Congenital afibrinogenemia, the most severe form of fibrinogen deficiency, is characterized by the complete absence of fibrinogen. The disease is caused by mutations in 1 of the 3 fibrinogen genes FGG, FGA, and FGB, clustered on the long arm of human chromosome 4. The majority of cases are due to null mutations in the FGA gene although one would expect the 3 genes to be equally implicated. However, most patients studied so far are white, and therefore the identification of causative mutations in non-European families is necessary to establish if this finding holds true in all ethnic groups. In this study, we report the identification of a novel nonsense mutation (Arg134Xaa) in the FGG gene responsible for congenital afibrinogenemia in 10 patients from Lebanon. Expression studies in COS-7 cells demonstrated that the Arg134Xaa codon, which is encoded by adjacent exons (TG-intron 4-A) affected neither mRNA splicing nor stability, but led to the production of an unstable, severely truncated fibrinogen γ chain that is not incorporated into a functional fibrinogen hexamer.
Introduction
Inherited disorders of fibrinogen are rare and affect either the quantity (hypofibrinogenemia and afibrinogenemia) or the quality (dysfibrinogenemia) of circulating fibrinogen. Fibrinogen is produced in the liver from 3 homologous polypeptide chains, Aα, Bβ, and γ, which assemble to form a hexamer containing 2 copies of each chain. Fibrinogen is the precursor of fibrin, the major protein constituent of the blood clot. Congenital afibrinogenemia (OMIM: online Mendelian Inheritance in Man; no. 202400),1 the most severe form of fibrinogen deficiency, is characterized by the complete absence of fibrinogen. The disease was originally described in 1920;2 it has an estimated prevalence of around 1 to 2 in 1 000 000. Umbilical cord hemorrhage is often the first sign of the disorder; gum bleeding, epistaxis, menorrhagia, gastrointestinal bleeding, and hemarthrosis occur with varying intensity, and spontaneous intracerebral bleeding and splenic rupture can occur throughout life.3,4
Congenital afibrinogenemia is inherited in an autosomal recessive manner: the condition exists only when both alleles are mutated, in homozygosity or compound heterozygosity; both sexes are affected. Although the disease was first described in 1920, the genetic locus responsible for the disorder was only recently determined. We identified the first causative mutations for this disorder in a nonconsanguineous Swiss family. The genetic defect was an apparently recurrent deletion of approximately 11 kb of DNA that eliminates the majority of the FGA gene and so leads to a complete absence of functional fibrinogen.5 The deletions were all identical to the base pair and probably resulted from nonhomologous (illegitimate) recombination, mediated by a direct 7 bp repeat, AACTTTT, and perhaps also by indirect repeats in the breakpoint region. Many families with this disorder have been studied since then, allowing the identification of numerous causative mutations (reviewed in Neerman-Arbez,6 Hanss and Biot,7 and Maghzal et al8 ). The disease is caused by mutations in any 1 of the 3 fibrinogen genes FGG, FGA, and FGB, clustered on the long arm of human chromosome 4 (region 4q28-31).9 Surprisingly, the majority of cases studied so far are due to null mutations in the FGA gene10 although one would intuitively expect the 3 genes to be equally implicated. However, the majority of patients studied are white; therefore, the identification of causative mutations in non-European families is necessary to establish if this interesting finding holds true in all ethnic groups. In this study, we report the identification of a novel nonsense mutation (Arg134Xaa) in the FGG gene responsible for congenital afibrinogenemia in 10 patients from a highly consanguineous Muslim community from the Bekaa valley in Lebanon. Interestingly, this premature termination codon (PTC) is split between exons 4 and 5 (TG-intron 4-A). Expression studies assaying mRNA splicing and stability and protein production and function were performed, to determine the molecular mechanism by which this mutation causes complete fibrinogen deficiency.
Patients, materials, and methods
Patients
Ten afibrinogenemic patients as well as 19 unaffected members of their families were investigated. They all belong to a Shiite community living in small villages in the Bekaa plateau, east of Lebanon. We distinguished 6 families (A to F) although some links between these families are quite conceivable. Indeed, even if it was not possible to determine a clear familial relationship between these families, the high rate of consanguineous marriage in this community11,12 suggests a common ancestor.
Coagulation tests
Following informed consent, blood samples were collected from the patients and some of their family members. The study was approved by the review board of the Swiss National Science Foundation. Coagulation studies included prothrombin time; activated partial thromboplastin time; as well as factor V, factor VIII, factors VII through X, and von Willebrand factor dosage; all were performed by means of standard methods.
Functional fibrinogen levels (Clauss method) and immunologic assays were determined as previously described.10 The sensitivity of the functional assay and of the immunoassay was 0.29 μM and 0.147 μM (0.1 and 0.05 g/L), respectively (normal range for both tests, 4.7-10.6/1 μM [1.6-3.5 g/L]).
Mutation screening
Mutation screening of FGA was as previously described.10 No mutation was identified in the coding sequences and intron-exon junctions. Polymerase chain reaction (PCR) amplification and sequencing of the FGG exons and intron-exon junctions were performed with the use of oligonucleotide primers shown in Table 1.
Oligonucleotide primers for PCR amplification and sequencing of the FGG exons and intron-exon junctions
Target . | Forward (5′ > 3′) . | Reverse (5′ > 3′) . |
|---|---|---|
| Exons 1-4 | FGG5′L: TCAGCTCCAGCCATTTGCAG | FGGi4R: TCAGGCATAATGTCACTGGG |
| Exons 5-6 | FGGx5L: TCAGGTCCACATTGTATTCC | FGGi6R: ATGAGCTACGGTTCACAAGG |
| Exon 7 | FGGx7L1: AGTTGATAGAACCAGTGCTC | FGGi7R: GACTCCTGGAGAAAATGGTG |
| Exon 8 | FGGx8L1: TTCCAAGGAAGCATCCTACG | FGGi8R: GTGGATTTCTTTAGAAGGGC |
| Exon 9-10 | FGGx9L: GTAACTGGCAATGCACTTCG | FGG3′R: GCTTTGCAAGTCCATTGTCC |
Target . | Forward (5′ > 3′) . | Reverse (5′ > 3′) . |
|---|---|---|
| Exons 1-4 | FGG5′L: TCAGCTCCAGCCATTTGCAG | FGGi4R: TCAGGCATAATGTCACTGGG |
| Exons 5-6 | FGGx5L: TCAGGTCCACATTGTATTCC | FGGi6R: ATGAGCTACGGTTCACAAGG |
| Exon 7 | FGGx7L1: AGTTGATAGAACCAGTGCTC | FGGi7R: GACTCCTGGAGAAAATGGTG |
| Exon 8 | FGGx8L1: TTCCAAGGAAGCATCCTACG | FGGi8R: GTGGATTTCTTTAGAAGGGC |
| Exon 9-10 | FGGx9L: GTAACTGGCAATGCACTTCG | FGG3′R: GCTTTGCAAGTCCATTGTCC |
Expression of wild-type and mutant genomic constructs in COS-7 cells
COS-7 cells were cultured in Dulbecco minimum essential medium (DMEM)–10% fetal calf serum (FCS) and passaged with the use of standard procedures. The wild-type and mutant FGG genomic constructs were generated by PCR amplification from a control and a homozygous Arg134Xaa DNA sample, respectively, with the use of oligonucleotides situated in FGG exon 1 and intron 6 (FGG5′L and FGGi6R) with the TaKaRa ExTaq enzyme (Axon Lab, Baden, Switzerland). PCR fragments (3.2 kb) were cloned into the pcDNA3.1/V5-His TOPO-TA mammalian expression vector (Invitrogen, Groningen, The Netherlands), and all coding sequences and intron-exon junctions verified by sequencing with the use of standard dye-terminator protocols (PE Biosystems, Foster City, CA).
mRNA analysis
The sequence-verified plasmids were transiently transfected into COS-7 cells with the use of Lipofectamin Plus (Invitrogen, Basel, Switzerland). RNA was extracted by means of the RNeasy kit (QIAGEN, Basel, Switzerland), and reverse-transcription PCR (RT-PCR) was performed with Ready-To-Go beads (AP Biotech, Dubendorf, Switzerland) and FGG exonic oligonucleotides situated in FGG exons 1 and 6 (FGG5′L and FGGx6R: 5′TCCATTTCCAGACCCATCGA3′). The RT-PCR products were separated on 1% or 1.5% agarose gels. A DNA size marker (1 kb ladder; Life Technologies, Basel, Switzerland) was loaded on each gel. Negative controls for the RT reaction with heat-inactivated Ready-To-Go beads were included for each RNA sample.
Mutant mRNA stability analysis by Pyrosequencing
To measure the mRNA stability of the FGG ARG134X mutant, COS-7 cells were transfected with a mixture of wild-type and mutant FGG genomic constructs. The relative proportion of wild-type versus mutant allele was quantified at both the DNA and mRNA levels by means of the Pyrosequencing method (Pyrosequencing, Uppsala, Sweden). Briefly, COS-7 cells were transfected with Lipofectin (Life Technologies), with 10 plasmid mixtures containing different ratios between the wild-type and mutant constructs (1:1, 1:3, 1:5, and 1:10). After 48 hours of culture, total DNA (QIAamp kit; Qiagen, Hilden, Germany) and RNA (RNeasy mini Kit; QIAGEN) was extracted from each of the transfected lines. The cDNA was produced with the use of the Superscript II enzyme (Invitrogen) and an oligo deoxythymidine (dT) primer. For DNA allele quantification, PCRs were performed with 30 ng DNA, and 5 pmol each primer (forward primer, 5′CGCTGCTACTTTGAAGTCC3′; reverse primer, 5′CTATGCTCAACATAATCAGGC3′). For mRNA allele quantification, we used cDNA-specific primers spanning an intron (forward, 5′ATGATAGACGCTGCTACTTT3′; reverse, 5′TTGGCAATGTCTTGACAATCT3′). In both cases, the reverse primers contained a 5′ biotin label.
For Pyrosequencing, an internal sequencing primer (AACACATGACTCAAGTATT) was designed one nucleotide 5′ to the mutation site. PCR products were immobilized with Dynabeads (Dynal, Oslo, Norway) by a 15-minute 65°C incubation in a buffer containing 10 mM Tris-HCl (tris(hydroxymethyl)aminomethane-HCl), 2 M NaCl, 1 mM EDTA (ethyl-enediaminetetraacetic acid), and 0.1% Tween 20. Immobilized PCR products were removed from solution by means of magnetic separation, denatured with 0.5 M NaOH, and neutralized with 200 mM Tris-acetate; 50 mM magnesium acetate. The remaining single-stranded DNA was then hybridized with the internal “sequencing” primer by heating the mix to 80°C and slowly cooling it to room temperature. Pyrosequencing analysis and peak quantification (5 replicates of each measurement) were performed according to the manufacturer's protocol.
Expression of wild-type and mutant cDNA constructs in COS-7 cells
The 3 human fibrinogen cDNAs, FGA, FGB, and FGG, were obtained by RT-PCR on HepG2 cells and cloned into the pcDNA3.1/V5-His TOPO mammalian expression vector (Invitrogen, Basel, Switzerland). The Arg134Xaa mutation was inserted in the FGG construct by means of the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Transient transfections of COS-7 cells were performed with the use of Fugene 6 reagent (Roche, Basel, Switzerland) according to the manufacturer's instructions. Transfections were performed in 100 mm dishes with 9 μg pcDNA3.1/V5-His TOPO vector (negative control) or with equal amounts (3 μg) of all expression vectors. At 48 hours after transfection, cells were washed twice with phosphate-buffered saline (PBS) and incubated for an additional 24 hours in media without serum. Conditioned medium with added protease inhibitors (Complete; Roche) and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) was centrifuged to remove cell debris, harvested, and concentrated with the use of Ultrafree-4 5K column (Millipore, Volketswil, Switzerland) before adding reducing or nonreducing 2 × concentrated Laemmli buffer. Cells were lysed in RIPA buffer: 150 mM NaCl; 50 mM Tris-HCl, pH 8.0; 1% NP-40; 0.1% sodium dodecyl sulfate (SDS); 0.5% Na-deoxycholate; and 0.5 mM PMSF; with protease inhibitors (Complete; Roche). After boiling at 95°C for 5 minutes, samples were analyzed by 7.5% or 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analysis was performed as previously described13 with the use of polyclonal rabbit antifibrinogen antibodies (Dako, Zug, Switzerland) or monoclonal mouse antiactin (Chemicon, distributed by Juro, Lucerne, Switzerland) and appropriate secondary horseradish-peroxidase conjugated antibodies (Amersham Biosciences, Otelfingen, Switzerland).
Results
Family characteristics, clinical history, and laboratory data
Lebanon's history explains the mosaic ethnic structure and the geographic distribution of the population, since for social, political, or religious reasons some groups can be physically isolated from the others, creating genetic isolates. This situation was exacerbated by the civil war between 1975 and 1990. A wide variety of genetic diseases have a high prevalence, favored by the close family relationships and the high rate of consanguineous marriages. The degree of “genetic impermeability” varies widely according to ethnic group and creates real clusters.11,12 Moreover, the prevalence of patrilateral parallel-cousin marriage is very common among Muslims.
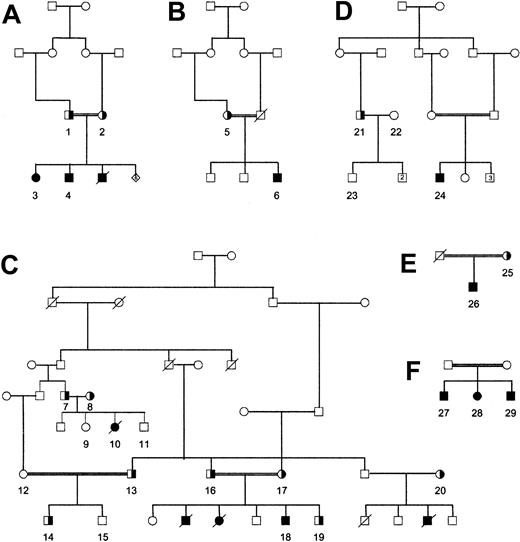
The 10 afibrinogenemic patients analyzed in this study were all born from Shiite consanguineous marriages (Figure 1). They all suffer from hemorrhagic complications (umbilical cord bleeding, menorrhagia, posttraumatic hematomas, hematuria, gastrointestinal bleeding, mouth and nose bleeding, hemarthroses, muscle hematomas), particularly because they only rarely benefit from treatment with cryoprecipitate or fibrinogen preparations. Some of them were anemic, one (D24), a 16-year-old girl, having 63 g/L (6.3 g/dL) hemoglobin the day the blood was drawn. Several members of these families died from bleeding problems, particularly from intracranial and gastrointestinal bleeding. Other family members, with intermediate or normal fibrinogen values, were asymptomatic.
Family trees. Ten afibrinogenemic patients as well as 19 family members, all belonging to a Shiite community living in the Bekaa plateau east of Lebanon, were studied. Six separate families (A-F) were distinguished, although links between these families are probable. Numbers refer to individuals listed in Table 2.
Family trees. Ten afibrinogenemic patients as well as 19 family members, all belonging to a Shiite community living in the Bekaa plateau east of Lebanon, were studied. Six separate families (A-F) were distinguished, although links between these families are probable. Numbers refer to individuals listed in Table 2.
Table 2 shows the prothrombin time and activated partial thromboplastin time as well as the fibrinogen values (coagulant and antigenic) for the 29 individuals who were genotyped. The numbers are identical to those used in the family trees (Figure 1). Patients with afibrinogenemia all had infinite prothrombin time and activated partial thromboplastin time. Heterozygous carriers all had intermediate fibrinogen values. There was a very good concordance between functional and immunologic measurements.
Laboratory data for the 6 families
Family . | Individual . | Fibrinogen activity, g/L . | Fibrinogen antigen, g/L . | PT, % . | aPTT, seconds . | Arg134 genotype* . |
|---|---|---|---|---|---|---|
| A | 1 | 1.1 | 1.26 | 81 | 30.7 | +/- |
| 2 | 0.88 | 1.2 | 85 | 30.1 | +/- | |
| 3 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 4 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| B | 5 | 1.01 | 1.25 | 91 | 29.1 | +/- |
| 6 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| C | 7 | 1.59 | 1.62 | 93 | 25.9 | +/- |
| 8 | 1.21 | 1.31 | 81 | 30.7 | +/- | |
| 9 | 3.09 | nd | 90 | 28.3 | +/- | |
| 10 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 11 | 2.73 | 2.51 | 91 | 27.1 | +/+ | |
| 12 | 3.22 | 2.8 | 95 | 29.3 | +/+ | |
| 13 | 1.04 | 1.3 | 88 | 27.2 | +/- | |
| 14 | 1.25 | 1.28 | 96 | 26.4 | +/- | |
| 15 | 3.3 | nd | 68 | 37.4 | +/+ | |
| 16 | 1.13 | 1.27 | 90 | 29.6 | +/- | |
| 17 | 0.92 | 1.03 | 80 | 30.6 | +/- | |
| 18 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 19 | 0.82 | 1.0 | 86 | 30 | +/- | |
| 20 | 0.94 | 1.1 | 88 | 27.2 | +/- | |
| D | 21 | 1.19 | 1.2 | 84 | 28.9 | +/- |
| 22 | 3.5 | 2.83 | 94 | 27.2 | +/+ | |
| 23 | 3.0 | nd | 93 | 31 | +/+ | |
| 24 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| E | 25 | 0.99 | 1.3 | 90 | 26.6 | +/- |
| 26 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| F | 27 | < 0.1 | < 0.05 | no clot | no clot | -/- |
| 28 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 29 | < 0.1 | < 0.05 | no clot | no clot | -/- |
Family . | Individual . | Fibrinogen activity, g/L . | Fibrinogen antigen, g/L . | PT, % . | aPTT, seconds . | Arg134 genotype* . |
|---|---|---|---|---|---|---|
| A | 1 | 1.1 | 1.26 | 81 | 30.7 | +/- |
| 2 | 0.88 | 1.2 | 85 | 30.1 | +/- | |
| 3 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 4 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| B | 5 | 1.01 | 1.25 | 91 | 29.1 | +/- |
| 6 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| C | 7 | 1.59 | 1.62 | 93 | 25.9 | +/- |
| 8 | 1.21 | 1.31 | 81 | 30.7 | +/- | |
| 9 | 3.09 | nd | 90 | 28.3 | +/- | |
| 10 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 11 | 2.73 | 2.51 | 91 | 27.1 | +/+ | |
| 12 | 3.22 | 2.8 | 95 | 29.3 | +/+ | |
| 13 | 1.04 | 1.3 | 88 | 27.2 | +/- | |
| 14 | 1.25 | 1.28 | 96 | 26.4 | +/- | |
| 15 | 3.3 | nd | 68 | 37.4 | +/+ | |
| 16 | 1.13 | 1.27 | 90 | 29.6 | +/- | |
| 17 | 0.92 | 1.03 | 80 | 30.6 | +/- | |
| 18 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 19 | 0.82 | 1.0 | 86 | 30 | +/- | |
| 20 | 0.94 | 1.1 | 88 | 27.2 | +/- | |
| D | 21 | 1.19 | 1.2 | 84 | 28.9 | +/- |
| 22 | 3.5 | 2.83 | 94 | 27.2 | +/+ | |
| 23 | 3.0 | nd | 93 | 31 | +/+ | |
| 24 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| E | 25 | 0.99 | 1.3 | 90 | 26.6 | +/- |
| 26 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| F | 27 | < 0.1 | < 0.05 | no clot | no clot | -/- |
| 28 | < 0.1 | < 0.05 | no clot | no clot | -/- | |
| 29 | < 0.1 | < 0.05 | no clot | no clot | -/- |
Normal laboratory levels are follows: fibrinogen activity, 1.6-3.5 g/L; fibrinogen antigen, 1.6-3.5 g/L; PT, 80%-100%; aPTT, 25-31 seconds.
PT indicates prothrombin time; aPTT, activated partial thromboplastin time; nd, not determined.
Arg134 genotypes are shown as follows: +/+, homozygous normal; +/-, heterozygous Arg134Xaa; -/-, homozygous Arg134Xaa
Factors VIII, V, VII, and X and von Willebrand factor (ristocetin cofactor) were normal in all individuals, except for individual C15 who had a prolonged prothrombin time with a prolonged aPTT (37.4 seconds; normal range, 25-31 seconds) and a decrease of factor VII and X (48%). This patient had normal fibrinogen levels, did not suffer from bleeding problems, but had liver cirrhosis.
Genetic analysis and genotype/phenotype correlation
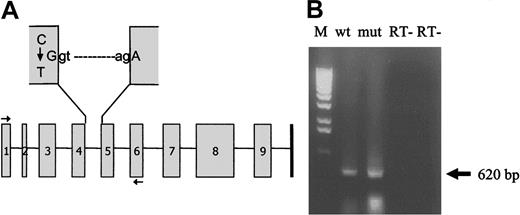
On the basis of our previous studies on the characterization of causative mutations for afibrinogenemia, which showed that the majority of cases are due to null mutations in the FGA gene, we chose to screen this gene first in patient samples. No mutations were identified in the FGA coding sequence or the intron-exon junctions. Screening of the FGG gene by PCR amplification followed by sequencing led to the identification of a novel nonsense mutation Arg134Xaa (codon numbering from the first ATG according to Antonarakis14 ) that was found in homozygosity in all affected individuals. The FGG Arg134 genotype correlated with the fibrinogen assays, both functional and antigenic, for all family members available for analysis: for example, individuals homozygous for the normal Arg134 had fibrinogen antigen levels greater than 7.35-8.3 μM/1 (2.5 g/L) (range, 2.9-4.76 μM/1 or 2.51-2.83 g/L); individuals heterozygous for Arg134Xaa had intermediate fibrinogen antigen levels (range, 2.9-4.76 μM/1 [1.0-1.62 g/L]) while all patients homozygous for Arg134Xaa had undetectable fibrinogen levels (Figure 1; Table 2). The mutation, a single base-pair change, g804C>T (numbering from the first base of the initiator codon, according to FGG genomic sequence M10 014) in the penultimate nucleotide of FGG exon 4 (Figure 2A), creates a premature stop codon (CGA>TGA) with the first base of FGG exon 5 and has not been previously described.
Splicing of wild-type and Arg134Xaa mutant mRNAs. (A) Schematic representation of the FGG gene showing the C>T change in the penultimate base of exon 4 that creates a premature termination codon (TGA) with the first base of exon 5. The horizontal arrows indicate the positions of the oligonucleotide primers used to produce the wild-type and mutant minigenomic constructs for mRNA analysis. (B) RT-PCR analysis of COS-7 cells transfected with wild-type (wt) and Arg134Xaa mutant (mut) minigenomic constructs followed by agarose gel electrophoresis indicates normal splicing for both constructs. The arrow indicates the normal mRNA product (expected size, 620 bp). Sequencing of these RT-PCR products confirmed normal splicing for both wild-type and mutant constructs. M indicates 1-kb DNA ladder marker; RT-, negative controls with heat-inactivated enzymes (“Patients, materials, and methods”).
Splicing of wild-type and Arg134Xaa mutant mRNAs. (A) Schematic representation of the FGG gene showing the C>T change in the penultimate base of exon 4 that creates a premature termination codon (TGA) with the first base of exon 5. The horizontal arrows indicate the positions of the oligonucleotide primers used to produce the wild-type and mutant minigenomic constructs for mRNA analysis. (B) RT-PCR analysis of COS-7 cells transfected with wild-type (wt) and Arg134Xaa mutant (mut) minigenomic constructs followed by agarose gel electrophoresis indicates normal splicing for both constructs. The arrow indicates the normal mRNA product (expected size, 620 bp). Sequencing of these RT-PCR products confirmed normal splicing for both wild-type and mutant constructs. M indicates 1-kb DNA ladder marker; RT-, negative controls with heat-inactivated enzymes (“Patients, materials, and methods”).
Because the nucleotide change occurs close to the exon 4–intron 4 junction in the donor splice site consensus sequence, we wished to determine if the mutation caused aberrant mRNA splicing. Alternatively, the PTC itself may induce abnormal mRNA splicing by a mechanism termed nonsense-mediated alternative splicing (NAS).15 To study mRNA splicing, genomic wild-type and Arg134Xaa constructs encompassing exons 1 to 6 were cloned as described in “Patients, materials, and methods” and were used to transiently transfect COS-7 cells. RNA was extracted after 2 days, and RT-PCR, followed by agarose gel electrophoresis and sequencing, demonstrated normal mRNA splicing for both wild-type and mutant constructs (Figure 2B).
In a second approach to investigate the molecular basis responsible for the afibrinogenemia in these patients, we assayed mutant mRNA stability in comparison with wild-type mRNA in transfected COS-7 cells. Indeed, mutant mRNA molecules containing PTCs can be degraded by a quality-control pathway known as nonsense-mediated decay (NMD). This mechanism has been shown to occur after splicing for PTCs situated inside exons followed by at least one downstream intron (reviewed in Byers,16 Frischmeyer and Dietz,17 and Hentze and Kulozik18 ) but also to occur for a PTC in the TCR-beta gene split by an intron.19 To our knowledge, however, this study is the first in which mRNA stability is investigated for a naturally occurring split PTC causing a human disease.
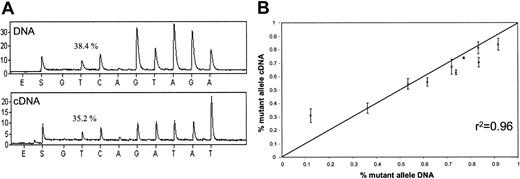
COS-7 cells were transfected with a mixture of wild-type and mutant FGG genomic constructs. The relative proportion of wild-type to mutant allele was determined at both the DNA and mRNA levels with the use of the Pyrosequencing method as described in “Patients, materials, and methods.” The quantification of the wild-type–to–mutant allele ratios at the DNA level serves to determine the actual proportion of constructs transfected for each cell line. Hence, the ratios measured at the mRNA (cDNA) level must be compared with the DNA values for the same cell line to determine if there is any significant difference. If the mutation results in an unstable mRNA, one would expect to see significant decreases in the mutant allele in mRNA with respect to the ratios observed at the DNA level. No difference was observed in the wild-type–mutant ratios obtained for DNA and for mRNA in 10 different transfection experiments: the correlation coefficient between the DNA and mRNA ratios was 0.96 (Figure 3).
Comparison of wild-type and Arg134Xaa mutant mRNA stability. (A) Example of DNA and cDNA allele quantifications for one of the transfected cell lines. The percentage above the first T peak refers to the proportion of the mutant allele as determined by the Pyrosequencing software. E indicates time of enzyme addition; S indicates time of substrate addition. (B) Average results of the DNA and cDNA allele quantifications for the 10 transfected cell lines. The units for the x- and y-axes refer to the proportion of mutant alleles. Four replicates for each transfected line were performed. Error bars indicate the standard deviations. The r2 value indicates the correlation coefficient between the DNA and cDNA quantifications.
Comparison of wild-type and Arg134Xaa mutant mRNA stability. (A) Example of DNA and cDNA allele quantifications for one of the transfected cell lines. The percentage above the first T peak refers to the proportion of the mutant allele as determined by the Pyrosequencing software. E indicates time of enzyme addition; S indicates time of substrate addition. (B) Average results of the DNA and cDNA allele quantifications for the 10 transfected cell lines. The units for the x- and y-axes refer to the proportion of mutant alleles. Four replicates for each transfected line were performed. Error bars indicate the standard deviations. The r2 value indicates the correlation coefficient between the DNA and cDNA quantifications.
Since the Arg134Xaa mutation had no effect on mRNA splicing or mRNA stability, at least under the experimental conditions and with the cell model used, we finally determined the effect of the mutation at the protein level. The predicted 107–amino acid fibrinogen γ chain (without the signal peptide) is missing 303 amino acids from the C-terminal portion and was expected either to be highly unstable or to impair fibrinogen assembly, since the C-terminal region of the fibrinogen γ chain has been shown to be necessary for intracellular hexamer assembly,20 in contrast to the C-terminal region of the β chain which is important for hexamer secretion.13,21-24
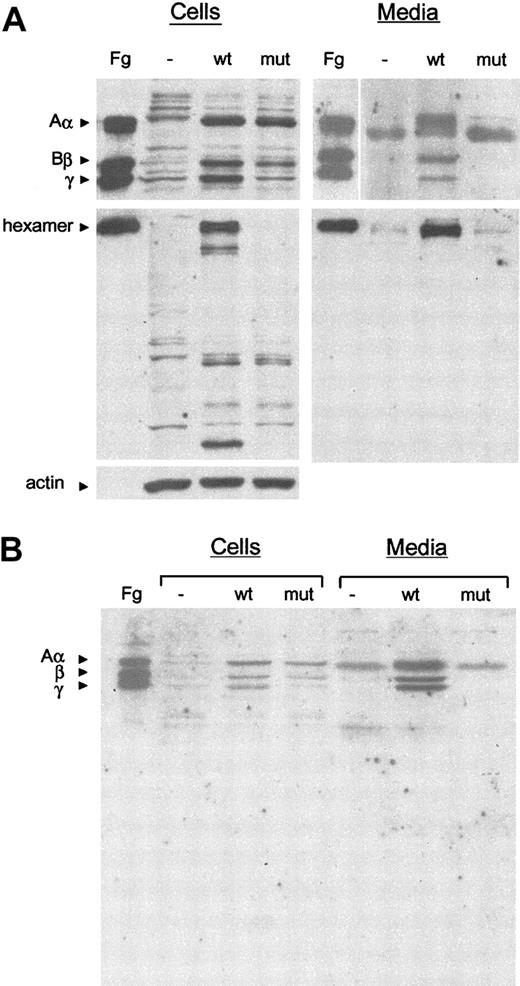
Arg134Xaa mutant and wild-type FGG cDNAs were transiently cotransfected with wild-type FGA and FGB cDNAs in COS-7 cells. Efficient expression of both FGG cDNAs was verified by RT-PCR (data not shown). At 18 hours after transfection, cells were lysed and the conditioned media harvested. Individual fibrinogen chains and assembled hexamers were detected by Western blot analysis with a polyclonal antifibrinogen antibody (Figure 4).
Western blot analysis of cell extracts and conditioned media of COS-7 cells transfected with fibrinogen cDNAs. (A) Samples of cell lysates and culture medium were subjected to 10% SDS-PAGE under reducing conditions (top panels) or 7.5% SDS-PAGE under nonreducing conditions (middle panels). The blots were incubated with polyclonal antihuman fibrinogen or monoclonal anti–β-actin antibodies (as a loading control for cell lysates in reducing conditions; bottom panel), and cross-reacting bands were revealed by chemiluminescence. Fg indicates purified fibrinogen control; -, COS-7 cells transfected with the empty vector; wt, COS-7 cells transfected with normal Aα, Bβ, and γ cDNAs; mut, COS-7 cells transfected with normal Aα, normal Bβ, plus mutant Arg134Xaa γ cDNAs. The positions of the hexameric complex and the normal Aα, Bβ, and γ chains are indicated. (B) Samples of cell lysates and culture medium were subjected to 12.5% SDS-PAGE under reducing conditions with shorter migration times. No truncated γ chain (predicted size, approximately 12 kD) is detectable.
Western blot analysis of cell extracts and conditioned media of COS-7 cells transfected with fibrinogen cDNAs. (A) Samples of cell lysates and culture medium were subjected to 10% SDS-PAGE under reducing conditions (top panels) or 7.5% SDS-PAGE under nonreducing conditions (middle panels). The blots were incubated with polyclonal antihuman fibrinogen or monoclonal anti–β-actin antibodies (as a loading control for cell lysates in reducing conditions; bottom panel), and cross-reacting bands were revealed by chemiluminescence. Fg indicates purified fibrinogen control; -, COS-7 cells transfected with the empty vector; wt, COS-7 cells transfected with normal Aα, Bβ, and γ cDNAs; mut, COS-7 cells transfected with normal Aα, normal Bβ, plus mutant Arg134Xaa γ cDNAs. The positions of the hexameric complex and the normal Aα, Bβ, and γ chains are indicated. (B) Samples of cell lysates and culture medium were subjected to 12.5% SDS-PAGE under reducing conditions with shorter migration times. No truncated γ chain (predicted size, approximately 12 kD) is detectable.
When COS-7 cells are transfected with the 3 normal fibrinogen cDNAs, all 3 chains are correctly expressed and assembled inside the cell, and the fibrinogen hexamers are secreted into the media. In contrast, when cells are transfected with normal FGA and FGB cDNAs and the Arg134Xaa FGG mutant cDNA, no fibrinogen hexamer is detected either in the cell lysates or in the media (Figure 4A). In addition, the mutant 107–amino acid γ chain (approximately 12 kD) was not detected in cell lysates or media, even when gel electrophoresis migration times were kept very short and acrylamide percentages increased (Figure 4B), strongly suggesting that the truncated protein is highly unstable.
Discussion
In this study, 29 individuals, including 10 afibrinogenemic patients from a highly consanguineous Muslim Shiite community in Lebanon, were analyzed. Screening of the FGG gene revealed a novel nonsense mutation, Arg134Xaa (numbering from the initiator ATG codon), which was found in homozygosity in all affected individuals and in heterozygosity in all obligate carriers studied. The FGG genotype of family members available for analysis correlated with the fibrinogen dosage in all cases.
The mutation, a single base-pair change C → T in the penultimate nucleotide of FGG exon 4, creates a premature stop codon (CGA>TGA) with the first base of FGG exon 5 and is the first split PTC mutation to be identified in afibrinogenemia. Expression of an Arg134Xaa mutant minigenomic construct encompassing exons 1 to 6 in transiently transfected COS-7 cells suggested that the mutation did not affect mRNA splicing or stability, at least under the experimental conditions used. Studies at the protein level in transfected cells showed that in all likelihood the mutation leads to the production of a highly unstable protein, since the truncated γ chain was not detected in either cell lysates or cell media. No assembled intermediate/hexamer containing the truncated γ chain was found, as expected, since the truncation at Arg134 (Arg108 of the mature γ chain) occurs in the coiled coil, which is essential for multichain intermediate assembly.
This study is, to our knowledge, the first in which the consequences of a naturally occurring split PTC identified in human disease are examined at the molecular level. It is therefore interesting to note that the split Arg134Xaa PTC eluded NAS and NMD under the experimental conditions used. A previous study investigating NMD for 5 mutations, 4 nonsense and 1 frameshift, all localized in the first 4 exons of the FGA gene,25 4 of which were in theory susceptible to be recognized and degraded by the NMD pathway, demonstrated that these mutations also escaped NMD. The experimental conditions used were similar to ours: that is, COS-1 cells were cotransfected with wild-type and mutant genomic constructs, and the expression levels of mutant and wild-type transcripts measured by semiquantitative RT-PCR. Although one cannot directly infer that the FGG Arg134Xaa split PTC we report here and the FGA mutations studied by Asselta et al would have the same outcome in the patient's hepatocytes, which were unavailable for study, these observations are at the least intriguing, and increase the number of mutations that are likely to bypass the NMD surveillance system.26-28
Prepublished online as Blood First Edition Paper, July 29, 2004; DOI 10.1182/blood-2004-06-2312.
Supported by grants from the Swiss National Science Foundation (SNF) (no. 31-064987), the Dr Henri Dubois Ferrière Dinu Lipatti Foundation, the Téléthon Action Suisse Foundation, and the Ernst and Lucie Schmidheiny Foundation. M.N.-A. is the recipient of an SNF Professorship (no. 631-66023).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all patients and family members for their cooperation and for donating blood samples. We are grateful to Peter Byers for stimulating discussions on NMD, Stylianos Antonarakis for helpful comments and suggestions, and Luciana Palumbo for expert technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal