Abstract
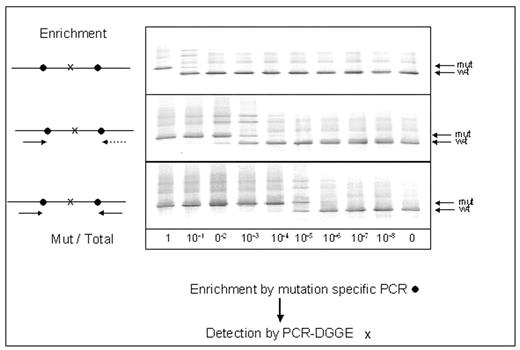
Measurement of minimal residual disease (MRD) is being increasingly used in haematological malignancies in order to assess prognosis and decide on treatment. However for some patients, including many patients with acute myeloid leukemia (AML), molecular techniques for MRD measurement cannot be performed owing to lack of a molecular marker. We have detected mitochondrial mutations (MM) in the D loop of the mitochondrial genome of the leukemic cells in approximately 40% of patients with AML and have investigated 2 techniques - denaturing gradient gel electrophoresis (DGGE) and single nucleotide primer extension (SNUPE) - to detect and quantify them. Mixing experiments showed that DGGE had a sensitivity of approximately 0.5% for detection of MM, and it was able to detect leukemia in remission marrow in 5 of 6 patients with AML. When present in a patient, MM are usually multiple. We therefore tested a 2-step strategy, as shown in the figure, which involved initial enrichment by PCR using primers directed at 1 or 2 flanking mutations followed by DGGE to detect an inner mutation. In 2 mixing experiments this 2-step strategy increased sensitivity of detection down to 0.0001% (this level of detection was evident in the original gel photo).
SNUPE was more sensitive than DGGE and in 3 mixing experiments it showed a sensitivity of 0.02–0.05%. Four patients with AML have now been studied by SNUPE and the levels of MRD in the marrow documenting morphological remission at the end of induction therapy were 2.8%,16.1%,40.8% and 15.7%. The relatively high levels of MRD as measured by PCR in 3 patients suggest that some of the leukaemic cells at the end of induction may be more mature than blasts and thus not identifiable by morphology. While questions remain to be answered, mitochondrial mutations are promising molecular markers for quantifying MRD in AML and possibly other haematological disorders
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal