently shown that ABCB1-expressing cells have lower intracellular concentrations of imatinib, which is again supported by our data in 2 cell lines, one of which was transfected with ABCB1 and has been used as a model to demonstrate the transport of many other drugs37 (Figures 1, 4). Our data have been produced using radiolabeled imatinib, which has thus far not been used in other studies of this type. This has an advantage in that it is easier to track the accumulation of imatinib and its movement across monolayers. This finding is also consistent with the mechanisms of drug resistance in other malignancies in which MDR1 gene amplification has been demonstrated.31,38 In contradiction to this study and other studies cited above,16,18,35,37 Ferrao et al39 have recently suggested that ABCB1 may not play a significant role in resistance to imatinib. However, only a single cell line, K562, was used, and it is possible that this cell line might have had a differential expression of influx transporters such as hOCT1, resulting in decreased uptake and, hence, a false-negative result regarding the role of ABCB1 in imatinib efflux. Indeed, we have analyzed the expression of hOCT1 and MDR1 in K562 cells and have found that the expression of hOCT1 and MDR1 was low compared with that of other cell lines and of primary CML cells (Figure 5).
It is, however, important to remember that ABCB1 is only one member of the ABC transporter family, which comprises 7 subfamilies (ABCA through ABCG), encoded by 48 genes.40 The substrate specificities of the individual transporter proteins are wide ranging, but there is also a great deal of overlap among transporters. With respect to imatinib, the only other ABC transporter studied has been ABCC1 (MRP1). One study demonstrates that imatinib is not a substrate for MRP1,41 and another suggests an interaction with MRP1 at supratherapeutic concentrations only.42
What are the clinical implications of our findings that imatinib undergoes uptake through hOCT1 and efflux by ABCB1? The net effect of these transport processes may be a decrease in the intracellular concentration of imatinib (Figure 6). The consequences of this are several-fold. First, the cell may be less likely to undergo apoptosis, thereby leading to the persistence of imatinib resistant subclones. An imatinib-resistant cell line produced by culturing the cells in suboptimal concentrations of imatinib8 showed a 4-fold increase in expression of the BCR-ABL protein. The BCR-ABL in these cells was still sensitive to imatinib, but, because of the increased protein levels, a higher intracellular imatinib concentration was required to induce apoptosis. Thus, the cells were effectively resistant at therapeutic concentrations.
Second, the decrease in intracellular imatinib concentration may also lead to selection and clonal expansion of cells harboring mutant copies of the BCR-ABL gene, which would render the protein insensitive to imatinib. Indeed, Ricci et al43 have reported the development of a resistant clone of the KBM5, BCR-ABL-positive, myeloid cell line. These cells were cultured in sublethal concentrations of imatinib, giving rise to the resistant subline that, unlike those produced by Le Coutre et al,8 only displayed a marginal increase in BCR-ABL protein expression. Imatinib resistance in these cells was caused by a point mutation in the adenosine triphosphate (ATP)-binding domain of the protein. This mutation was not detectable in the parental cell line, suggesting that suboptimal drug exposure caused the mutation to arise and the cells to become insensitive to imatinib.
To date, there have been no studies on the role of the OCT in hematologic malignancies. Here we show that hOCT1 is expressed at varying levels in CML cell lines and in primary CML cells (Figures 4, 5). However, the clinical implications of this novel finding need further investigation in patient samples collected longitudinally in relation to cellular resistance, prognosis, and interactions. By contrast, the clinical implications of the overexpression of ABCB1 are better understood. For example, ABCB1 expression has been shown to be a prognostic indicator in acute lymphocytic leukemia,44 though its role in CML is unclear. Apart from the effects on blast cells, ABCB1 expression in the gastrointestinal tract may reduce the absorption and bioavailability of imatinib. It may be possible to overcome resistance to imatinib in these instances by the use of transport inhibitors.40,41 A further clinical implication for the transport of imatinib by ABCB1 is in relation to drug interactions. For instance, rifampicin, a broad-spectrum antibacterial, is known to induce intestinal expression of ABCB1 and to decrease the bioavailability of digoxin, a known ABCB1 substrate.45 Furthermore, rifampicin also induces the expression of ABCB1 in lymphocytes46 and may exert similar effects in myeloid cells. Thus, the net consequence of the coprescription of imatinib with rifampicin would be reduced bioavailability and reduced intracellular concentrations in leukemic cells.
In summary, we have shown that the anti-CML drug imatinib is transported out of cells by ABCB1 (MDR1) and into cells by an active uptake mechanism that is most likely to be hOCT1. Our findings indicate that the differential expression or function of these transporters will lead to a reduction in intracellular imatinib concentrations. These pharmacokinetic mechanisms of resistance to imatinib may occur in addition to, or may induce, the well-described pharmacodynamic mechanisms involving the BCR-ABL gene in patients with CML. These data should provide a launching point from which further studies in BCR-ABL-positive cell lines and patient samples can be carried out. These may reveal any correlation between expression and function of these transporters and clinical prognosis and may allow the development of strategies to overcome imatinib resistance and increase its clinical usefulness.
Prepublished online as Blood First Edition Paper, August 17, 2004; DOI 10.1182/blood-2003-12-4276.
Supported by the Royal Liverpool University Hospital Research and Development Fund.
J.T. and L.W. contributed equally to this paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Novartis Pharmaceuticals for supplying radiolabeled imatinib.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal