In most myeloid leukemias induced in mice by γ-radiation, one copy of chromosome 2 has suffered a deletion. To search for a potential tumor suppressor gene in that region, we have delineated the deletions in a panel of these tumors. A commonly deleted region of 2 megabase pairs (Mbp) includes the gene encoding the PU.1 transcription factor, a powerful inducer of granulocytic/monocytic differentiation. Significantly, in 87% of these tumors the remaining PU.1 allele exhibited point mutations in the PU.1 DNA binding domain. Surprisingly, 86% of these mutations altered a single CpG, implicating deamination of deoxycytidine, a common mutational mechanism, as the origin of this lesion. The “hot spot” resides in the codon for a contact residue essential for DNA binding by PU.1. In keeping with a tumor suppressor role for PU.1, enforced expression of wild-type PU.1 in the promyelocytic leukemia cells inhibited their clonogenic growth, induced monocytic differentiation, and elicited apoptosis. The mutant PU.1 found in tumors retained only minimal growth suppressive function. The results suggest that PU.1 normally suppresses development of myeloid leukemia by promoting differentiation and that the combination of gene deletion and a point mutation that impairs its ability to bind DNA is particularly leukemogenic.
Introduction
The hallmark of acute myeloid leukemia (AML) is impaired differentiation, and the cells frequently exhibit chromosomal translocations involving transcription factors that govern normal hematopoiesis.1,2 Nevertheless, the genetic basis for most cases of human AML remains unknown. Further insight should emerge from elucidating the cytogenetic alterations in myeloid leukemia of mice. For nearly 3 decades it has been known that one copy of mouse chromosome 2 has suffered a deletion in most radiation-induced myeloid leukemias.3-7 Because no common breakpoint is involved, the deletions presumably involve loss of a tumor suppressor gene rather than activation of an oncogene.8 The search for the relevant gene(s) has been greatly hampered, however, by the large regions deleted (typically about 40 megabase pairs [Mbp]). To narrow the window in which the putative tumor suppressor lies, we and others have previously used loss of heterozygosity (LOH) to delineate a commonly deleted region (cdr), located about 100 Mbp from the centromere.8,9
We have now further refined the cdr. Within it, the most plausible candidate for a suppressor of myeloid leukemogenesis is the gene encoding PU.1, a member of the Ets family of transcription factors. PU.1 is a pivotal regulator of hematopoiesis, promoting or inhibiting differentiation in different lineages.2 In the erythroid lineage, its overexpression inhibits maturation and leads to erythroid leukemia.10,11 On the other hand, in the granulocytic, monocytic, and B-lymphoid lineages, PU.1 is instead essential for differentiation.12-14 Loss of PU.1 function might therefore be leukemogenic in one or more of those lineages by impeding terminal differentiation. In human AML, however, attempts to detect mutations in the PU.1 gene have led to conflicting results (see “Discussion”).
Notwithstanding important exceptions, 15 classic features of a tumor suppressor gene include frequent deletion or mutation of both its alleles in a given tumor type and inhibition of tumor cell growth with restored gene function. We show by these criteria that PU.1 is a potent tumor suppressor for myelocytic cells. Intriguingly, the second PU.1 allele in the myeloid leukemias is typically inactivated by point mutation at a specific CpG sequence in its DNA binding domain, reminiscent of mutational “hot spots” in the p53 tumor suppressor gene.16 We demonstrate that reintroduction of wild-type PU.1 into the leukemia cells impairs their clonogenic growth and provokes terminal differentiation and concomitant apoptosis. Our evidence that specific point mutations of the PU.1 gene are particularly leukemogenic complements the very recent finding17 that reduced PU.1 expression can also provoke leukemia (see “Discussion”).
Materials and methods
Generation and analysis of tumors and cell lines
Myeloid leukemia was induced in (CBA X SJL) F1 male mice by treatment with hydrocortisone (to reduce the incidence of T-cell lymphomas) and γ-irradiation.3,5,6,8 Tumors were adapted to culture and cloned as described.8 Cell lines designated with the prefix S derived from the previously described radiation-induced splenic tumors of SJL mice, whereas an X prefix denotes splenic tumors or derived cell lines from SJL X CBA F1 mice.8
DNA was extracted as described.8 Total RNA was prepared by Trizol (Invitrogen, Carlsbad, CA) and chloroform extraction, followed by isopropanol precipitation. Random primed cDNAs were generated from 1 μg of each RNA preparation using SUPERSCRIPT-II reverse transcriptase (Invitrogen). The cDNA was serially diluted in four 5-fold increments. Polymerase chain reaction (PCR) was performed using 0.15 ng of each primer, 0.2 mM deoxyribonucleoside triphosphates (dNTPs), 1.5 mM MgCl2, and 2 units Taq polymerase (Sigma, St Louis, MO). Amplification was performed for 25 to 30 cycles and determined to be in the exponential range for each primer pair. Primers were as published for mpo, mcsfr, gcsfr, and hprt18 for gmcsfrb and CD11b19 and gp91.20 For immunoblot analysis of proteins, cells were lysed in RIPA buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl [pH 7.4], 50 mM NaCl, 10 mM EDTA [ethylenediaminetetraacetic acid], 0.25% [wt/vol] sodium deoxycholate, 1% [vol/vol] Nonidet P-40 [NP40], 0.1% sodium dodecyl sulfate [SDS]) and the proteins electrophoresed through 14% Tris/glycine/SDS polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) filters, which were probed with anti-PU.1 (Santa Cruz Biotechnology, Santa Cruz, CA) and developed using the Amersham Biosciences (Freiburg, Germany) enhanced chemiluminescence (ECL) system.
Mapping the commonly deleted region
Construction of a YAC and BAC contig across the cdr
Libraries of WI/MIT820 YACs and RPCI-23 BACs were obtained from Research Genetics (Invitrogen) and the Australian Genome Research Facility (AGRF) (Melbourne, Victoria, Australia), respectively. YACs were screened by acrylamide electrophoresis of PCR products generated using microsatellite primers from Research Genetics. YAC-end sequences were obtained by a vector-hexamer PCR technique.21 Gridded BAC filters (BACPAC Resources, Roswell Park Cancer Institute, Buffalo, NY) were screened by hybridization with γ-32P–labeled primers and the identities of selected BACs confirmed by PCR. Sublibraries of BAC fragments digested with HaeIII and RsaI, constructed in pBluescriptII(KS+) (Stratagene, La Jolla, CA), were screened for CA repeats using a (CA)15 oligomer. Positive clones were sequenced to allow the design of new primers. Six novel polymorphisms were identified, one of which (WC200/1) showed retention of heterozygosity in the centromeric breakpoint-defining tumor (X78), thus reducing the cdr by approximately 250 kilobase pairs (kbp).22 Transcribed sequences within the cdr were identified from the Celera, European Molecular Biology Laboratory (EMBL), and University of California, Santa Cruz (UCSC) genomic sequences and expressed sequence tag (EST) databases.
Analysis of genomic and cDNA sequences
DNA was amplified by 40 cycles of PCR using High-Fidelity Platinum Taq, prepared for sequencing using an ABI BigDye v3.1 Kit (Perkin Elmer, Boston, MA), and sequenced by AGRF. All sequences were determined from 2 primers in each direction. The PU.1 mRNA and amino acid sequences (from National Center for Biotechnology Information [NCBI] entries X17463 and NP_035485) are numbered from the start of the open reading frame defined by the first of 2 potential ATG initiation codons.
Retroviral vector and infection
The murine stem cell virus (MSCV)–based retroviral vector used contains an enhanced green fluorescent protein (EGFP) cDNA downstream from an internal ribosome entry site (IRES).23 Coding sequences of wild-type PU.1 cDNA or a mutant (703C → T) were amplified by PCR and inserted in-frame after a hemagglutinin (HA) tag sequence upstream from the IRES. Construct integrity was confirmed by sequencing through the PU.1 and flanking sequences. To generate retroviruses, DNA of the vector and an amphotropic packaging virus23 were cotransfected into 293T cells via lipofection. Supernatants, passed through 0.22-μm filters, were used to infect leukemia cell lines by spinoculation.24 Within 24 hours, EGFP-positive cells were selected by flow cytometric sorting using Diva (Becton Dickinson, San Jose, CA) or MoFlo (Cytomation, Freiburg, Germany) instruments and assayed for growth in soft agar or liquid culture or extracted for protein or RNA. Cells were later harvested for fluorescence-activated cell sorter (FACS) and reverse transcription (RT)–PCR analysis or cytospin preparations.
Cytospin preparations were stained as described, 19 and photographed using an Axiocam color camera (Carl Zeiss, Oberkochen, Germany) and an Optiphot 2 microscope (Nikon Instruments, Shanghai, China) fitted with a Plan Apo 40/1.0 (Figures 5A, 6C) or 100/1.40 (Figure 4A) objective immersed in Zeiss Immersol 518N imaging medium at 25°C. Images were acquired using Axiovision 3.1 software.
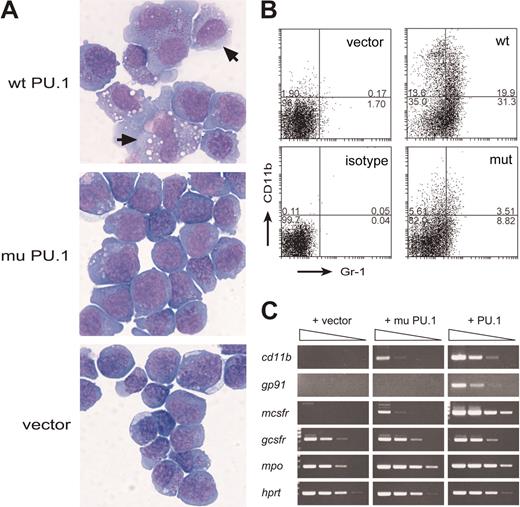
Exogenous wt PU.1 promotes differentiation of the PU.1-deficient leukemia cells. (A) Morphology of S1.1 cells infected with wild-type PU.1, mutant PU.1, or the vector and cultured for 4 days with GM-CSF. Cells in which PU.1 has induced a macrophage-like appearance are indicated (arrows). (B) Flow cytometric profiles showing that, in the presence of GM-CSF, wt PU.1 induced more marked expression of the myeloid differentiation markers CD11b (Mac-1) and Gr-1. Numbers indicate percentages of cells in each quadrant. (C) Semiquantitative RT-PCR (5-fold steps) showing that wt PU.1 induced a more striking increase in expression of the mRNAs for CD11b, gp91, M-CSF receptor, and myeloperoxidase (mpo) than the mutant PU.1 and that neither affected expression of G-CSF receptor or hypoxanthine phosphoribosyl transferase (HPRT), a loading control.
Exogenous wt PU.1 promotes differentiation of the PU.1-deficient leukemia cells. (A) Morphology of S1.1 cells infected with wild-type PU.1, mutant PU.1, or the vector and cultured for 4 days with GM-CSF. Cells in which PU.1 has induced a macrophage-like appearance are indicated (arrows). (B) Flow cytometric profiles showing that, in the presence of GM-CSF, wt PU.1 induced more marked expression of the myeloid differentiation markers CD11b (Mac-1) and Gr-1. Numbers indicate percentages of cells in each quadrant. (C) Semiquantitative RT-PCR (5-fold steps) showing that wt PU.1 induced a more striking increase in expression of the mRNAs for CD11b, gp91, M-CSF receptor, and myeloperoxidase (mpo) than the mutant PU.1 and that neither affected expression of G-CSF receptor or hypoxanthine phosphoribosyl transferase (HPRT), a loading control.
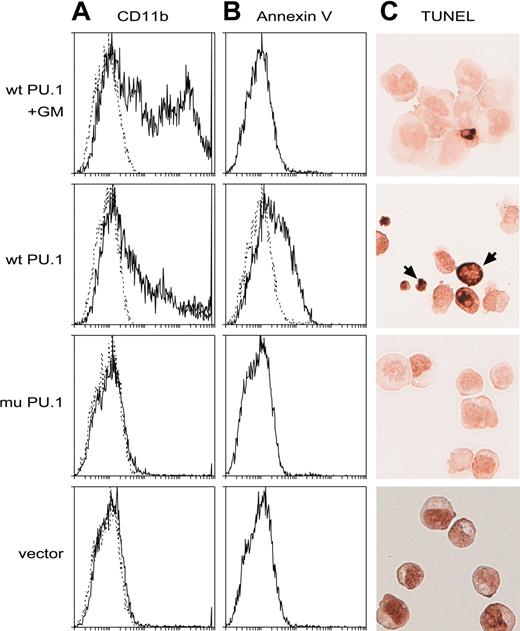
Introduced PU.1 promotes apoptosis of the leukemia cells. (A-B) Flow cytometric profiles showing that wt but not mutant PU.1 provokes apoptosis in the deleted lines, as assessed by the binding of annexin V (B), and that inclusion of GM-CSF in the medium (top panels only) is protective, allowing more differentiated (CD11b+) cells to develop or survive (A). Dotted lines indicate staining with isotype control antibodies. (C) Apoptotic cells (brown, arrows) visualized on cytospins by the TUNEL assay for DNA degradation. Cells shown in the top panel only had been cultivated in M-CSF.
Introduced PU.1 promotes apoptosis of the leukemia cells. (A-B) Flow cytometric profiles showing that wt but not mutant PU.1 provokes apoptosis in the deleted lines, as assessed by the binding of annexin V (B), and that inclusion of GM-CSF in the medium (top panels only) is protective, allowing more differentiated (CD11b+) cells to develop or survive (A). Dotted lines indicate staining with isotype control antibodies. (C) Apoptotic cells (brown, arrows) visualized on cytospins by the TUNEL assay for DNA degradation. Cells shown in the top panel only had been cultivated in M-CSF.
Phenotype of the myeloid leukemia cells. (A) Cytospins of cells in a representative tumor cell line (S1.1) with one PU.1 allele deleted and Arg235 mutated, showing the absence of differentiated cells but the presence in some cells of Sudan Black B–stained granules (arrows), a characteristic of the granulocyte lineage. (B) Flow cytometric plots showing expression on the leukemic cells of the granulocytic marker Gr-1 and the c-Kit cell surface receptor, a marker of early hematopoietic cells. Dotted lines indicate staining with isotype control antibodies. (C) Semiquantitative RT-PCR showing expression in the cells of the mRNA for myeloperoxidase and the receptors for G-CSF and GM-CSF.
Phenotype of the myeloid leukemia cells. (A) Cytospins of cells in a representative tumor cell line (S1.1) with one PU.1 allele deleted and Arg235 mutated, showing the absence of differentiated cells but the presence in some cells of Sudan Black B–stained granules (arrows), a characteristic of the granulocyte lineage. (B) Flow cytometric plots showing expression on the leukemic cells of the granulocytic marker Gr-1 and the c-Kit cell surface receptor, a marker of early hematopoietic cells. Dotted lines indicate staining with isotype control antibodies. (C) Semiquantitative RT-PCR showing expression in the cells of the mRNA for myeloperoxidase and the receptors for G-CSF and GM-CSF.
Liquid and agar cultures
The medium was Dulbecco modified Eagle medium supplemented with 2 mM glutamine, 330 μM asparagine, 50 μM β-mercaptoethanol, and 10% iron-supplemented calf serum (Hyclone, Logan, UT). Where indicated, this was supplemented with recombinant mouse granulocyte-macrophage colony-stimulating factor (GM-CSF) (5 ng/mL) or M-CSF (100 ng/mL) (Chemicon, Temecula, CA). For colony assays, sorted cells were plated in quadruplicate in 36-mm plates containing medium with 0.3% Agar (Bacto Laboratories, Liverpool, Australia), at 50 and 100 cells per 2 mL of agar medium per plate. Another 1 mL agar medium was added on day 4, and colonies were counted on day 8 to 10. For liquid culture, sorted cells were plated at 1 × 105 cells per milliliter.
Results
Candidate suppressors of myeloid leukemia in a refined commonly deleted region
To narrow the chromosome 2 region in which deletions occur in myeloid leukemia, we extended our previous analysis8 to a larger panel of radiation-induced myeloid tumors. F1 mice (CBA X SJL) were used so that we could delineate the deletions in different tumors by testing for LOH with microsatellite markers that distinguish the parental alleles. With the more than 100 microsatellite markers analyzed, none of the tumors showed the instability of such markers observed after loss of caretaker genes.25 In the F1 tumors with a deletion, SJL and CBA alleles were lost at similar frequencies (30 versus 20; P < .2), so the chromosome 2 region is unlikely to contain a strain-specific tumor susceptibility locus. In addition to 53 tumors with a chromosome 2 deletion (denoted hereinafter as “deleted”), 20 tumors had no deletion, consistent with reports that tumors can be induced in SJL mice by activation of the endogenous mouse mammary tumor virus.26
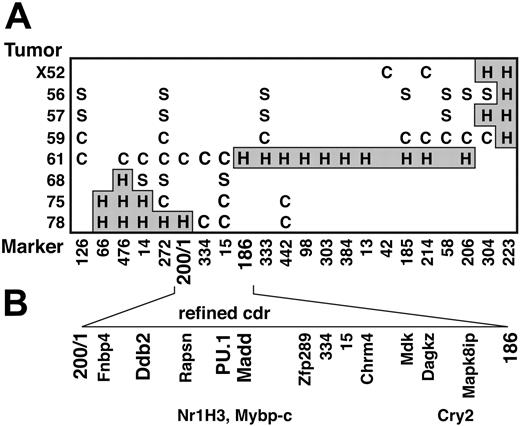
Figure 1A shows the genotypes of the relevant chromosome 2 region in 8 representative deleted tumors. No tumor exhibited loss of both alleles. The region deleted on one chromosome in all the tumors we have analyzed (the cdr) was about 2 Mbp long. The distal marker, D2Mit186, resides on the provisional Ensembl mouse chromosome 2 sequence (www.ensembl.org) at 92.8 Mb from the centromere, and the proximal marker, WC200/1, was a novel one identified by us 250 kbp distal to D2Mit272 (ie, at 90.65 Mb).
Refinement of the cdr on chromosome 2 and identification of candidate tumor suppressor genes within the cdr. (A) Genotype of tumors (listed on left) with polymorphic D2Mit and WC200/1 markers. C indicates CBAallele retained; S, SJL allele retained; H and gray shading, heterozygous. (B) The cdr, which represents the minimal region where all the tumors with deletions exhibit LOH. The positions of 13 genes that we have mapped,22 including 3 candidate tumor suppressors (bold), are indicated. Their positions largely agree with those in the provisional database sequence for chromosome 2, but the 3 genes in the lower row are ambiguously mapped with respect to those immediately above them in the upper row.
Refinement of the cdr on chromosome 2 and identification of candidate tumor suppressor genes within the cdr. (A) Genotype of tumors (listed on left) with polymorphic D2Mit and WC200/1 markers. C indicates CBAallele retained; S, SJL allele retained; H and gray shading, heterozygous. (B) The cdr, which represents the minimal region where all the tumors with deletions exhibit LOH. The positions of 13 genes that we have mapped,22 including 3 candidate tumor suppressors (bold), are indicated. Their positions largely agree with those in the provisional database sequence for chromosome 2, but the 3 genes in the lower row are ambiguously mapped with respect to those immediately above them in the upper row.
According to the provisional database genome sequences, the cdr contains 24 known genes and, as summarized in Figure 1B, we have mapped 13 of them to individual BACs within a contig spanning the cdr.22 We considered that only 3 of the 24—namely PU.1, Ddb2,27 and Madd28 —had known functions that might qualify them as tumor suppressors. To determine whether any of these 3 genes had been inactivated in the tumors, we sequenced exons spanning the coding region of the remaining allele of each from 20 to 25 deleted tumor cell lines. Only PU.1 exhibited mutations. Therefore, PU.1 exons and/or cDNA from an additional 14 deleted and 17 nondeleted tumors were amplified and sequenced.
A CpG hot spot for PU.1 mutation
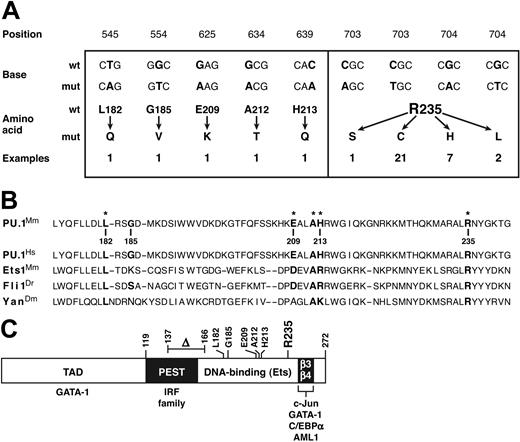
In all 17 nondeleted tumors, both PU.1 alleles were wild type (wt). In stark contrast, in 34 of 39 hemizygously deleted tumors and cell lines examined (87%), the remaining PU.1 allele was mutated. Aside from a single in-frame deletion in a cell line (Figure 2C), all the alterations were point mutations confined to exon 5 (Figure 2A), which encodes the DNA binding Ets domain (Figure 2C).29 The 6 transversions were far exceeded by 30 transitions (83%), all of which were C → TorG → A.
Missense mutations identified in PU.1. (A) The 36 missense mutations identified in 34 myeloid tumors and/or derived cell lines. The mRNA sequence is numbered from the first AUG of the open reading frame. Two synonymous changes (C → T at bp 700 and G → C at bp 702) found in tumors that also displayed an adjacent Arg235 missense mutation are not shown. (B) Point mutations affect conserved positions in the Ets domain. On the relevant portion of the mouse PU.1 Ets domain, the 6 residues in which mutations were found are indicated in bold, with an asterisk marking the 5 that reside at well-conserved positions, illustrated by the corresponding Ets sequences from human PU.1, mouse Ets1, zebrafish Fli1, and Drosophila Yan.30 Boldfacing indicates nucleotides and amino acids affected by mutations. (C) Diagram showing the domains of the PU.1 protein (TAD denotes the transcription activation domain), the location of the mutations found, and the regions (including the β3-β4 region of the Ets domain30 ) through which PU.1 engages other transcription factors.2
Missense mutations identified in PU.1. (A) The 36 missense mutations identified in 34 myeloid tumors and/or derived cell lines. The mRNA sequence is numbered from the first AUG of the open reading frame. Two synonymous changes (C → T at bp 700 and G → C at bp 702) found in tumors that also displayed an adjacent Arg235 missense mutation are not shown. (B) Point mutations affect conserved positions in the Ets domain. On the relevant portion of the mouse PU.1 Ets domain, the 6 residues in which mutations were found are indicated in bold, with an asterisk marking the 5 that reside at well-conserved positions, illustrated by the corresponding Ets sequences from human PU.1, mouse Ets1, zebrafish Fli1, and Drosophila Yan.30 Boldfacing indicates nucleotides and amino acids affected by mutations. (C) Diagram showing the domains of the PU.1 protein (TAD denotes the transcription activation domain), the location of the mutations found, and the regions (including the β3-β4 region of the Ets domain30 ) through which PU.1 engages other transcription factors.2
Surprisingly, 21 of the mutations were 703C → T, and another 7 were 704G → A (Figure 2A). Three of the transversions also alter this CpG hot spot: a single 703C → A and two 704G → T mutations. Thus, 86% of all the missense mutations changed the same CpG sequence and thereby replaced arginine 235, a highly conserved contact residue considered essential for DNA binding, 29,30 with cysteine (in 21 cases), histidine (7), leucine (2), or serine (1) (Figure 2A).
Artifactual explanations for the prevalent CpG alterations were ruled out. All were confirmed to be somatic mutations by verifying that normal cells from the parental CBA and SJL strains, as well as all the nondeleted tumors, had the expected CpG sequence. Cross-contamination between tumor DNAs was excluded by confirming that 15 samples of genomic DNA bearing 703C → T had the expected distinct LOH patterns. Moreover, in the 7 tumors for which both the gene and the cDNA were sequenced, the same mutation was found. Finally, the mutations were not simply derived during in vitro culture, because 9 of the samples displaying 703C → T and 5 having 704G → A were from primary tumors.
Ets domains in proteins from species as diverse as Drosophila, zebrafish, birds, and mammals are highly conserved, 30 and 5 of the 6 PU.1 amino acid residues where we found mutations (Figure 2B) represent conserved positions. Indeed, Arg235, targeted by the hot spot mutations, is invariant among Ets domains and is considered essential for DNA binding by PU.1, because it contacts the first base in the conserved core GGAA recognition sequence for Ets domain proteins, and 5 different replacements of it all fail to bind DNA.29,30 Similarly, Leu182, which resides in the hydrophobic core of the domain, is also invariant and Ala212 almost so. Moreover, Glu209 occupies a largely conserved acidic site and His213 a conserved basic position, 30 so the replacement of the former by lysine and the latter by glutamine represent disruptive charge changes. Finally, even though Gly185 resides in a variable position, no Ets domain has a bulky hydrophobic residue such as the observed valine substitution at that position. Hence, most, if not all, of the point mutations identified here would be expected to disrupt or impair PU.1 DNA binding function.
Remarkably, in 2 tumors, the DNA binding domain exhibited 2 independent missense mutations. The Leu182Gln mutation was coupled with Arg235His (in tumor S7), while the His213Gln was found with Arg235Cys (in X19.1). Similarly, another tumor harboring the common Arg235Cys mutation (X18) gave rise to a cell line that had acquired the sole deletion observed, an in-frame excision of 29 amino acid residues (137 to 166) removing the C-terminal moiety of the PEST domain and the N-terminus of the DNA binding domain (Figure 2C). This deletion presumably represents a transformation progression event selected during in vitro culture. These double mutations argue that Arg235 mutation alone may not eliminate all PU.1 functions (Figure 5).
Enforced wt PU.1 expression in the tumor-derived cell lines reduces their clonogenic growth
If impaired PU.1 function allowed tumor development, restoring that function might impair tumor cell growth. To test this hypothesis, we introduced sequences from wt PU.1 or the most common mutant (703C → T), each bearing an epitope tag to distinguish it from endogenous PU.1, into an MSCV-based retrovirus expressing EGFP from the same transcript. Two representative deleted tumor cell lines (S1.1 and X30.1), as well as a nondeleted line (X31.1), were then infected, and the infected cells sorted by flow cytometry for expression of EGFP and hence of PU.1. Expression of the exogenous PU.1, at a level comparable to that of the endogenous protein, was confirmed by Western blotting (Figure 3D).
Wild-type PU.1 inhibits growth of leukemic cell colonies in agar. (A-C) Colony growth of cells from the indicated cell lines infected with vector expressing EGFP alone or EGFP plus either wild-type PU.1 or mutant PU.1. At 16 to 20 hours after infection, EGFP-expressing cells were sorted and plated at 100 and 50 cells per dish, as described in “Materials and methods.” Values shown are mean ± SD of quadruplicate plates and are representative of 2 to 4 experiments. (D) Western blot showing exogenous and/or endogenous PU.1 expressed in a leukemia cell line (X31.1) that had been infected with each of the indicated 3 viruses.
Wild-type PU.1 inhibits growth of leukemic cell colonies in agar. (A-C) Colony growth of cells from the indicated cell lines infected with vector expressing EGFP alone or EGFP plus either wild-type PU.1 or mutant PU.1. At 16 to 20 hours after infection, EGFP-expressing cells were sorted and plated at 100 and 50 cells per dish, as described in “Materials and methods.” Values shown are mean ± SD of quadruplicate plates and are representative of 2 to 4 experiments. (D) Western blot showing exogenous and/or endogenous PU.1 expressed in a leukemia cell line (X31.1) that had been infected with each of the indicated 3 viruses.
We first tested whether enforced PU.1 expression affected clonogenic growth of the leukemic cells in agar. As shown in Figure 3B-C, the introduced wt PU.1 inhibited clonogenic growth in both the deleted cell lines, although the extent of suppression was reproducibly more marked in the S1.1 line, possibly due to different secondary tumorigenic mutations in the lines. In the nondeleted line X31.1, on the other hand, exogenous PU.1 expression had no significant effect (Figure 3A), showing that wt PU.1 was not simply toxic. In this experiment, the mutant PU.1 did not significantly impede clonogenic growth of the deleted lines, but a significant degree of inhibition has been observed in some other experiments.
PU.1 promotes differentiation and apoptosis of the PU.1-deficient leukemia cells
Cells in the lines derived from the deleted tumors had a homogeneous promyelocytic morphology with no evidence of differentiation in the culture (Figure 4A). Consistent with the myeloid appearance, a few of the cells exhibited Sudan Black B–stained granules (Figure 4A), and most expressed surface markers expected for early myeloid cells, such as c-Kit and a variable level of Gr-1 (Figure 4B). Moreover, RT-PCR analysis revealed expression of the mRNA for myeloperoxidase and the receptors for G-CSF and GM-CSF (Figure 4C). Although expression of GM-CSFRα chain was low, the cells responded to GM-CSF with a 5-fold increase in growth rate (data not shown). The cells lacked the mRNA, however, for CD11b (Mac1), a marker of late-stage differentiation that requires PU.1 for its transcription (Figure 4C). Accordingly, flow cytometry revealed no CD11b on the surface of either the X30.1 or the S1.1 cells (Figure 5B and data not shown).
Histologic and cell surface analysis revealed that the introduced wt PU.1 had induced marked changes. By day 4 after infection, many of the cells had acquired a macrophage-like morphology (Figure 5A) and expressed CD11b (Figure 5B). Consistent with increased maturation, RT-PCR revealed that wt PU.1 had induced expression of the mRNAs for CD11b, for the gp91 component of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, and for the M-CSF receptor as well as modestly up-regulating myeloperoxidase mRNA.
Interestingly, compared with the vector control, the mutant PU.1 also elicited some evidence of differentiation (Figure 5B). In 6 independent experiments involving 2 deleted cell lines, the proportion of CD11b-expressing cells produced by overexpressed mutant PU.1 ranged from less than 1% to 22% (mean, 12% ± 11% SD) of that promoted by wt PU.1. The intensity of anti-CD11b staining was also much lower—on average only about 20% of that observed on those expressing wt PU.1. Thus, the impetus for differentiation conveyed by the mutant protein was only a small percentage of that promoted by its wt counterpart. Accordingly, the mutant PU.1 promoted limited expression of the mRNAs for CD11b and M-CSF receptor but at a level 20- to 100-fold less than wt PU.1. Notably, gp91 mRNA was not induced by the mutant. Hence, the Arg235 mutant seems to retain partial PU.1 function (see “Discussion”).
Significantly, the differentiation elicited by wt PU.1 was accompanied by substantial apoptosis, as assessed by morphology (Figure 6C), by the binding of annexin V to phosphatidylserine exposed on the cell surface (Figure 6B) and by the transferase-mediated deoxyuridine triphosphate nick-end labeling TUNEL assay, which measures internucleosomal DNA cleavage (Figure 6C). Mutant PU.1 elicited no detectable apoptosis (1.2% ± 0.6%, compared with 4.0% ± 2.4% for the vector).
The survival and differentiation of myeloid cells is promoted by cytokines such as G-CSF, M-CSF, or GM-CSF, and the effects of PU.1 on hematopoiesis are mediated in part by up-regulation of the transcription of the cognate receptors.19,31 Accordingly, when GM-CSF was included in the culture medium, the proportion of differentiated cells induced by wt PU.1 increased markedly (Figure 6A) and the proportion of apoptotic cells dropped correspondingly (Figure 6B-C). The death imposed by the introduced PU.1 was also partially suppressed by culture with M-CSF (Figure 6C), apparently by its rescue of the differentiated progeny (data not shown).
Discussion
Our results establish that PU.1, a key regulator of hematopoietic lineage commitment and differentiation, 2 is a suppressor of myelocytic leukemia. In 87% of the γ-radiation–induced tumors with one PU.1 allele deleted, the remaining allele was mutated (Figure 2). The missense mutations were all confined to the Ets domain, and most, if not all, would be expected to impair or ablate its DNA binding function.29,30 Consistent with a tumor suppressor function, reintroduction of the wt PU.1 gene into lines bearing only a mutant allele inhibited the growth of tumor cell colonies in agar (Figure 3B-C).
Nature of the mutations
No tumor had both alleles of PU.1 deleted; nor did any have a frameshift or nonsense mutation in the second allele. The conspicuous absence of null mutants argued that many of the missense mutants (Figure 2) might retain some PU.1 function, and the detection of 3 “double mutant” lines is consistent with that notion. Indeed, the most common mutant exhibited limited differentiation ability (Figure 5B-C). That residual activity may rely upon association with other proteins. PU.1 function is thought to require its physical interaction, in a synergistic or antagonistic fashion, with several other key transcription factors, including GATA-1, c-Jun, interferon regulatory factor-4 (IRF-4), IRF-8, CCAAT/enhancer binding protein alpha (C/EBPα), and AML1 (Figure 2C), and those associations involve PU.1 regions2 that the most common mutations should not affect. Due to such interactions, point mutants of key hematopoietic regulators can be more disruptive than null mutants. An Ikaros point mutation that perturbs its DNA binding has a more devastating impact on hematopoiesis, and is more leukemogenic, than gene disruption.32 Similarly, when the C/EBPα gene, which is critical for granulocytic differentiation, is mutated in human AML, point mutants that impair its DNA binding function are strongly preferred over null mutants.33 Thus, crippled DNA binding function in a master regulator can be leukemogenic.
Remarkably, most of the point mutations (86%) targeted a single CpG sequence in the codon for an arginine residue (Arg235) considered critical for DNA binding (Figure 2). Indeed, because 2 of the substitutions at other positions were found together with an Arg235 mutation, that residue was replaced in 31 of the 34 tumors (91%) where a missense mutation was found. Because the PU.1 coding region contains 41 CpG sequences, including 18 within the DNA binding domain, the hot spot must reflect intense selection for its effect on PU.1 function. The prevalent transitions at this CpG can be ascribed to deamination of deoxycytidine, leading to thymidine, on the top or bottom DNA strand. Despite the reduced frequency of CpG sequence in vertebrate DNA, its conversion to TpG or CpA is the most common disease-causing mutation, accounting for one third of all germ-line and somatic point mutations, 34 including the 3 most frequent tumorigenic mutations of the p53 tumor suppressor.35
In the coding regions of genes, CpG sequences are usually at least partially methylated, and that appears to be the case for those in the PU.1 coding region (our unpublished data WDC, 2003). Although cytosine methylation has been thought to facilitate mutation by allowing direct conversion of 5-methyl deoxycytidine to thymidine, 16,36 demethylated CpG sequences remain comparably prone to transition mutations, 37,38 probably because they can be deaminated by the methyltransferases that recognize these sequences.38,39
In some (13%) of the tumors with deletions spanning the cdr we have defined, the coding region of PU.1 on the remaining allele is not mutated. Whether PU.1 is down-regulated in some other fashion in those tumors remains to be determined. Although the PU.1 gene clearly is a major target for the chromosome 2 deletions, their large size—commonly about 40 Mb—suggests that hemizygous deletion of other genes in that vicinity contributes to the leukemogenesis. Indeed, some radiation-induced myeloid tumors exhibit a second cdr, telomeric to that described here.40 Further scrutiny of this portion of chromosome 2 may reveal genes whose loss or reduced expression acts together with impaired PU.1 function in leukemia development.
Roles of PU.1 in suppression of myeloid leukemia
Analysis of myeloid cells from PU.1–/– embryos has clearly established that PU.1 is essential for terminal differentiation in both the granulocytic and monocytic lineages.12-14,19,31 In accord with those findings, we show that impaired PU.1 function promotes myeloid leukemia primarily by blocking maturation of the cells. Restoration of wild-type PU.1 by retroviral infection induced monocytic differentiation, as assessed by morphology, surface markers, and gene expression (Figure 5). The maturation role of PU.1 is mediated in part by up-regulated expression of cytokine receptors such as M-CSF and GM-CSF.19,31 Indeed, restored wt PU.1 function induced their expression (Figure 5C), and addition of GM-CSF to the cultures markedly increased the extent of differentiation, with half the cells becoming CD11b+ (Figures 5B and 6A). Evidence of full granulocytic maturation has not yet been obtained, however, perhaps due to secondary mutations in the tumors or because the level of introduced wt PU.1 obtained favored monocytic over granulocytic differentiation.41
The introduced wt PU.1 also induced considerable apoptosis in the leukemia cells bearing mutant PU.1 (Figure 6). The cell death might be an indirect consequence of PU.1 maturation function: survival of the mature cells induced by wt PU.1 expression might require a cytokine signal (eg, GM-CSF) not required by the leukemic progenitor cells. Consistent with that notion, GM-CSF precluded most of the death induced by enforced expression of PU.1 in tumor-derived cell lines (Figure 6). On the other hand, PU.1 might directly induce expression of proapoptotic molecules that must be countered by prosurvival molecules elicited by the cytokine signals. The survival of hematopoietic cells is governed by opposing members of the Bcl-2 protein family, 42,43 and impaired apoptosis represents a central step in tumor development.44 Thus, loss of PU.1 may contribute to leukemogenesis by impairing apoptosis as well as by precluding differentiation.
No neoplasia has been reported in PU.1+/– mice and seemingly none in mice devoid of PU.1 function due to a conditional knock-out.45 Nevertheless, during preparation of this paper, Rosenbauer and coworkers17 reported the intriguing finding that AML develops regularly in mice with an engineered distal enhancer deletion that markedly reduces the expression of PU.1 (to about 20% of the wt level). As in our mice, the leukemia reflected impaired maturation of myeloid progenitor cells.
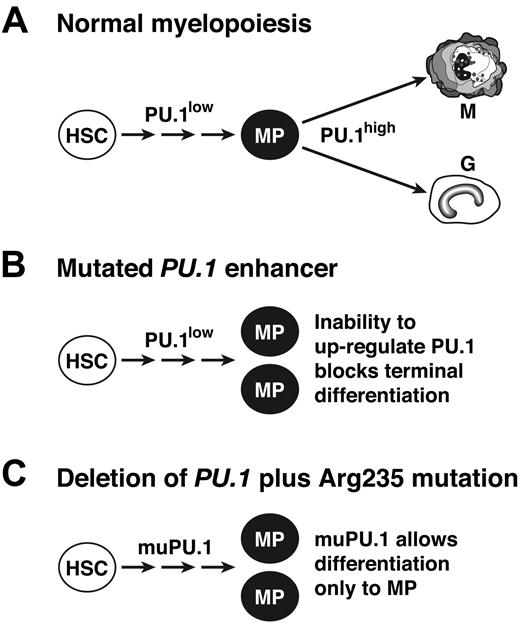
Because PU.1 level influences differentiation fate, 41,46 most likely a high level is required for the terminal granulocytic/monocytic differentiation step, whereas minimal PU.1 function is sufficient to generate the myeloid progenitors (MPs) from hematopoietic stem cells (Figure 7A). We propose that this threshold PU.1 function can be met not only by a very low level of wt PU.1 (Figure 7B)17 but also, in the absence of the wt protein, by Arg235-mutated PU.1 (Figure 7C). Thus, we suggest that both the engineered enhancer mutation and the tumor-derived mutant protein are hypomorphs unable to drive terminal differentiation.
Model for the role of PU.1 as a suppressor of myeloid leukemia. (A) In normal hematopoiesis, a low level of PU.1 is sufficient for the generation of a pool of myeloid progenitor cells (MP), 17 but a high level is needed for terminal maturation.41 (B) In mice with an engineered deletion of a PU.1 enhancer element, up-regulation above the basal level is precluded, so myeloid progenitors accumulate, providing the pool from which leukemia develops.17 (C) In cells that lack wt PU.1 but express PU.1 with a mutated DNA binding domain (eg, altered Arg235), residual activity of the mutant PU.1 is proposed to allow accumulation of myeloid progenitors but to prevent terminal differentiation, thereby stimulating leukemogenesis.
Model for the role of PU.1 as a suppressor of myeloid leukemia. (A) In normal hematopoiesis, a low level of PU.1 is sufficient for the generation of a pool of myeloid progenitor cells (MP), 17 but a high level is needed for terminal maturation.41 (B) In mice with an engineered deletion of a PU.1 enhancer element, up-regulation above the basal level is precluded, so myeloid progenitors accumulate, providing the pool from which leukemia develops.17 (C) In cells that lack wt PU.1 but express PU.1 with a mutated DNA binding domain (eg, altered Arg235), residual activity of the mutant PU.1 is proposed to allow accumulation of myeloid progenitors but to prevent terminal differentiation, thereby stimulating leukemogenesis.
Due to the multiple partners of PU.1, the mutant protein might not only retain residual ability to activate transcription from certain promoters but also to inhibit transcription from other genes. Partners whose function mutant PU.1 might inhibit include C/EBPα and IRF-8 (interferon consensus sequence–binding protein [ICSBP]), which have central roles in granulocytic differentiation and can suppress myeloid leukemia.33,47 For example, the complex of PU.1 and IRF-8 can induce transcription of the INK4B gene, which encodes the p15 cell cycle inhibitor.48 The mutant PU.1 protein might form an inactive complex with IRF-8 and thereby preclude the cell cycle exit associated with terminal differentiation. Thus, alteration of Arg235 could represent a gain-of-function as well as a loss-of-function mutation.
Relevance to human AML
Although our findings suggest that PU.1 sequences in human AML should be scrutinized for mutation, particularly in the DNA binding domain, the available data suggest that PU.1 mutation (particularly biallelic) in human AML is rare. Mueller et al49 reported mutations in 9 of 126 AML cases, but 7 of them retained a wt allele. No mutations have been found in 4 other studies, involving 60 AML cases, 50 77 cases, 51 112 cases, 52 and 140 cases, 53 and only one other mutation appeared among another 113 AML patients studied by Mueller et al (see response to Dohner et al52 ). Thus, analysis of 628 AML cases has revealed only 10 mutations (about 1.5%) and only 3 that are biallelic.
Interestingly, the 2 missense mutations observed in AML that affect the PU.1 DNA binding domain49 do not involve the residues found to be mutated in our study, even though the human gene possesses the pertinent CpG in an identical sequence context. Hence, the mouse tumors may arise through a different mutagenic mechanism, probably related to their induction by γ-irradiation. We therefore speculate that human PU.1 point mutations, particularly in CpG sequences within its DNA binding domain, might be most common in the secondary leukemias that occasionally arise following chemotherapy or radiation treatment for cancer.
Even if the PU.1 gene is only infrequently mutated in human AML, PU.1 may well often have a central role. For example, the gene might be silenced epigenetically (eg, by methylation). Alternatively, other oncogenic changes may act through PU.1. In support of that notion, the AML1-ETO fusion product of the t(8;21) can bind to PU.1 and inhibit its activity.54 Moreover, mutations in the juxtamembrane region of the Flt3 cytokine receptor, which are found in 25% of AML cases, lead to a several-fold depression in PU.1 mRNA.55
In conclusion, our findings and those of Rosenbauer et al17 make it likely that several of the common oncogenic alterations in human AML will prove to act at least in part by either reducing the expression of PU.1 or compromising the function of the protein. Therefore, the demonstration that PU.1 is a potent suppressor of induced myeloid leukemia suggests that pharmacologic agents that augment its expression or activity could provide a new approach to the treatment of this intractable malignancy.
Prepublished online as Blood First Edition Paper, August 10, 2004; DOI 10.1182/blood-2004-06-2234.
Supported by grants from the Cancer Council of Victoria, National Health and Medical Research Council (Program Grant ID 257502 and Project Grant ID 107510), the Leukemia & Lymphoma Society (Specialized Center of Research [SCOR] grant), and the U.S. National Cancer Institute (CA80188).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Andreas Strasser, Donald Metcalf, Suzanne Cory, Simon Foote, Glenn Begley, and Stephen Jane for discussions; Frank Battye, Catherine Tarlinton, Viki Lapatis, Christine Clark, and Carley Young for expert cell sorting and FACS analysis; Meagan James for animal husbandry; William Bong for geno-typing; and Simon Taplin and Peter Maltezos for preparation of figures.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal