Shwachman-Diamond Syndrome (SDS) is a rare multisystem disorder characterized by exocrine pancreatic insufficiency, bone marrow dysfunction, and metaphyseal chondrodysplasia. Recent studies show that mutations of SBDS, a gene of unknown function, are present in the majority of patients with SDS. In the present study, we show that most, but not all, patients classified based on rigorous clinical criteria as having SDS had compound heterozygous mutations of SBDS. Full-length SBDS protein was not detected in leukocytes of SDS patients with the most common SBDS mutations, consistent with a loss-of-function mechanism. In contrast, SBDS protein was expressed at normal levels in SDS patients without SBDS mutations. These data confirm the absence of SBDS mutations in this subgroup of patients and suggest that SDS is a genetically heterogeneous disorder. The presence (or absence) of SBDS mutations may define subgroups of patients with SDS who share distinct clinical features or natural history.
Introduction
Shwachman-Diamond Syndrome (SDS) is a rare multisystem disorder characterized by exocrine pancreatic insufficiency, bone marrow dysfunction, and metaphyseal chondrodysplasia.1-5 SDS is the second most common cause of congenital exocrine pancreatic insufficiency, after cystic fibrosis. Bone marrow dysfunction is present in nearly all patients with SDS.3-6 In the largest published series (88 patients), chronic or intermittent neutropenia was present in 97%.5 Anemia and thrombocytopenia were present in more than a third of patients. Patients with SDS have a marked propensity to develop myelodysplasia or acute myeloid leukemia.6-8
SDS is inherited in an autosomal recessive fashion.9 Recently, Boocock et al10 reported that compound heterozygous mutations of the SBDS (Shwachman-Bodian-Diamond syndrome) gene on chromosome 7 were present in the majority of patients with SDS. Most of these mutations resulted from gene conversion with a neighboring pseudogene (SBDSP). Similar data were reported in a smaller series of patients of Japanese ancestry.11 Most of the SBDS mutations are predicted to truncate a substantial portion of the SBDS protein, suggesting that they act in a loss-of-function manner. Herein, we show that most, but not all, patients classified prospectively based on clinical criteria as having probable SDS had SBDS gene mutations. Moreover, a strong correlation between SBDS genotype and expression of full-length SBDS protein was observed.
Study design
Human subjects
There were 33 families who attended the Second International Conference on Shwachman-Diamond Syndrome in St Louis in 1999 and who were invited to participate in a study to elucidate the genetic basis of SDS. After obtaining informed consent, a thorough history, physical examination, review of medical records, and selected laboratory testing were performed. In addition, genomic DNA was extracted from peripheral blood leukocytes using standard protocols. The diagnosis of SDS was based on the presence of the following 2 criteria:12 bone marrow failure (neutrophils < 1500 × 106/L, hemoglobin < 10 g/dL, or platelets < 150 × 109/L) or exocrine pancreatic insufficiency (serum trypsinogen < 16.7 μg/L, abnormal pancreatic stimulation test, low fat-soluble vitamin levels, or abnormal 72-hour fecal fat study together with a characteristic pancreatic abnormality in an imaging study). A diagnosis of SDS was considered probable when both criteria were documented in the medical record or by the on-site evaluation. A diagnosis of SDS was considered possible if only one criterion was documented and a second was suggested by an answer in the questionnaire completed by families. As controls, genomic DNA from 48 ethnically matched, healthy volunteers was sequenced. The human studies committee at Washington University approved this study.
Sequence analysis
A polymerase chain reaction (PCR)–based strategy to sequence all 5 exons, splice junctions, and 500 base pairs of 5′- and 3′-untranslated regions of the SBDS gene was developed. Primers were designed to specifically amplify the SBDS gene but not the SBDSP pseudogene (Supplemental Table S1 available on the Blood web site; see the Supplemental Table link at the top of the online article). Genomic DNA (5 ng) was amplified using AccuTaq (Sigma, St Louis, MO) and MJ Research (Cambridge, MA) PTC-225 thermal cyclers with the following parameters: hot start, 96°C for 2 minutes, 30 seconds; and 96°C for 20 seconds, 60°C for 15 minutes, for 35 cycles. Amplicons were sequenced with Big Dye version 3.1 chemistry and analyzed using ABI 3730 capillary sequencers. Sequence analysis was done in a blinded fashion with respect to the clinical diagnosis.
Immunoblotting
Rabbits were immunized with a mixture of 2 peptides from the extreme carboxy-termini of the murine (VLSLKDVEEGDEKFE) or human (VLNLKDVEEGDEKFE) SBDS protein (both with a cysteine at their aminotermini). The resulting antiserum recognized an approximately 29 kDa protein from both human and murine cell lysates (Figure 1 and data not shown). Cyropreserved leukocytes were lysed in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA [ethylenediaminetetraacetic acid], 0.5% sodium deoxycholate, 1% Nonidet P40 [NP40], 0.1% sodium dodecyl sulfate, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM diisopropylfluorophosphate, and 10 μg/mL aprotinin and leupeptin), and immunblotted with SBDS or actin antiserum (sc-1615; Santa Cruz Biotechnology, Santa Cruz, CA) or immunoprecipitated with SBDS or control antiserum.
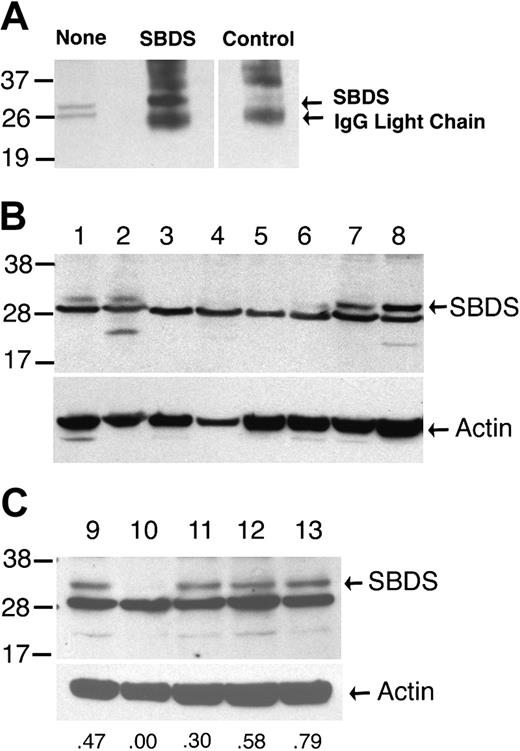
SBDS Western blot. (A) Proteins extracted from normal cyropreserved blood leukocytes were analyzed by immunoblotting with the SBDS antiserum either directly (none) or after immunoprecipitation with SBDS or control antiserum. There are 2 bands of approximately 29 kDa that are detected in whole-cell extracts. The upper band but not the lower band is efficiently immunoprecipitated by the SBDS antiserum, suggesting that the upper band represents the SBDS protein. (B,C) Protein extracts of cyropreserved blood leukocytes from family members in the registry were analyzed by immunoblotting with the SBDS antiserum (upper panels) or an actin antibody (lower panels). SBDS genotypes are lanes 1, 2, and 9: normal; lanes 3, 4, 5, and 10: 183-184TA>CT × 258+2T>C; lane 6: 258+2T>C × 505C>T; lanes 7 and 8: normal with clinical diagnosis of possible SDS; lanes 11 and 13: normal × 258+2T>C; and lane 12: normal × 183-184TA>CT. The relative densitometry signal for the SBDS band compared with the actin band is shown for lanes 9 to 13. Molecular size markers are indicated at the left in kilodaltons. These data are representative of 2 independent experiments using the same protein extracts.
SBDS Western blot. (A) Proteins extracted from normal cyropreserved blood leukocytes were analyzed by immunoblotting with the SBDS antiserum either directly (none) or after immunoprecipitation with SBDS or control antiserum. There are 2 bands of approximately 29 kDa that are detected in whole-cell extracts. The upper band but not the lower band is efficiently immunoprecipitated by the SBDS antiserum, suggesting that the upper band represents the SBDS protein. (B,C) Protein extracts of cyropreserved blood leukocytes from family members in the registry were analyzed by immunoblotting with the SBDS antiserum (upper panels) or an actin antibody (lower panels). SBDS genotypes are lanes 1, 2, and 9: normal; lanes 3, 4, 5, and 10: 183-184TA>CT × 258+2T>C; lane 6: 258+2T>C × 505C>T; lanes 7 and 8: normal with clinical diagnosis of possible SDS; lanes 11 and 13: normal × 258+2T>C; and lane 12: normal × 183-184TA>CT. The relative densitometry signal for the SBDS band compared with the actin band is shown for lanes 9 to 13. Molecular size markers are indicated at the left in kilodaltons. These data are representative of 2 independent experiments using the same protein extracts.
Results and discussion
Genomic DNA suitable for sequencing was available for 29 of the 33 families. Clinical characteristics of the probands in these families at the time of their participation in this study are shown in Table 1. Based on clinical criteria outlined in “Study design,” 18 of these families were classified as having one or more affected individuals with probable SDS, 6 with possible SDS, and 3 were judged not to have SDS. In 2 families, insufficient data were available to classify the probands (“unclassified”).
Patient characteristics and SBDS genotypes
Proband . | Sex . | Age, years* . | WBC, × 10-9/L . | Hgb, gm/dL . | Plt, × 10-9/L . | ANC, × 10-9/L† . | IRT, ng/mL‡ . | SDS classification . | SBDS genotype . |
|---|---|---|---|---|---|---|---|---|---|
| 001 | M | 2 | 5 | 12.5 | 315 | 0.169 | 28.7 | Probable | normal × normal |
| 004 | F | 4 | 7.2 | 11.3 | 239 | 0.189 | 8.2 | Probable | 183/184 × 258 + 2 |
| 006 | F | 10 | 2.6 | 11.8 | 105 | 0.816 | 26.9 | Probable | 652C>T × 258 + 2 |
| 019 | M | 12 | 2.6 | 12.1 | 110 | 0.276 | 22.1 | Probable | 505C>T × 258 + 2 |
| 020 | M | 8 | 6.2 | 12.5 | 234 | (G) 3.782 | NA | Probable | 183/184 × 258 + 2 |
| 022 | F | 3 | 4.6 | 11.8 | 176 | 0.156 | 24.3 | Probable | 183/184 × 258 + 2 |
| 023 | M | 10 | 5.5 | 11.5 | 197 | 1.776 | NA | Probable | 183/184 × 258 + 2 |
| 026 | M | 6 | 4.8 | 11.7 | 325 | (G) 12.097 | 17.4 | Probable | normal × normal |
| 028 | F | 3 | 5 | 11.6 | 130 | 1.655 | 5.6 | Probable | 183/184 × 258 + 2 |
| 048 | M | 13 | 2.7 | 12.2 | 90 | 0.251 | NA | Probable | 183/184 × 258 + 2 |
| 050 | F | 35 | 1.7 | 11.6 | 118 | (G) 0.262 | 0.6 | Probable | 183/184 × 258 + 2 |
| 053 | F | 6 | 4.8 | 12.6 | 145 | 0.379 | 15.4 | Probable | 183/184 × 258 + 2 |
| 058 | F | 8 | 3.8 | 11.4 | 198 | 0.901 | 3.8 | Probable | 183/184 × 258 + 2 |
| 060 | M | 8 | 3 | 12.5 | 134 | 1.035 | NA | Probable | 183/184 × 258 + 2 |
| 075 | M | 9 | 2.3 | 12.4 | 72 | 0.619 | 0 | Probable | 183/184 × 258 + 2 |
| 076 | M | 15 | 4.4 | 13.4 | 217 | 2.182 | 0.8 | Probable | 183/184 × 258 + 2 |
| 077 | M | 5 | 7.7 | 11.7 | 234 | 0.400 | 42.2 | Probable | 183/184 × 258 + 2 |
| 078 | M | 24 | 1.3 | 6.8 | 20 | NA | 14.4 | Probable | 183/184 × 258 + 2 |
| 042 | M | 3 | 5.2 | 12.1 | 367 | 1.550 | 26.4 | Possible | normal × normal |
| 056 | F | 5 | 4.4 | 12.9 | 224 | 4.488 | 3.8 | Possible | 183/184 × 258 + 2 |
| 059 | M | 14 | 3.4 | 12.7 | 194 | 1.486 | 2.4 | Possible | 183/184 × 258 + 2 |
| 073 | F | 7 | 5.9 | 12.4 | 265 | NA | 1.8 | Possible | normal × normal |
| 074 | F | 2 | 5.8 | 11.6 | 385 | 1.334 | 22.7 | Possible | normal × normal |
| 082 | M | 10 | 5.2 | 13.3 | 96 | 1.976 | 12.8 | Possible | 258 + 1 × 258 + 2 |
| 017 | M | 8 | 2.9 | 12.8 | 255 | 1.450 | 11.3 | Not | normal × normal |
| 055 | M | 7 | 4.5 | 13.2 | 363 | 2.349 | 20.1 | Not | normal × normal |
| 057 | M | 2 | 3.6 | 9.9 | 323 | 0.788 | 17 | Not | normal × normal |
| 079 | M | 10 | 3.1 | 10.9 | 121 | 0.738 | NA | Unclassified | 183/184 × 258 + 2 |
| 081 | F | 29 | 3 | 12.5 | 136 | 0.951 | NA | Unclassified | 183/184 × 258 + 2 |
Proband . | Sex . | Age, years* . | WBC, × 10-9/L . | Hgb, gm/dL . | Plt, × 10-9/L . | ANC, × 10-9/L† . | IRT, ng/mL‡ . | SDS classification . | SBDS genotype . |
|---|---|---|---|---|---|---|---|---|---|
| 001 | M | 2 | 5 | 12.5 | 315 | 0.169 | 28.7 | Probable | normal × normal |
| 004 | F | 4 | 7.2 | 11.3 | 239 | 0.189 | 8.2 | Probable | 183/184 × 258 + 2 |
| 006 | F | 10 | 2.6 | 11.8 | 105 | 0.816 | 26.9 | Probable | 652C>T × 258 + 2 |
| 019 | M | 12 | 2.6 | 12.1 | 110 | 0.276 | 22.1 | Probable | 505C>T × 258 + 2 |
| 020 | M | 8 | 6.2 | 12.5 | 234 | (G) 3.782 | NA | Probable | 183/184 × 258 + 2 |
| 022 | F | 3 | 4.6 | 11.8 | 176 | 0.156 | 24.3 | Probable | 183/184 × 258 + 2 |
| 023 | M | 10 | 5.5 | 11.5 | 197 | 1.776 | NA | Probable | 183/184 × 258 + 2 |
| 026 | M | 6 | 4.8 | 11.7 | 325 | (G) 12.097 | 17.4 | Probable | normal × normal |
| 028 | F | 3 | 5 | 11.6 | 130 | 1.655 | 5.6 | Probable | 183/184 × 258 + 2 |
| 048 | M | 13 | 2.7 | 12.2 | 90 | 0.251 | NA | Probable | 183/184 × 258 + 2 |
| 050 | F | 35 | 1.7 | 11.6 | 118 | (G) 0.262 | 0.6 | Probable | 183/184 × 258 + 2 |
| 053 | F | 6 | 4.8 | 12.6 | 145 | 0.379 | 15.4 | Probable | 183/184 × 258 + 2 |
| 058 | F | 8 | 3.8 | 11.4 | 198 | 0.901 | 3.8 | Probable | 183/184 × 258 + 2 |
| 060 | M | 8 | 3 | 12.5 | 134 | 1.035 | NA | Probable | 183/184 × 258 + 2 |
| 075 | M | 9 | 2.3 | 12.4 | 72 | 0.619 | 0 | Probable | 183/184 × 258 + 2 |
| 076 | M | 15 | 4.4 | 13.4 | 217 | 2.182 | 0.8 | Probable | 183/184 × 258 + 2 |
| 077 | M | 5 | 7.7 | 11.7 | 234 | 0.400 | 42.2 | Probable | 183/184 × 258 + 2 |
| 078 | M | 24 | 1.3 | 6.8 | 20 | NA | 14.4 | Probable | 183/184 × 258 + 2 |
| 042 | M | 3 | 5.2 | 12.1 | 367 | 1.550 | 26.4 | Possible | normal × normal |
| 056 | F | 5 | 4.4 | 12.9 | 224 | 4.488 | 3.8 | Possible | 183/184 × 258 + 2 |
| 059 | M | 14 | 3.4 | 12.7 | 194 | 1.486 | 2.4 | Possible | 183/184 × 258 + 2 |
| 073 | F | 7 | 5.9 | 12.4 | 265 | NA | 1.8 | Possible | normal × normal |
| 074 | F | 2 | 5.8 | 11.6 | 385 | 1.334 | 22.7 | Possible | normal × normal |
| 082 | M | 10 | 5.2 | 13.3 | 96 | 1.976 | 12.8 | Possible | 258 + 1 × 258 + 2 |
| 017 | M | 8 | 2.9 | 12.8 | 255 | 1.450 | 11.3 | Not | normal × normal |
| 055 | M | 7 | 4.5 | 13.2 | 363 | 2.349 | 20.1 | Not | normal × normal |
| 057 | M | 2 | 3.6 | 9.9 | 323 | 0.788 | 17 | Not | normal × normal |
| 079 | M | 10 | 3.1 | 10.9 | 121 | 0.738 | NA | Unclassified | 183/184 × 258 + 2 |
| 081 | F | 29 | 3 | 12.5 | 136 | 0.951 | NA | Unclassified | 183/184 × 258 + 2 |
WBC indicates white blood cell count; Hgb, hemoglobin; Plt, platelets.
Age at time of entry into registry and laboratory evaluation.
The absolute neutrophil count (ANC) for patients receiving G-CSF is indicated by G.
Normal value for immunoreactive serum trypsinogen (IRT) is greater than 16.7 ng/mL. Not available (NA), 183-184CT>TA (183/184), 258 + 2T>C (258 + 2), 258 + 1G>C (258 + 1).
Compound heterozygous mutations of the SBDS gene were detected in the majority of patients with probable or possible SDS (Table 1). In agreement with previous reports, the 258+2T>C and 183-184TA>CT mutations were the most common.10,11 The 258+2T>C mutation results in the disruption of the donor splice site of intron 2 and the use of a cryptic splice donor site at position 251-252. The resulting abnormally spliced mRNA results in a frameshift and the premature truncation of the 250 amino acid SBDS protein at amino acid 84.10 A single patient had a 258+1G>C mutation that results in a similar splicing defect. The 183-184TA>CT mutation introduces a premature stop codon, resulting in the truncation of the SBDS protein at amino acid 62. Point mutations resulting in single amino acid substitutions were detected in 2 families. The 505C>T mutation has been reported previously and results in a cysteine-for-arginine substitution at amino acid 169.10 The 652C>T mutation is novel and generates a premature stop codon at amino acid 218. None of these mutations were detected in 48 healthy controls.
In 18% (5 of 28) of patients classified with probable or possible SDS, SBDS mutations were not detected (Table 1). Likewise, no SBDS mutations were detected in 11% (17 of 158)10 and 29% (2 of 7)11 of previously studied patients with SDS. Since sequence analysis in all 3 studies was limited to exons, splice junctions, and the immediate 5′- and 3′-untranslated regions, it is possible that mutations outside of the sequenced regions may have been missed that effect SBDS expression. To address this possibility, SBDS protein expression in blood leukocytes from these patients was examined. In samples from a healthy individual, 2 bands of approximately 29 kDa (the predicted size for the SBDS protein) were detected by immunoblotting with the SBDS antiserum. Immunoprecipitation with the SBDS antiserum showed that the upper band represents the SBDS protein (Figure 1A). The SBDS protein was detected in samples from family members with normal SBDS alleles (Figure 1B, lanes 1 and 2) but not in patients containing the most common SBDS genotype (compound heterozygous mutations for 183-184TA>CT plus 258+2T>C). The SBDS antiserum was raised against a carboxy-terminal peptide, thus these data do not exclude the presence of an amino-terminal protein fragment. A preliminary study by Leary et al13 also showed the absence of detectable SBDS protein in cell lines derived from patients with SDS who had SBDS mutations. Reduced SBDS protein expression in the sample containing the 258+2T>C and 505C>T mutations was observed; based on densitometry, the relative intensity of the SBDS band in the 505C>T sample was 40% of that observed in normal samples, suggesting that the 505C>T allele results in the production of full-length SBDS protein (Figure 1B, lane 6). Unfortunately, no protein was available from the 2 patients with probable SDS and normal SBDS alleles. However, SBDS protein was readily detected in 2 patients classified as having possible SDS with normal SBDS alleles (Figure 1B, lanes 7 and 8). Of note, SBDS protein expression in family members heterozygous for a SBDS mutation was comparable to family members with normal SBDS alleles (Figure 1C).
Together with the sequencing data, the SBDS protein expression data provide strong evidence that there is a subgroup of patients with clinical features of SDS who do not have SBDS mutations. These data suggest at least 2 possibilities. It is possible that patients with normal SBDS alleles have been misclassified and actually have a distinct clinical syndrome. Alternatively, SDS may be a genetically heterogenous disorder. Mutation of a gene or genes that disrupt a pathway shared by SBDS may result in disease with identical clinical features. Distinguishing between these possibilities will require the elucidation of the biologic functions of SBDS.
Prepublished online as Blood First Edition Paper, July 29, 2004; DOI 10.1182/blood-2004-04-1516.
Supported by grants from the Washington University Digestive Diseases Research Core Center (P30 DK52574; D.C.L.), National Institutes of Health (NIH PO1 CA101937 [D.C.L.]; NIH T32 HL07088 [A.S.R.]; NIH RO1 CA105312 [M.B., P.J.M.]), the Wellcome Trust (P.J.M.) and from a Bursary Award from the Aplastic Anemia and MDS International Foundation, Inc (M.B.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Michael R. DeBaun for his assistance in organizing the Second International Conference on Shwachman-Diamond Syndrome in St Louis, and for his expertise in classifying patients with SDS. We also thank Hui Sun, Hui Du, and Mike McLellan for their expert technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal