Abstract
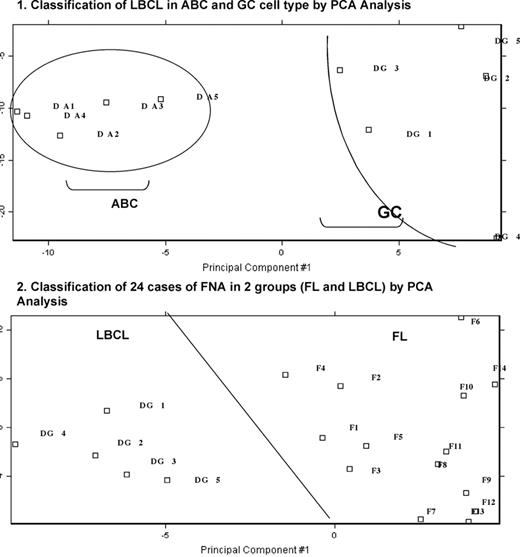
We tested the feasibility of performing gene expression profiling using amplified RNA from Fine Needle Aspiration (FNA) obtained from lymph nodes. Twenty-four samples from patients with a diagnosis of Follicular Lymphoma (FL) or Large B-Cell Lymphoma (LBCL) were obtained after informed consent. The diagnosis were performed by two pathologists and classified into two groups (10 LBCL and 14 FL) using conventional morphology and immunophenotyping. Total RNA was extracted using RNaqueous (Ambion, Austin, TX). Total RNA (100ng /per sample) was subjected to 2 cycles of standard double-stranded cDNA synthesis using Superscript Choice System (Invitrogen, Grand Island, NY) and in vitro transcription for target amplification as per GeneChip’s Eukaryotic Small Sample Target Labeling protocol (Affymetrix, Inc, Santa Clara, CA). The biotinylated cRNA from each sample was hybridized to Affymetrix HG-U133A chips. Gene expression profiling results were first analyzed by Principal Component Analysis (PCA) using a list of 146 probe sets representing 62 genes characteristic of Acivated B-Cell (ABC) or Germinal Center (GC) signature. This analysis identified 5 LBCL cases with ABC cell signature (Fig 1). Using a list of 207 probe sets representing 113 genes involved in FL transformation (K Elenitoba-Johnson, PNAS 2003), PCA analysis identified two overlapping clusters corresponding to FL and GC-DLBCL. To further improve this classification, we generated a list of 82 genes differentially expressed between FL and GC-LBCL. Using this list of genes, PCA analysis demonstrated a clear separation between FL and GC-LBCL (Fig 2). These results demonstrate that comprehensive transcription profiles can be performed in patients with NHL using RNA obtained from FNA and that one can distinguish among FL and LBCL lymphoma genetic subtypes.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal