Abstract
Two upstream regions of the human urokinase (uPA) gene regulate its transcription: the minimal promoter (MP) and the enhancer element. The activity of the minimal promoter is essential for basal uPA transcription in prostate adenocarcinoma PC3 cells. Binding of a phosphorylated Sp1 transcription factor is, in turn, essential for the activity of the MP. Here we report that the Jun kinase (JNK) pathway is required for the basal activity of the MP and for the expression of the endogenous uPA gene in PC3 cells and for activated transcription in LNCaP cells. On the other hand, the p42/p44 mitogen-activated protein kinase (MAPK) pathway activates uPA gene expression through Sp1 phosphorylation in HeLa, LNCaP, and CCL39-derivative cells that do not typically express uPA in basal conditions. In HeLa cells the dominant-negative form of JNK interferes with the p42/p44 MAPK activation of the uPA-MP. The results suggest that the stress-activated protein kinase (SAPK)/JNK pathway plays an important role in the phosphorylation of Sp1, which, in turn, leads to basal or activated transcription from the uPA-MP element.
Introduction
In cancer cells, different pathways may be activated and/or different transcription factors may be expressed, resulting in the specific expression of a repertoire of genes in distinct cell types. A good example is the urokinase-type plasminogen activator (uPA) gene, which is expressed at high basal levels in PC3 cells but is not expressed in HeLa or LNCaP cells.1-3
uPA is a serine protease involved in cell migration in nonpathological (eg, tissue remodeling) and pathological (eg, tumor invasion and metastases formation) events.4,5 Many human cancers, in fact, overexpress uPA, and reduction of the invasive and metastatic phenotype has been obtained by down-regulating the expression of the gene or by inhibiting the enzymatic activity of this protease.4,5 The minimal promoter (MP) of the human uPA gene extends approximately 86 bp upstream of the transcription start site1,6 and contains 5 (high, 3 × GGGCGG; low, 2 × GGGAGG) affinity binding sites for the Sp1-family transcription factors immediately upstream of the TATA box. This cis-acting element binds the transcription factor Sp1 in vivo in PC3 cells, which constitutively express the uPA gene. In this cell line, a uPA minimal promoter–driven reporter construct is 10-fold transcriptionally more active than in HeLa cells.1 This result correlates with the presence of the phosphorylated form of Sp1 in PC3 cells and with the absence of the phosphorylated transcription factor1 and the lack of transcription of the endogenous uPA gene in HeLa cells. Thus, the uPA minimal promoter plays a very important role in uPA gene expression, invasion, and metastasis formation in cancer cells.
The expression of the uPA gene is inducible in several cells by stimulating the enhancer, located about 2 kb upstream of the transcription start site,6 which binds transcription factors belonging to different families.7-27 Despite its relevance in these cells, the uPA enhancer appears to have no role (or a very modest one) in uPA expression in PC3 cells, where, instead, transcriptional activity is dependent on the binding of the transcription factor Sp1 to the minimal promoter.1 Phosphorylation of Sp1 seems to be an absolute requirement, because HeLa cells, which express but do not phosphorylate Sp1, are unable to activate the MP.
Recently, it has been shown that in a CCL39 (Chinese hamster fibroblast)–derivative cell line, which expresses a chimeric, estrogen-inducible Raf:ER chimera, vascular endothelial growth factor (VEGF) expression is under the control of the p42/p44 mitogen-activated protein (MAP) kinase pathway,28 which targets the transcription factor Sp1 and promotes its phosphorylation at residues Thr453 and Thr739.29 The phosphorylated form of Sp1, in turn, drives transcription from the VEGF minimal promoter, which contains 2 binding sites for this transcription factor, which bracket a binding site for the transcription factor AP-2.28 The role of VEGF-MP in the overall expression of VEGF is not known. Interestingly, the constitutive levels of VEGF and uPA mRNA in PC3 and HeLa cells are, in fact, “mirror images,” because PC3 cells have high levels of uPA mRNA and low levels of VEGF mRNA, while HeLa cells have a substantial level of VEGF mRNA but do not express uPA mRNA.1,30,31
Because both the VEGF and uPA genes appear to depend on their Sp1-driven minimal promoters for expression,1,29 we have investigated whether phosphorylation of the Sp1 transcription factor was dependent on the same signal transduction pathways.
In this report we show that Sp1 phosphorylation is achieved through the p42/p44 MAP kinase pathway in HeLa cells, through the stress-activated protein kinase/Jun kinase (SAPK/JNK) pathway in PC3 cells, and through both in LNCaP cells. Cotransfection of a dominant negative form of JNK in HeLa and LNCaP cells drastically decreases the transcriptional induction of the MP observed with the kinases of the p42/p44 MAPK and JNK pathways, respectively. Also, transfection of a dominant negative JNK in uPA-producing PC3 cells decreases the basal transcriptional activity of the uPA promoter. Transfected MEK/ERK induces phosphorylation of endogenous Sp1 in HeLa cells, whereas dominant negative JNK causes its dephosphorylation in PC3 cells. In parallel, increased levels of endogenous uPA mRNA in HeLa cells and decreased levels in PC3 cells were also observed. The results indicate that the SAPK/JNK pathway plays a major role both in basal and activated uPA gene expression in these cells.
Materials and methods
Cell culture and transient transfections
HeLa, PC3, and HT-1080 cells were grown in Dulbecco modified Eagle medium (DMEM) with the addition of 10% (vol/vol) fetal calf serum (FCS), 100 mM sodium pyruvate, 200 mM glutamine, and 50 000 units of penicillin and streptomycin. LNCaP cells were grown in RPMI with 10% (vol/vol) FCS, 200 mM glutamine, and 50 000 units of penicillin and streptomycin.
Chinese hamster fibroblast (CCL39)–derivative cells have been previously described.29
Transient transfections on HeLa, PC3, and HT1080 cells were performed using Lipofectamin 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions using a total 1 μg DNA per 80 000 cells in 24-well plates. Transfections on LNCaP cells were performed using 12-well plates with 150 000 cells per well.
Luciferase reporter vectors have been previously described.1,28 Expression plasmids for the p42/p44 MAP kinase pathway mutants were as described29 ; the expression plasmid for the dominant negative JNK (JNKDN) mutant was a kind gift from Dr Roger Davis (Case Western Reserve University, Cleveland, OH), and the dominant negative SEK (SEKDN) was a gift from Dr Dennis Templeton (University of Massachusetts, Worcester). Cells were also transfected with the empty vectors hosting the kinase mutants (pECE; Stratagene, La Jolla, CA) as control. The experiments in Figures 4 and 5B and in Tables 1 and 2 were performed with the GAL4-cJun and GAL4-Elk1 fusion protein expression plasmids, pFC-MEKK, pFC-MEK1, and the GAL4 luciferase reporter vector of the PathDetect Trans Reporting System kit (Stratagene). The MEK1 and 2 (extracellular signal-regulated kinase 1/2 [ERK1/2]) kinase inhibitor UO126 (Sigma, St Louis, MO) was dissolved in dimethyl sulfoxide (DMSO) and was added to the cells at a final concentration of 50 μM or 100 μM 2 hours before the preparation of extracts. Control cells received 5 μLor 10 μL of DMSO alone for the same time.
A MEK dominant negative mutant or treatment with the kinase inhibitor UO126 does not affect transcription from the uPA minimal promoters in PC3 cells. PC3 cells were cotransfected with the reporter plasmids used in Figure 1 and with increasing amounts of a plasmid constitutively expressing a MEK dominant negative mutant (MEKDN). The uPA minimal promoter construct (A) was only marginally affected by MEKDN. Error bars indicate SD. (B) Quantitation of the data in panel A is as in Figure 1 but expressed as fold decrease. (C) Quantitation of the results obtained with the UO126 treatment of uPA-MP–transfected PC3 cells. The cells were given an amount of DMSO, as a control, equivalent to that containing 50 or 100 μM UO126. Also in this case, the results are expressed as fold decrease.
A MEK dominant negative mutant or treatment with the kinase inhibitor UO126 does not affect transcription from the uPA minimal promoters in PC3 cells. PC3 cells were cotransfected with the reporter plasmids used in Figure 1 and with increasing amounts of a plasmid constitutively expressing a MEK dominant negative mutant (MEKDN). The uPA minimal promoter construct (A) was only marginally affected by MEKDN. Error bars indicate SD. (B) Quantitation of the data in panel A is as in Figure 1 but expressed as fold decrease. (C) Quantitation of the results obtained with the UO126 treatment of uPA-MP–transfected PC3 cells. The cells were given an amount of DMSO, as a control, equivalent to that containing 50 or 100 μM UO126. Also in this case, the results are expressed as fold decrease.
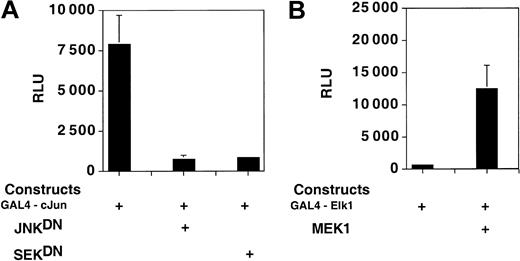
A GAL4-cJun, but not a GAL4-Elk1 fusion protein, drives transcription from a reporter construct in PC3 cells in the absence of cotransfected, kinase-expressing plasmids. PathDetect (Stratagene) GAL4-cJun– and GAL4-Elk1–expressing constructs were cotransfected in PC3 cells with plasmids expressing the wild-type or mutant forms of various kinases and with a luciferase reporter plasmid containing 5 GAL4 binding sites (PathDetect; Stratgene). The Jun-driven construct (A) shows high levels of expression in PC3 cells, which are reduced by cotransfection with the dominant negative forms of JNK and SEK. The Elk-driven construct (B) displays background levels of transcription in the absence of cotransfected kinase MEK1. Error bars indicate SD.
A GAL4-cJun, but not a GAL4-Elk1 fusion protein, drives transcription from a reporter construct in PC3 cells in the absence of cotransfected, kinase-expressing plasmids. PathDetect (Stratagene) GAL4-cJun– and GAL4-Elk1–expressing constructs were cotransfected in PC3 cells with plasmids expressing the wild-type or mutant forms of various kinases and with a luciferase reporter plasmid containing 5 GAL4 binding sites (PathDetect; Stratgene). The Jun-driven construct (A) shows high levels of expression in PC3 cells, which are reduced by cotransfection with the dominant negative forms of JNK and SEK. The Elk-driven construct (B) displays background levels of transcription in the absence of cotransfected kinase MEK1. Error bars indicate SD.
Effect of MEKK and MEK1 kinases on the transcription of a GAL4-luc reporter plasmid elicited by GAL4-cJun and GAL4-Elk1 fusion proteins in LNCaP cells
Constructs . | Fold increase . |
|---|---|
| GAL4-luc + GAL4-cJun | 1 |
| GAL4-luc + GAL4-cJun + pFC-MEKK | 2.5 |
| GAL4-luc + GAL4-cJun + pFC-MEK | 1 |
| GAL4-luc + GAL4-Elk1 | 1 |
| GAL4-luc + GAL4-Elk1 + pFC-MEKK | 19.6 |
| GAL4-luc + GAL4-Elk1 + pFC-MEK | 15.6 |
Constructs . | Fold increase . |
|---|---|
| GAL4-luc + GAL4-cJun | 1 |
| GAL4-luc + GAL4-cJun + pFC-MEKK | 2.5 |
| GAL4-luc + GAL4-cJun + pFC-MEK | 1 |
| GAL4-luc + GAL4-Elk1 | 1 |
| GAL4-luc + GAL4-Elk1 + pFC-MEKK | 19.6 |
| GAL4-luc + GAL4-Elk1 + pFC-MEK | 15.6 |
Quantitation of the dose-dependent effect of JNKDN on the transcription driven by the uPA-MP LNCaP cells cotransfected with MEKK (400 ng) or MEK1 (400 ng)
Constructs . | Fold decrease . |
|---|---|
| uPA-MP + MEKK | 1 |
| uPA-MP + MEKK + 4 ng JNKDN | 0.13 |
| uPA-MP + MEKK + 40 ng JNKDN | 0.02 |
| uPA-MP + MEKK + 400 ng JNKDN | 0.01 |
| uPA-MP + MEK1 | 1 |
| uPA-MP + MEK1 + 50 ng JNKDN | 0.71 |
| uPA-MP + MEK1 + 100 ng JNKDN | 1.12 |
Constructs . | Fold decrease . |
|---|---|
| uPA-MP + MEKK | 1 |
| uPA-MP + MEKK + 4 ng JNKDN | 0.13 |
| uPA-MP + MEKK + 40 ng JNKDN | 0.02 |
| uPA-MP + MEKK + 400 ng JNKDN | 0.01 |
| uPA-MP + MEK1 | 1 |
| uPA-MP + MEK1 + 50 ng JNKDN | 0.71 |
| uPA-MP + MEK1 + 100 ng JNKDN | 1.12 |
In all cases, cells were transfected also with a β-galactosidase reporter vector (pCMV-βgal; Clontech, Palo Alto, CA) or with PhRL or pGL3-Promoter vector (Promega, Madison, WI) for normalization. The results shown are the average of at least 2 experiments in triplicate.
Nuclear extracts and Western blots
Nuclear extracts were prepared by the method of Dignam et al.32 Aliquots were frozen and kept at –80° C. Phosphatase treatment of nuclear extracts was as described.1
For Western blots, 10% (wt/vol) polyacrylamide–sodium dodecyl sulfate gels were loaded with 50 μg of nuclear extracts from the different cell lines. Transfer to polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA) was performed in a semidry transfer apparatus (Sigma) for 2 hours at 0.8 mA/cm2. Overlay of the blot with the anti-Sp1 antibody (sc-59X; Santa Cruz Biotechnology, Santa Cruz, CA) was performed overnight at 4° C in phosphate-buffered saline (PBS) containing 3% (wt/vol) dried milk as quencher. Secondary antibody reaction (antirabbit immunoglobulin G [IgG]; Amersham, Arlington Heights, IL) was for 1 hour at 4° C in the same buffer. Bands were revealed by chemoluminescence using the SuperSignal kit (Pierce, Rockford, IL).
Total RNA preparation, real-time PCR, and Northern blots
Total RNA was extracted from HeLa and PC3 cells either transfected with empty vectors (pECE) or with the expression vectors for the MEK/ERK chimeric kinase or for JNKDN, respectively, with TRIZOL Reagent (Invitrogen), according to the maufacturer's instructions.
Four micrograms of total RNA was used in the reverse transcription step using SuperScript First-Strand Synthesis System for reverse transcriptase–polymerase chain reaction (RT-PCR) (Invitrogen), according to the maufacturer's instructions, using oligo(dT) to prime first-strand synthesis in a total volume of 20 μL.
The product of the 4 reverse transcription reactions (HeLa + empty vector; HeLa + MEK/ERKA; PC3 + empty vector; PC3 + JNKDN) were diluted 1:10, and 1 μL of each reaction was used for quantitative real-time PCR reaction with either 500 nM uPA primers (uPA forward [fwd]: 5′-ATCgAACTgTgACTgTCTAAATgg-3′; uPA reverse [rev]: 5′-TCTgTgggCATggTACgTTTgC-3′; spanning nucleotides 140 to 400 of the uPA sequence NM_002658) or 500 nM β-actin primers (β-actin fwd: 5′-ggCATCCTgACCCTgAAgT-3′; β-actin rev: 5′-CggATgTCAACgTCACACTT-3′; spanning nucleotides 260 to 942 of the β-actin sequence NM_001101). The final 20 μL reaction mix also contained 1 × SYBR-green1 mix (inclusive of deoxyribonucleoside triphosphates [dNTPs] and FastStart Taq DNA polymerase; Roche, Milan, Italy) and 4 mM MgCl2.
Amplifications were run in the Light Cycler 1.0 (Roche) apparatus on triplicate reactions with the following protocol: hot start (denaturation), 95° C for 10 minutes; and amplification (3 steps): denaturation, 95° C for 10 seconds; annealing, 63° C for 0 seconds (for uPA reverse-transcribed samples) or 60° C for 0 seconds (for β-actin samples); extension, 72° C for 20 seconds.
For each sample, the average crossing-point value (number of cycles at which each sample reaches a threshold fluorescence value) was calculated from triplicate reactions and is indicated as Ct. The formula DCtkinase = Ctk-uPA – Ctk-β-actin estimates the difference (expressed as the number of PCR cycles) between uPA and β-actin samples, from kinase-transfected cells, at a fixed fluorescence value, and DCtempty vector = Ctev-uPA – Ctev-β-actin indicates the difference between the samples in empty vector–transfected cells. Thus, the formula DCt = DCtkinase – DCtempty vector represents the difference between the normalized value of the uPA sample in kinase-versus empty vector–transfected cells and is used in the formula 2–DDCt, which indicates the fold increase or decrease of endogenous uPA mRNA in the different transfection conditions and is reported in Table 4. The experiment was repeated twice with triplicate samples.
Endogenous uPA mRNA levels in MEK/ERK-transfected HeLa cells and JNKDN-transfected PC3 cells, as estimated by real-time PCR
. | HeLa + empty vector . | HeLa + MEK/ERKA . | PC3 + empty vector . | PC3 + JNKDN . |
|---|---|---|---|---|
| Endogenous uPA mRNA | 1 | 3.2 | 1 | 0.8 |
. | HeLa + empty vector . | HeLa + MEK/ERKA . | PC3 + empty vector . | PC3 + JNKDN . |
|---|---|---|---|---|
| Endogenous uPA mRNA | 1 | 3.2 | 1 | 0.8 |
Endogenous uPA mRNA levels in MEK/ERK-transfected HeLa cells and JNKDN-transfected PC3 cells as estimated by real- time PCR. The values are calculated as described in “Materials and methods.”
Results
Members of the p42/p44 MAP and the SAPK/JNK kinase pathway can activate expression from the uPA-MP
We tested whether the p42/p44 pathway affects transcription of a luciferase reporter gene driven by the uPA minimal promoter element. HeLa cells were cotransfected with reporter constructs containing the luciferase gene driven by the uPA minimal promoter and with the constitutively active mutant of MEK (MEKA) or with a MEK/ERK chimeric kinase,29 and luciferase activity was monitored.
Figure 1A,C shows that transcription of the uPA minimal promoter was stimulated by the MEKA construct in a dose-dependent manner. The effect observed, however, was modest, increasing only 2-fold.
The uPA minimal promoter element responds to a constitutively active MEK kinase and to a MEK/ERK chimeric kinase. HeLa cells were transfected with a reporter construct in which the luciferase gene was driven by the minimal promoter element of the uPA gene. The cells were also cotransfected with increasing amounts of a plasmid constitutively expressing the active form of MEK (MEKA; panel A) or the MEK/ERK chimeric kinase (MEK/ERKA; panel B). The uPA minimal promoter element displays a dose-dependent response to both the constitutively active kinases. RLU indicates relative light units. Error bars indicate SD. (C-D) Quantitation of the data in panels A and B, respectively. Variation in transcriptional activation is expressed as fold induction relative to the minimal promoter construct alone.
The uPA minimal promoter element responds to a constitutively active MEK kinase and to a MEK/ERK chimeric kinase. HeLa cells were transfected with a reporter construct in which the luciferase gene was driven by the minimal promoter element of the uPA gene. The cells were also cotransfected with increasing amounts of a plasmid constitutively expressing the active form of MEK (MEKA; panel A) or the MEK/ERK chimeric kinase (MEK/ERKA; panel B). The uPA minimal promoter element displays a dose-dependent response to both the constitutively active kinases. RLU indicates relative light units. Error bars indicate SD. (C-D) Quantitation of the data in panels A and B, respectively. Variation in transcriptional activation is expressed as fold induction relative to the minimal promoter construct alone.
The use of a chimeric construct in which the active peptides of both the MEK and ERK kinases were fused (MEK/ERKA) and in which the residues subject to phosphorylation on the MEK portion of the chimera were mutated to aspartic acid to mimic a constitutive activation produced an even more substantial effect of the construct on the the uPA minimal promoter (Figure 1B-D). The values reach an almost 20-fold activation of the reporter constructs at a 10-fold lower concentration of MEK/ERKA-expressing plasmid with respect to MEKA.
The results indicate that transcription from the uPA minimal promoter can be induced in HeLa cells by overexpression of constitutively active kinases of the p42/p44 MAPK pathway; however, the effects of the kinases of the p42/p44 kinase pathway on the transcription of the reporter construct were substantially reduced in a dose-dependent fashion by cotransfection with the dominant negative form of JNK, JNKDN (Figure 2).
The dominant negative form of JNK reduces the transcriptional activation of the uPA-MP reporter construct by interfering with the constitutively active MEK/ERK (chimeric kinase). (A) The uPA-MP reporter construct was cotransfected with 50 ng of the constitutively active MEK/ERK chimeric kinase (see Figure 1B) in HeLa cells. Cells were further cotransfected with increasing amounts of the construct expressing JNKDN, and luciferase activity was monitored as described in “Materials and methods.” In HeLa cells, JNKDN strongly decreases the transcriptional activity of the uPA-MP construct, elicited by MEK/ERKA. Error bars indicate SD. (B) Quantitation of the results in panel A is as for Figure 1.
The dominant negative form of JNK reduces the transcriptional activation of the uPA-MP reporter construct by interfering with the constitutively active MEK/ERK (chimeric kinase). (A) The uPA-MP reporter construct was cotransfected with 50 ng of the constitutively active MEK/ERK chimeric kinase (see Figure 1B) in HeLa cells. Cells were further cotransfected with increasing amounts of the construct expressing JNKDN, and luciferase activity was monitored as described in “Materials and methods.” In HeLa cells, JNKDN strongly decreases the transcriptional activity of the uPA-MP construct, elicited by MEK/ERKA. Error bars indicate SD. (B) Quantitation of the results in panel A is as for Figure 1.
We also tested LNCaP cells, in which the uPA gene is not expressed.2 We used the GAL4-cJun and GAL4-Elk1 fusion protein expression plasmids, pFC-MEKK, pFC-MEK1, and the GAL4 luciferase reporter vector of the PathDetect Trans Reporting System kit (Stratagene) to test whether the p42/p44 MAPK and SAPK/JNK pathways were active in this cell line. Table 1 shows that the transcription from the GAL4-luc reporter vector is activated by the GAL4-cJun and GAL4-Elk1 fusion proteins only in the presence of cotransfected MEKK and MEK1 kinases, indicating that both pathways are inactive in this cell line, as previously reported.33 Interestingly, the GAL4-Elk1 fusion protein was responsive to both cotransfected kinases.
LNCaP cells were therefore transfected with the uPA-MP reporter construct and cotransfected with MEKK and MEK1 constructs, and luciferase activity was monitored as above.
The results in Figure 3A,C show that transcription from the uPA minimal promoter responds in a dose-dependent manner to cotransfection with the MEKK construct. The uPA-MP construct also responds to cotransfection with the MEK1 construct in this cell line (Figure 3B,D). The former was completely inhibitable by cotransfection of JNKDN, while the latter was not significantly inhibited (Table 2), suggesting that both pathways are able to activate the MP in these cells.
The uPA-MP construct responds to the MEKK and MEK1 kinases in LNCaP cells. LNCaP cells, transfected with reporter plasmids as for Figure 1, were cotransfected with increasing amounts of plasmids constitutively expressing MEKK or MEK1. As in Figure 1, the uPA-MP reporter construct shows a significant dose-dependent response to the cotransfected MEKK (A) and MEK1 (B). Error bars indicate SD. (C-D) Quantitation of the data in panels A and B, respectively, is as in Figure 1.
The uPA-MP construct responds to the MEKK and MEK1 kinases in LNCaP cells. LNCaP cells, transfected with reporter plasmids as for Figure 1, were cotransfected with increasing amounts of plasmids constitutively expressing MEKK or MEK1. As in Figure 1, the uPA-MP reporter construct shows a significant dose-dependent response to the cotransfected MEKK (A) and MEK1 (B). Error bars indicate SD. (C-D) Quantitation of the data in panels A and B, respectively, is as in Figure 1.
The JNK, but not the p42/p44 MAPK, pathway is constitutively active in PC3 cells
We tested whether the p42/p44 signal transduction pathway was involved in the transcriptional activity of the uPA minimal promoter in the PC3 adenocarcinoma cell line. We expected that a cotransfected dominant negative mutant of MEK (MEKDN) or treatment of the cells with a specific metabolic inhibitor of this pathway (UO126) would substantially reduce transcription of the reporter construct driven by the uPA promoter element. As shown in Figure 4A-B, the effect of the MEKDN construct is very modest on the uPA minimal promoter, where transcriptional inhibition reaches 25%. A comparable result was obtained by treating PC3 cells with the MAP kinase inhibitor UO126 (Figure 4C).
The results indicate that, unlike HeLa cells, the basal transcriptional activity of the uPA minimal promoter in PC3 cells line is not, or only minimally, affected by the p42/p44 MAP kinase pathway, suggesting the involvement of a different signal transduction cascade.
Since the transcriptional activity of the uPA minimal promoter in PC3 cells relies on Sp1 phosphorylation,1 a signaling cascade that is constitutively active in this cell line, different from the p42/p44 MAP kinase pathway must be present. Therefore, fusion proteins containing the GAL4 DNA binding domain and either the Elk1 or cJun activation domains were expressed from cytomegalo-virus (CMV)–driven plasmids, which were cotransfected with a luciferase reporter plasmid carrying 5 GAL4 binding sites in PC3 cells. Transcriptional activation through the GAL4-Elk1 fusion protein is expected to occur if the p42/p44 MAPK pathway is active in PC3 cells, whereas transcriptional activation through the GAL4-cJun fusion protein would occur if the JNK pathway were active.
Figure 5A shows that the GAL4-cJun fusion protein was very active in PC3 cells, and when cotransfected with 2 different dominant negative mutants of the SAPK/JNK pathway, transcription was substantially reduced (Figure 5A). Conversely, the construct expressing the GAL4-Elk1 fusion protein did not activate transcription from the reporter construct unless the cells were cotransfected with a plasmid expressing MEK1 (Figure 5B). Thus, the p42/p44 MAP kinase pathway is not constitutively active in PC3 cells and hence is unlikely to constitutively affect Sp1 phosphorylation.
We conclude that the SAPK/JNK signaling cascade is constitutively active in PC3 cells33 and may be responsible for Sp1 phosphorylation.
In light of these results, we tested whether the cotransfection in PC3 cells of the expression vector coding for the dominant negative form of JNK (JNKDN) with the uPA-MP reporter construct would affect its transcriptional activity. Indeed, the uPA minimal promoter was strongly affected by the JNKDN (Figure 6A), which reduced transcription by approximately 70% (Figure 6B).
Transcription from the uPA reporter construct is affected by cotransfection of the dominant negative form of JNK in PC3 cells. A uPA-MP reporter plasmid, as for Figure 1, was cotransfected in PC3 cells with increasing amounts of a plasmid constitutively expressing the dominant negative form of JNK. The uPA minimal promoter element displays a substantial, dose-dependent reduction of transcription in the presence of JNKDN (A). Error bars indicate SD. (B) Quantitation of the results in panel A is as in Figure 1.
Transcription from the uPA reporter construct is affected by cotransfection of the dominant negative form of JNK in PC3 cells. A uPA-MP reporter plasmid, as for Figure 1, was cotransfected in PC3 cells with increasing amounts of a plasmid constitutively expressing the dominant negative form of JNK. The uPA minimal promoter element displays a substantial, dose-dependent reduction of transcription in the presence of JNKDN (A). Error bars indicate SD. (B) Quantitation of the results in panel A is as in Figure 1.
Similar results were obtained in HT-1080 cells (Table 3), which also constitutively express the uPA gene (see, for instance, Medcalf 34 ).
Quantitation of the dose-dependent effect of JNKDN on the transcription of a reporter construct driven by the uPA minimal promoter in HT-1080 cells
Constructs . | Fold decrease . |
|---|---|
| uPA-MP | 1 |
| uPA-MP + 4 ng JNKDN | 0.41 |
| uPA-MP + 40 ng JNKDN | 0.39 |
Constructs . | Fold decrease . |
|---|---|
| uPA-MP | 1 |
| uPA-MP + 4 ng JNKDN | 0.41 |
| uPA-MP + 40 ng JNKDN | 0.39 |
We conclude that in PC3 and HT-1080 cells the SAPK/JNK signal transduction pathway is constitutively active and affects the activity of the uPA promoter element, possibly through phosphorylation of Sp1.
Sp1 phosphorylation is modulated by the p42/p44 MAP kinase pathway and the JNK pathway
In PC3 cells the transcriptional activator Sp1 in its phosphorylated form is required for the function of the uPA minimal promoter element.1,28 To unveil a direct link between the p42/p44 MAPK and JNK signal transduction pathways and Sp1 phosphorylation, we established the state of Sp1 phosphorylation in HeLa and PC3 cells.
The results in Figure 7A show that in HeLa cells a phosphorylated band of Sp1 appears only in the presence of MEK/ERKA; in PC3 cells (Figure 7B) phosphorylated Sp1 disappears in the presence of JNKDN.
The transfected MEK/ERK chimera increases Sp1 phosphorylation in HeLa cells, whereas JNKDN causes its dephosphorylation in PC3 cells. HeLa and PC3 cells were transfected with the constitutively active MEK/ERK chimera (MEK/ERKA; see Figure 2) or with JNKDN, respectively. Equal amounts of nuclear extract from transfected and untransfected cells were treated (or not) with alkaline phosphatase prior to fractionation on a 10% polyacrylamide–sodium dodecyl sulfate (polyacrylamide-SDS) gel and transfer to a nylon membrane. The Western blots were overlain with anti-Sp1 polyclonal antibodies that recognize both the phosphorylated and unphosphorylated forms of the transcription factor. Transfection of HeLa cells with the active MEK/ERK chimera induced phosphorylation of Sp1 (A), whereas transfection of PC3 cells with the dominant negative form of JNK abolished it (B).
The transfected MEK/ERK chimera increases Sp1 phosphorylation in HeLa cells, whereas JNKDN causes its dephosphorylation in PC3 cells. HeLa and PC3 cells were transfected with the constitutively active MEK/ERK chimera (MEK/ERKA; see Figure 2) or with JNKDN, respectively. Equal amounts of nuclear extract from transfected and untransfected cells were treated (or not) with alkaline phosphatase prior to fractionation on a 10% polyacrylamide–sodium dodecyl sulfate (polyacrylamide-SDS) gel and transfer to a nylon membrane. The Western blots were overlain with anti-Sp1 polyclonal antibodies that recognize both the phosphorylated and unphosphorylated forms of the transcription factor. Transfection of HeLa cells with the active MEK/ERK chimera induced phosphorylation of Sp1 (A), whereas transfection of PC3 cells with the dominant negative form of JNK abolished it (B).
MEK/ERK and JNK expression affect the steady-state level of endogenous uPA mRNA
Our results indicate that Sp1 phosphorylation (or the lack of it) affects the activity of reporter genes driven by the uPA minimal promoter. We asked then whether transfection of HeLa and PC3 cells with the MEK/ERK constitutively active kinase and with the JNKDN mutant, respectively, would also affect the level of endogenous uPA mRNA in these cell lines.
Total mRNA from mock-transfected or MEK/ERKA-transfected HeLa cells and JNKDN-transfected PC3 cells was extracted, reverse transcribed, and subjected to real-time PCR with specific uPA and β-actin mRNA primers.
The variation of endogenous uPA mRNA was calculated from the number of PCR cycles necessary for each template to reach the same fluorescence value, normalized to β-actin values, used as internal standard, and expressed as fold increase or decrease as compared with the values obtained from cells transfected with an empty vector.
The MEK/ERKA-transfected HeLa cells had a 3.5-fold higher level of endogenous uPA mRNA over the level detected in mock-transfected cells (Table 4). In PC3 cells, where the uPA gene is constitutively expressed and has a high basal level of mRNA,1 the transfection of the JNKDN kinase reduced the level of endogenous uPA mRNA by 20% as compared with mock-transfected cells (Table 4).
Thus, the endogenous uPA promoter promptly responds to the p42/p44 MAPK and JNK signal transduction pathways in HeLa and PC3 cells, respectively.
The endogenous uPA gene in Chinese hamster lung fibroblast (CCL39)–derivative cells responds to stimulation through the p42/p44 kinase pathway and depends on Sp1 phosphorylation
To establish a causal connection between uPA gene expression and Sp1 phosphorylation, we have used CCL39-derivative cells stably transfected with a Raf:ER chimera under the control of an estradiol-inducible promoter. In these cells, VEGF expression depends on the p42/p44 MAP kinase pathway,28 which targets Thr453 and Thr739 of the transcription factor Sp1.29
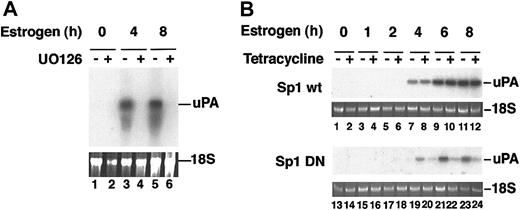
We first established the involvement of the p42/p44 pathway in endogenous uPA expression by treating CCL39 derivatives with estradiol in the presence or absence of UO126, a specific inhibitor of the MAPK pathway. Total RNA from estradiol- and/or UO126-treated CCL39 Raf:ER cells was extracted, fractionated, and a Northern blot was probed with the murine uPA cDNA. As shown in Figure 8A, induction of Raf:ER induced uPA mRNA, and treatment with the kinase inhibitor completely abolished uPA mRNA transcription.
Transcriptional induction of uPA is stimulated through the p42/p44 MAP kinase pathway. CCL39-derivative cells were induced to express the Raf:ER chimera by estrogen treatment for the times indicated. (A) Estrogen-induced and noninduced cells were treated with UO126, as described, and total RNA was assayed by Northern blot using a murine urokinase probe. (B) The cells were also transfected with plasmids expressing wild-type Sp1 or mutant Sp1 upon tetracycline treatment, and total RNA was assayed as for panel A. The uPA mRNA increases after 4 hours of estrogen treatment in the presence of wild-type Sp1, whereas in the presence of mutant Sp1 uPA mRNA levels are barely above background and are absent from UO126-treated cells.
Transcriptional induction of uPA is stimulated through the p42/p44 MAP kinase pathway. CCL39-derivative cells were induced to express the Raf:ER chimera by estrogen treatment for the times indicated. (A) Estrogen-induced and noninduced cells were treated with UO126, as described, and total RNA was assayed by Northern blot using a murine urokinase probe. (B) The cells were also transfected with plasmids expressing wild-type Sp1 or mutant Sp1 upon tetracycline treatment, and total RNA was assayed as for panel A. The uPA mRNA increases after 4 hours of estrogen treatment in the presence of wild-type Sp1, whereas in the presence of mutant Sp1 uPA mRNA levels are barely above background and are absent from UO126-treated cells.
We then asked whether Sp1 phosphorylation was the effector for Raf:ER induction of uPA transcription, as for VEGF.29 CCL39 Raf:ER cells were transfected with a wild-type or dominant negative (mutated in the phosphorylatable residues) Sp1 transcription factor under the control of a tetracycline-inducible promoter.29 Following estradiol treatment and tetracycline induction, total RNA was extracted, fractionated, and probed as above. The uPA mRNA was detected after 4 hours of estradiol treatment and remained high after 6 and 8 hours (Figure 8B, lanes 8-12). When a mutant Sp1, unable to undergo phosphorylation by p42/p44 MAP kinases,28 was overexpressed upon tetracycline treatment, the induced endogenous level of uPA mRNA was drastically reduced (Figure 8B, compare lanes 20, 22, and 24 with lanes 19, 21, and 23). However, tetracycline induction of the wild-type form of Sp1 did not affect the endogenous uPA mRNA levels (Figure 8B, compared lanes 8, 10, and 12 with lanes 7, 9, and 11).
We conclude that the p42/p44 pathway affects uPA gene expression through phosphorylation of the transcription factor Sp1.
Discussion
Transcription of the uPA gene in human cells is controlled by a large 5′ regulatory region, in which 2 elements, the minimal promoter (–86/+1) and the enhancer (–1976/–1880), have been thoroughly studied.1,6-18,21-24,26,35-37 The intervening sequence (–1879/–87) is not well characterized, although it is known to contain a putative negative regulatory element.38 By comparing a uPA-nonproducing, primary tumor cell line (HeLa) with a constitutively high-level uPA-producing, metastatic cell line (PC3), we discovered that the uPA minimal promoter was active only in the latter.1 This difference could be attributed mainly to the transcription factor Sp1, which has 5 binding sites within the minimal promoter region.1 The Sp1 protein in both cell lines is expressed at approximately equal levels, but only in PC3 cells is it phosphorylated and bound to the uPA minimal promoter chromatin.1
Here we have analyzed the signaling pathways leading to uPA minimal promoter activation and Sp1 phosphorylation. An unanticipated result was that cotransfection of the dominant negative form of JNK with the constitutively active kinases of the p42/p44 pathway substantially diminished, in a dose-dependent fashion, the activity of the transfected, uPA minimal promoter–driven reporter construct both in HeLa and LNCaP cells. Evidence that the JNK pathway is involved in the transcriptional up-regulation of the uPA gene has been gathered mostly in murine cells and point to the enhancer (specifically the AP-1 sites) as the key element through which activation takes place,19,27,39-42 whereas this element has drawn attention for its responsiveness to the protein kinase C (PKC) pathway in human cell lines.12,43,44 Our results indicate that the minimal promoter of the uPA gene may also be a target of the SAPK/JNK pathway and suggest a model in which coregulation of enhancer and minimal promoter may promote gene activation.
Our findings also suggest that a “crosstalk” occurs between the p42/p44 and JNK pathways in HeLa cells, because the dominant negative form of JNK decreases in a dose-dependent manner the uPA-MP–activated response elicited by MEK/ERK. This is in line with recent findings that have shown the negative modulation of the p42/p44 MAP kinase pathway by effectors of the JNK pathway.45 This opens the possibility that crosstalk between the p42/p44 MAPK and the SAPK/JNK may represent a further level of regulation of gene expression, suggesting that a tight control on uPA transcription may be achieved upstream of the protein-DNA interaction level.
In PC3 cells we have observed that the use of the dominant negative form of a kinase of the p42/p44 MAPK pathway only minimally affected the activity of the reporter construct. Indeed, the transcriptional activation of the uPA-MP reporter construct is elicited by the constitutively active SAPK/JNK pathway in this cell line. This molecular mechanism is not limited to PC3 cells, because we find that also in another uPA-producing, HT-1080 fibrosarcoma cell line, transcriptional activation of the uPA-MP construct is affected by cotransfection with the dominant negative form of JNK.
In the uPA-nonproducing LNCaP cells, the uPA-MP element is responsive both to the p42/p44 MAPK and to the SAPK/JNK pathways. However, differently from what is observed in HeLa cells, the pathways do not seem to merge, because JNKDN has no effect on the cotransfected MEK1, as it does on MEK/ERK in HeLa cells.
Phosphorylation of Sp1 seems to be the key element in the transcription of the uPA gene, at least from the minimal promoter element, as also suggested by the results in CCL39-derivative cells. Thus, signals leading to uPA gene expression through the minimal promoter element are channeled to their Sp1 target by at least 2 signal transducing pathways: the p42/p44 and the SAPK/JNK. Whether these pathways perform separate actions or merge seems to depend strongly on the cell line context. However, it seems likely that the SAPK/JNK pathway plays a dominant role at least in prostate cancer–derived cell lines.
Prepublished online as Blood First Edition Paper, March 18, 2004; DOI 10.1182/blood-2003-08-2661.
Supported by the Italian Association for Cancer Research (AIRC), the Italian Ministry of Research (MIUR), Cofinanziamento (COFIN), the European Union Integrated Project “Cancer Degradome” (contract no. 503297), and the National Institute of Environmental Health Sciences (NIEHS) (Department of Health and Human Services [DHHS] 5 P01 ES10535).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Charlotte Kilstrup-Nielsen for critically reading the manuscript.
This work was carried out in the framework of the MIUR Center of Excellence in Physiopathology of Cell Differentiation.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal