Abstract
The diagnostic procedure of chronic pulmonary opacities may envisage the search for non-Hodgkin lymphoma (NHL). Previous retrospective studies have shown that clonality analysis of bronchoalveolar B lymphocytes could reflect the clonality of pulmonary lymphocytes. Our objective was to define the diagnostic usefulness of bronchoalveolar lavage (BAL) B-lymphocyte clonality analysis in the setting of a clinical suspicion of both primary and secondary pulmonary lymphoma. A prospective BAL fluid B-cell clonality analysis was performed by polymerase chain reaction (PCR) in 106 consecutive patients presenting with a clinical suspicion of pulmonary NHL. Diagnosis was pulmonary B-cell lymphoma for 22 patients (13 primary and 9 secondary). When compared, pulmonary biopsy and BAL fluid have clonal identity. The detection of a strong B-cell clonal population in BAL fluid was associated with the diagnosis of pulmonary NHL (P < .0001), with a 97% specificity and a 95% negative predictive value. Thus, the absence of a dominant B-cell clone detection in BAL fluid could help to dismiss invasive investigations of pulmonary lesions. The detection of a dominant B-cell clone would lead to the performance of a pulmonary biopsy to get histologic diagnosis in primary pulmonary lymphoma and, by contrast, would avoid the need for biopsy in the setting of a secondary pulmonary lymphoma. (Blood. 2004;103: 3208-3215)
Introduction
B-cell non-Hodgkin lymphomas (B-NHLs) are clonal lymphoproliferative malignancies primarily involving lymph nodes and the spleen.1 Pulmonary localization of NHL can develop in 2 clinical settings, as primary pulmonary lymphoma or as pulmonary localization of a known extended lymphoma. Primary pulmonary lymphoma is an extranodal NHL defined by the involvement of the pulmonary parenchyma and the absence of any other tissue localization for 3 months following the diagnosis.2 When the lung is the main tumor site, this definition also includes multifocal mucosa-associated lymphoid tissue (MALT) NHL and pulmonary involvement with satellite nodes (hilar or mediastinal).3 Diagnosis requires biopsies by invasive methods such as transbronchial, percutaneous, or even surgical lung biopsies4-6 for both histologic analysis and immunohistologic staining. More than 95% of primary pulmonary lymphomas are B-NHL7,8 with a predominance of low-grade lymphomas (75%-80%), most of which consist of MALT lymphomas. High-grade malignant lymphomas represent 20% to 25% of primary pulmonary lymphomas.7,9 A secondary involvement of the lung from a known B-NHL is encountered2 either at diagnosis, leading to a stage IV disease,10 or during the course of the disease as progression or relapse.
The clinical and radiologic abnormalities observed in pulmonary B-NHL are not specific11 even if peribronchovascular interstitial thickenings on a computerized tomography (CT) scan may suggest the diagnosis.12,13 Atypical lymphocytes can be detected by cell morphology in bronchoalveolar lavage (BAL) fluid,14 but their detection is inconstant. Because lymphocytes could spread to the bronchoalveolar space, it has been suggested that the repertoire of lymphocytes recovered by BAL could reflect the pulmonary infiltration repertoire. The phenotypic analysis of the lymphocytes in BAL fluid can detect a monotypic B-cell population,15 but CD8+ T cells are more frequently dominant even in the presence of a pulmonary B-NHL,16,17 leading to unhelpful conclusions.
The development of molecular biology techniques, Southern blotting,18 and polymerase chain reaction (PCR)19 has provided the ability to detect a monoclonal population of B lymphocytes on the basis of the detection of rearrangements of the genes that encode B-cell immunoglobulin receptor proteins. This detection has been applied to support the diagnosis of nodal20,21 or extranodal, for example, gastric22 and pulmonary23-25 B-NHL. The clonality of pulmonary lymphocyte infiltrates has been appreciated by clonality analysis of alveolar lymphocytes. Detection of the same dominant B-cell clone in BAL fluid and lymphomatous pulmonary biopsy has been reported by Southern blot15,16,26 and, in our more recent work, by PCR,17 suggesting that alveolar lymphocytes clonality analysis could help in the diagnosis of pulmonary B-NHL involvement.
The aim of this study was to determine the feasibility of this procedure in routine practice, and the sensitivity, specificity, and predictive value of B-cell clonality in alveolar lymphocytes recovered by BAL for the diagnosis of pulmonary B-NHL. We studied a prospective cohort of 106 patients presenting with a clinical suspicion of pulmonary NHL.
Patients and methods
Patients and study design
From January 1, 1994, to December 31, 1999, patients followed in the Department of Respiratory Diseases at Foch Hospital (Suresnes) and Tenon Hospital (Paris) were included in this bicentric prospective study. Patient management was as follows: Patients admitted for the diagnosis of chronic pulmonary opacity (more than 3 weeks) underwent a first-line standardized diagnostic procedure. It consisted of clinical and radiologic evaluation, as well as bacteriologic assessment of sputum. Thereafter, patients with a specific diagnosis were treated accordingly. Otherwise, patients had bronchoscopy, bronchoalveolar lavage (BAL), and bronchial biopsies. In 1 of the 2 departments, transbronchial biopsies were obtained. An aliquot of 10 mL lavage fluid was cultured for bacteria, mycobacteria, fungi, and viruses. Different stainings of BAL smears were performed for evaluation of cytologic and opportunistic pathogens. In the absence of diagnosis within the first 72 hours, 10 mL BAL fluid, which was kept at 4°C, was analyzed for B-cell clonality.
Inclusion criteria were the following: (1) one of the 2 following clinical situations: (a) chronic alveolar focal or diffuse, unique or multiple, opacity lasting for more than 3 weeks or (b) appearance of a pulmonary opacity in a patient affected by a known extrapulmonary B-NHL; and (2) absence of pulmonary infectious disease or cancer diagnosed by routine endoscopic examination and bronchoalveolar fluid analysis within the first 72 hours following sampling.
Patients suffering from a previously known leukemic form of a B-cell lymphoproliferative disease (ie, mostly chronic lymphocytic leukemia [CLL]), were not included in the study. Indeed, in CLL, a dominant B-cell clone is usually detected in BAL fluid because of the presence of a highly dominant clone in circulating B-cell repertoire.
The purpose of this study was to define the sensitivity, the specificity, and the predictive value of the analysis of lymphocyte clonality in the BAL of patients suspected of pulmonary lymphoma. The analysis of clonality was interpreted without knowledge of the diagnosis as concluded by the clinicians. Conversely, the clinical diagnosis relied on usual strategies (description to follow) without taking into account the PCR results. The pulmonary diagnosis that was recorded for the study was established within 6 months after admission of the patient.
The patients were separated in 2 groups: group I, patients without previously diagnosed lymphoproliferative disorder, and group II, patients having B-cell lymphoproliferative disease previously diagnosed before the pulmonary disease occurrence. The diagnosis of primary pulmonary lymphoma was made histologically and in the absence of any other localization of the lymphoma during the 3 months following the diagnosis.2 When the lung was the principal tumor site, multifocal MALT NHL was also classified as primary pulmonary lymphoma.3 The diagnosis of secondary pulmonary lymphoma could be made by (1) a pulmonary biopsy or (2) a lymph node biopsy when the pulmonary lesions waned under lymphoma treatment or progressed and/or relapsed with progression and/or relapse of the lymphoma in other sites. The diagnostic process of other lung diseases was made according to the recommendations of the American Thoracic and European Respiratory Societies.26
Bronchoalveolar lavage procedure
Bronchoalveolar lavage was done by using a fiberoptic bronchoscope wedged into the most extensively involved pulmonary segment as seen on thoracic CT scan. Sterile saline was instilled in 4 aliquots of 50 mL, and the fluid was recovered by gentle aspiration.
Specific staining and culture for viruses, bacteria, mycobacteria, and fungi were routinely set up for all samples. Cytologic analysis, with differential cell count, including lymphocytes, was performed on Giemsa-stained cytocentrifuged preparations. The presence of atypical lymphocytes in the cytologic analysis was recorded. Lymphocyte subsets were determined by flow cytometry (EPICS ELITE; Beckman Coulter, Villepinte, France). Lymphocytes were stained by anti-CD3–, anti-CD4–, and anti-CD8–specific monoclonal antibodies for T lymphocytes and by anti-CD19 for B lymphocytes (Beckman Coulter, Villepinte, France).
Samples
Fresh bronchoalveolar fluid (10 mL) was referred to the immunologic department for PCR analysis. Cells were centrifuged into pellets and kept at 4°C until final inclusion (described in inclusion criteria in “Patients and study design”).
Blood and bronchoalveolar fluid samples were analyzed simultaneously.
Frozen biopsies of diagnostic tissue were retrospectively analyzed when available.
Only specimens already acquired for routine diagnosis were used in the study. As recommended by the French governmental Agence Nationale d'Accréditation et d'Evaluation en Santé (ANAES) in its recommendations for cryopreserved cell and tissue libraries for molecular analyses (http://www.anaes.fr/ANAES/SiteWeb.nsf/wRubriquesID/APEH-3ZMHJP), patients were informed that part of the specimens could be used for molecular analysis provided that all routine examinations have been performed.
Histologic studies
Bronchial and transbronchial pulmonary biopsies were obtained during bronchoscopy as described in “Patients and study design.” Alternatively, mediastinal lymph node or open lung biopsies were obtained after completion of the BAL fluid analysis. They were referred to the Pathology Department at Foch Hospital or Tenon Hospital, either fresh or frozen or fixed. Non-Hodgkin lymphomas were classified by using the World Health Organization (WHO) classification.27
PCR methodology
DNA was obtained by a standard proteinase K digestion and a phenol-chloroform extraction of either bronchoalveolar fluid cells, frozen biopsies, or peripheral blood mononuclear cells. For immunoglobulin gene rearrangement study, 2 PCR analyses were performed as previously described28 (Figure 1A). In one PCR, the 5′ sense oligonucleotide matched homologous sequences within the framework 2 region29 ; in the second PCR it matched homologous sequences within the framework 3 region.21 In the two PCRs, the 3′ antisense oligonucleotide was hybridized to the JH region.30
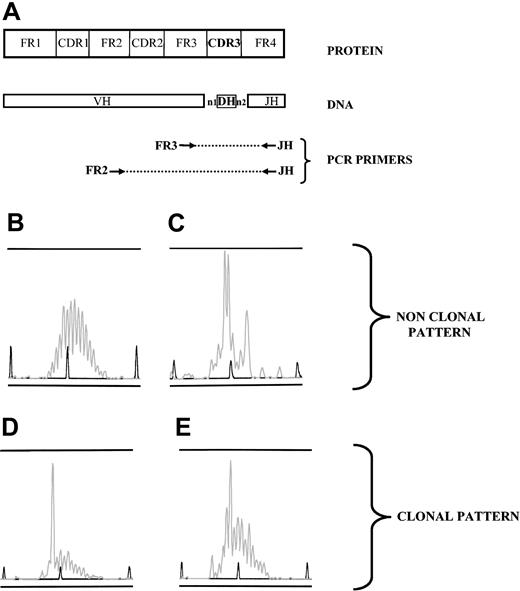
Clonality pattern of immunoglobulin gene rearrangements, analyzed by PCR. (A) DNA was extracted from samples, and 2 PCRs were performed using 2 sets of primers (FR3/JH and FR2/JH) to amplify all the size-variable CDR3 rearranged regions (FR2, GC(C/T) (C/T)CC GG(A/G) AA(A/G) (A/G)GT CTG GAG TGG; FR3, ACA CGG C(C/T)(G/C) TGT ATT ACT GT; JH, ACCTGAGGAGACGGTGACC). (B-E) PCR products were loaded on an ABI prism fragment size gel analysis. Four patterns were observed; when a polyclonal B-cell population was present in the sample, random sizes of all CDR3 regions resulted in a gaussian distribution of peak sizes (B). Several peaks could dominate in a sample without individualization of one peak, reflecting an oligoclonal pattern (C). One peak could be clearly individualized, either with a strongly dominant pattern, ie, with a low polyclonal background (D) or as a weak dominant peak, that is, with an intense polyclonal background (E). Patterns such as those in panels B and C were considered nonclonal. When a pattern such as that in panel D or E was observed either from FR3/JH or FR2/JH PCR, the sample was considered clonal.
Clonality pattern of immunoglobulin gene rearrangements, analyzed by PCR. (A) DNA was extracted from samples, and 2 PCRs were performed using 2 sets of primers (FR3/JH and FR2/JH) to amplify all the size-variable CDR3 rearranged regions (FR2, GC(C/T) (C/T)CC GG(A/G) AA(A/G) (A/G)GT CTG GAG TGG; FR3, ACA CGG C(C/T)(G/C) TGT ATT ACT GT; JH, ACCTGAGGAGACGGTGACC). (B-E) PCR products were loaded on an ABI prism fragment size gel analysis. Four patterns were observed; when a polyclonal B-cell population was present in the sample, random sizes of all CDR3 regions resulted in a gaussian distribution of peak sizes (B). Several peaks could dominate in a sample without individualization of one peak, reflecting an oligoclonal pattern (C). One peak could be clearly individualized, either with a strongly dominant pattern, ie, with a low polyclonal background (D) or as a weak dominant peak, that is, with an intense polyclonal background (E). Patterns such as those in panels B and C were considered nonclonal. When a pattern such as that in panel D or E was observed either from FR3/JH or FR2/JH PCR, the sample was considered clonal.
Clonality analysis
For CDR3 length diversity analysis, the amplified products were run on an ABI prism 310 genetic Analyzer (Applied Biosystems, Courtaboeuf, France). Results were analyzed by using Genscan analysis software (Applied Biosystems). Samples with a unique dominant peak within the polyclonal background (Figure 1D-E) were considered as clonal, and the clonal population was characterized by its CDR3 size. A peak was “strong” when it was associated with a low or absent polyclonal background (Figure 1D). A peak was “weak” when it was included within a high polyclonal background (Figure 1D). Samples with multiple dominant peaks (oligoclonal) (Figure 1C), or samples with a polyclonal repertoire (Figure 1B) were considered as nonclonal. When samples were available, the B-cell repertoire pattern in diagnostic biopsies and/or blood samples was compared with that in alveolar lymphocytes. In a patient, 2 dominant B-cell populations were considered identical when they displayed the same CDR3 size (Figure 2). When a dominant peak was detected in a lymphoma biopsy, it was assumed to characterize the tumoral population.
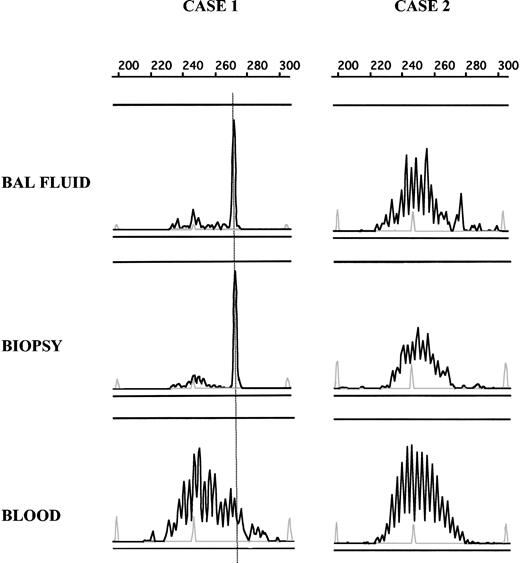
Comparison of B-cell clonality in BAL fluid, pulmonary biopsy, and blood. DNA from BAL fluid, biopsy, and blood samples was amplified, and PCR products were loaded on an ABI prism fragment size gel analysis. By using size marker (gray peak) and Genscan software, CDR3 size of the dominant peak could be determined and compared between different samples in a single patient. In case 1, the same dominant peak can be seen in PCR products obtained from BAL fluid and pulmonary biopsy but not from a blood sample, showing the presence of the same clonal B-cell population in both biopsy lymphocyte infiltrate and BAL fluid. In case 2, no dominant peak was detected in any analyzed tissue.
Comparison of B-cell clonality in BAL fluid, pulmonary biopsy, and blood. DNA from BAL fluid, biopsy, and blood samples was amplified, and PCR products were loaded on an ABI prism fragment size gel analysis. By using size marker (gray peak) and Genscan software, CDR3 size of the dominant peak could be determined and compared between different samples in a single patient. In case 1, the same dominant peak can be seen in PCR products obtained from BAL fluid and pulmonary biopsy but not from a blood sample, showing the presence of the same clonal B-cell population in both biopsy lymphocyte infiltrate and BAL fluid. In case 2, no dominant peak was detected in any analyzed tissue.
When indicated, heteroduplex analysis or sequence analysis was performed. For heteroduplex analysis, PCR products were denatured at 95°C for 5 minutes followed by rapid random renaturation at 4°C for 1 hour and electrophoresed in an 8% polyacrylamide gel. This process enforces duplex formation results in many different heteroduplexes with different migration speeds in case of polyclonal B-cell repertoire, but results in homoduplexes with identical rapid migration in case of dominant monoclonal B-cell population. Thus, heteroduplex analysis took advantage not only of the CDR3 length diversity (as Genscan does) but also of the nucleotidic junctional region diversity.31 For nucleotide sequence analysis, strong dominant PCR products were extracted from 6% polyacrylamide gel by elution. Big Dye Terminator cycle sequencing reaction kit (Applied Biosystems) was used prior to electrophoresis on an automated sequencer (ABI 310; Applied Biosystems). The sequences were aligned to immunoglobulin sequences from the international ImmunoGeneTics database (IMGT, http//imgt.cines.fr:8104; initiator and coordinator, Marie-Paule Lefranc, Montpellier, France), and the CDR3 region was identified.
Statistical analysis
Student t test was used to compare means. Chi-square or Fisher exact test was used to compare categorical data when appropriate. Significance was set at P < .05. Analysis was done by using Statview 5.0 (SAS Institute, Cary, NC).
Sensitivity (%) = true positive × 100/true positive + false negative; specificity (%) = true negative × 100/true negative + false positive. Negative predictive value (NPV) of a test is the proportion of times that a patient will be confirmed negative for disease when the test is negative and is estimated by using the following formula: NPV = true negative × 100/true negative + false negative. To determine this parameter, only patients with unpreviously diagnosed B-NHL were considered. In this group, positive diagnosis of pulmonary lymphoma was made by the histologic examination of pulmonary biopsy, the gold standard criteria. For other diagnosis, biopsy was made as recommended.26
Results
Patient characteristics
One hundred six patients with a clinical suspicion of pulmonary NHL were included in the study. The diagnosis of pulmonary lesions in these 106 patients, as obtained by usual strategies without taking into account the PCR results, is shown in Table 1. Eighty-two patients had no previous history of lymphoproliferative disorder before the pulmonary symptoms occurred. A thoracic biopsy was performed to establish the diagnosis in 43 patients (41 pulmonary and 2 bronchial biopsies), including all cases with a diagnosis of pulmonary lymphoma. Thirteen patients had a primary pulmonary B-NHL, 10 of whom with a MALT histology, either with pulmonary involvement alone (n = 6) or associated with asymptomatic gastric (n = 2), salivary and/or parotid gland localization (n = 2). Three patients had diffused large B-NHL. Infectious and autoimmune pulmonary diseases were otherwise the 2 main diagnoses. Six patients lost from follow-up are considered as having undetermined diagnosis (Table 1).
Patient classification and diagnosis of pulmonary lesions
Patient characteristics . | No. patients . |
|---|---|
| Patients with unpreviously diagnosed B-NHL | 82 |
| Primary pulmonary lymphomas | 13 |
| MALT lymphomas | 10 |
| DLBCL | 3 |
| Nonlymphomatous pulmonary lesions | 63 |
| Diffuse interstitial pneumoniae | 35 |
| Fibrosis/BOOP/LIP/drug-induced | 16 |
| Associated with autoimmune disease | 13 |
| Granulomatosis/sarcoidosis | 6 |
| Infectious pneumoniae | 20 |
| Miscellaneous* | 8 |
| Undetermined diagnosis | 6 |
| Patients with previously diagnosed B-cell lymphoproliferative disorder† | 24 |
| Secondary pulmonary B-cell NHL | 9 |
| DLBCL | 2 |
| Mantle cell lymphoma | 1 |
| Follicular lymphoma | 1 |
| Diffuse small cell lymphoma | 4 |
| MALT lymphoma | 1 |
| Nonlymphomatous pulmonary lesions | 12 |
| Diffuse interstitial pneumoniae | — |
| Fibrosis/drug-induced | 5 |
| Infectious pneumoniae | 5 |
| Miscellaneous cancer | 2 |
| Undetermined diagnosis | 3 |
Patient characteristics . | No. patients . |
|---|---|
| Patients with unpreviously diagnosed B-NHL | 82 |
| Primary pulmonary lymphomas | 13 |
| MALT lymphomas | 10 |
| DLBCL | 3 |
| Nonlymphomatous pulmonary lesions | 63 |
| Diffuse interstitial pneumoniae | 35 |
| Fibrosis/BOOP/LIP/drug-induced | 16 |
| Associated with autoimmune disease | 13 |
| Granulomatosis/sarcoidosis | 6 |
| Infectious pneumoniae | 20 |
| Miscellaneous* | 8 |
| Undetermined diagnosis | 6 |
| Patients with previously diagnosed B-cell lymphoproliferative disorder† | 24 |
| Secondary pulmonary B-cell NHL | 9 |
| DLBCL | 2 |
| Mantle cell lymphoma | 1 |
| Follicular lymphoma | 1 |
| Diffuse small cell lymphoma | 4 |
| MALT lymphoma | 1 |
| Nonlymphomatous pulmonary lesions | 12 |
| Diffuse interstitial pneumoniae | — |
| Fibrosis/drug-induced | 5 |
| Infectious pneumoniae | 5 |
| Miscellaneous cancer | 2 |
| Undetermined diagnosis | 3 |
MALT indicates mucosa-associated lymphoid tissue; DLBCL, diffuse large B-NHL; BOOP, bronchiolitis obliterans organizing pneumonia; LIP, lymphoid interstitial pneumonitis; and —,.
Five cancer, 1 Hodgkin disease, 1 alveolar proteinosis, 1 Castleman disease.
Seven myeloma/monoclonal gammapathy, 8 DLBCL, 1 mantle cell lymphoma, 1 follicular lymphoma, 7 diffuse small cell lymphoma, 1 MALT lymphoma.
Twenty-four patients had a previously diagnosed B-cell lymphoproliferative disorder (Table 1), either ongoing when pulmonary lesions occurred (n = 13) or previously treated with a suspicion of relapse (n = 11). A thoracic biopsy (8 pulmonary, 1 bronchi, and 1 mediastinal lymph node) was performed in 10 of these patients. Nine patients were considered as having a secondary pulmonary localization, demonstrated either histologically in 6 patients or clinically in 3 patients (described in criteria in “Patients and study design”). Three patients were considered to have an undetermined diagnosis.
None of the clinical symptoms (cough, dyspnea, chest pain) or radiologic patterns (alveolar opacities, interstitial opacities, pleural effusion) was significantly associated with pulmonary NHL diagnosis, in accordance with previously reported data (reviewed by Cadranel et al3 ).
Cytology in BAL fluid was available in 97 patients, and the percentage of lymphocytes is shown in Table 2.
Cytologic analysis of BAL fluid
. | Primary pulmonary NHL; n = 12 . | Secondary pulmonary NHL; n = 8 . | Nonlymphomatous pulmonary lesions; n = 71 . | Undetermined pulmonary diagnosis; n = 6 . |
|---|---|---|---|---|
| Cell count, mean (range) | ||||
| Total cell count, × 109/L | 0.542 (0.18-2.3) | 0.396 (0.152-1.08) | 0.481 (0.051-3.6) | 0.450 (0.09-0.92) |
| Neutrophils, % | 4 (0-20) | 17 (0-50) | 13 (0-85) | 36 (1-77) |
| Eosinophils, % | 0.8 (0-2) | 0.38 (0-1) | 2.3 (0-19) | 3.3 (0-17) |
| Macrophages, % | 58 (18-93) | 48 (10-81) | 47 (5-92) | 42 (10-74) |
| Lymphocytes, % | 36 (3-81) | 30 (3-61) | 40 (1-89) | 18 (6-39) |
| Cytology | ||||
| No. samples with atypical lymphocytes | 6 | 3 | 1 | 0 |
| Lymphocyte phenotype | ||||
| Number of studied samples | 7 | 2 | 29 | 1 |
| CD3, % (range) | 58 (27-90) | 98-79 | 91 (52-98) | 87 |
| CD4, % (range) | 35 (11-77) | 28-21 | 58 (2-93) | 17 |
| CD8, % (range) | 23 (8-56) | 71-24 | 40 (4-92) | 69 |
| CD19, % (range) | 37 (9-62) | 0-18 | 1.8 (0-12) | 0 |
. | Primary pulmonary NHL; n = 12 . | Secondary pulmonary NHL; n = 8 . | Nonlymphomatous pulmonary lesions; n = 71 . | Undetermined pulmonary diagnosis; n = 6 . |
|---|---|---|---|---|
| Cell count, mean (range) | ||||
| Total cell count, × 109/L | 0.542 (0.18-2.3) | 0.396 (0.152-1.08) | 0.481 (0.051-3.6) | 0.450 (0.09-0.92) |
| Neutrophils, % | 4 (0-20) | 17 (0-50) | 13 (0-85) | 36 (1-77) |
| Eosinophils, % | 0.8 (0-2) | 0.38 (0-1) | 2.3 (0-19) | 3.3 (0-17) |
| Macrophages, % | 58 (18-93) | 48 (10-81) | 47 (5-92) | 42 (10-74) |
| Lymphocytes, % | 36 (3-81) | 30 (3-61) | 40 (1-89) | 18 (6-39) |
| Cytology | ||||
| No. samples with atypical lymphocytes | 6 | 3 | 1 | 0 |
| Lymphocyte phenotype | ||||
| Number of studied samples | 7 | 2 | 29 | 1 |
| CD3, % (range) | 58 (27-90) | 98-79 | 91 (52-98) | 87 |
| CD4, % (range) | 35 (11-77) | 28-21 | 58 (2-93) | 17 |
| CD8, % (range) | 23 (8-56) | 71-24 | 40 (4-92) | 69 |
| CD19, % (range) | 37 (9-62) | 0-18 | 1.8 (0-12) | 0 |
The presence of atypical lymphocytes in the bronchoalveolar fluid was noted in 9 (45%) of 20 patients with pulmonary B-NHL and in 1 (1%) of 71 patients without lymphoma. Lymphocyte phenotypic analysis was performed in 39 cases and showed a predominant T-cell infiltrate except in 4 cases of primary pulmonary lymphoma in which B cells were in excess (Table 2).
B-cell clonality in BAL fluid relates to that in pulmonary parenchyma and not merely to that in blood lymphocytes
B-cell lymphomatous tissues are infiltrated by monoclonal malignant B cells. Immunoglobulin gene rearrangement analysis of tumoral biopsies by PCR enabled us to molecularly define the malignant B cells by the peak size of the dominant clonal population in the PCR product. Then, we could look for this peak, ie, these malignant B cells, in other tissues or fluids.
As indicated earlier, at least one thoracic biopsy was performed in 53 of 106 patients (43 of 82 + 10 of 24). In 19 patients, it was possible to analyze by PCR either pulmonary (n = 12) or bronchial (n = 5) tissue, or a mediastinal lymph node (n = 2). Eleven had lymphoma, including 9 primary and 2 secondary NHL. The presence of a monoclonal B-cell population could be demonstrated in 10 of 11 lymphoma biopsies by the detection of a strong dominant peak in the PCR products. By contrast, a clonal peak of weak type was observed in 1 of 8 nonlymphomatous biopsies. In 18 patients, the pattern of the B-cell clonality in BAL fluid could be compared with that in the biopsy in which the diagnosis was histologically established (Figure 2; Table 3). The detection of a dominant B-cell clone in BAL fluid was highly correlated with the detection of a dominant clone in the diagnostic biopsy (P < .001). When detected, the CDR3 size of the dominant peaks was identical in the 2 samples. Furthermore, in 6 of the 8 patients with a same sized dominant peak in BAL fluid and biopsy, we determined the CDR3 nucleotide sequences of the 2 peaks. For 2 patients, BAL fluid DNA was no longer available. As shown in Table 4, the CDR3 nucleotide sequences were identical between the 2 samples in 6 of 6 studied cases.
Comparison of clonality in BAL fluid and diagnostic biopsy in 18 patients
. | Clonal BAL fluid . | Nonclonal BAL fluid . |
|---|---|---|
| Clonal biopsy | 6 primary pulmonary B-NHLs; 2 secondary B-NHLs | 1 primary pulmonary B-NHL* ; 1 bronchiolitis obliterans organizing pneumoniae |
| Nonclonal biopsy | 1 vasculitis | 1 primary pulmonary lymphoma; 6 nonlymphomatous samples† |
. | Clonal BAL fluid . | Nonclonal BAL fluid . |
|---|---|---|
| Clonal biopsy | 6 primary pulmonary B-NHLs; 2 secondary B-NHLs | 1 primary pulmonary B-NHL* ; 1 bronchiolitis obliterans organizing pneumoniae |
| Nonclonal biopsy | 1 vasculitis | 1 primary pulmonary lymphoma; 6 nonlymphomatous samples† |
The BAL fluid lymphocytes were oligoclonal. Despite the fact that a peak with an identical size to the “tumoral” peak was seen in the oligoclonality, it was not possible to assume that the tumoral clone was present in the BAL fluid.
Pulmonary fibrosis (n = 1), drug-related pneumoniae (n=1), bronchiolitis obliterans organizing pneumonia (n=1), granulomatosis (n=2), viral infection (n = 1).
CDR3 nucleotide sequences of dominant peaks
. | . | CDR3 Sequences . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | . | VH . | . | n1 . | DH . | n2 . | JH . | . | ||||||
| 1 | BAL | VH3 - 23 | TGTGCGAAAG | CCTTGCGGC | GGATTACGATTTTTGGAGTGGGTAT | CGGGAAGGATGGGGG | CTTTGAGTACTGG | JH4 | ||||||
| Tumor | VH3 - 23 | TGTGCGAAAG | CCTTGCGGC | GGATTACGATTTTTGGAGTGGGTAT | CGGGAAGGATGGGGG | CTTTGAGTACTGG | JH4 | |||||||
| 2 | BAL | VH3 - 23 | TGTGCGAAA | TTGGTG | GGAGGCAGCTGCT | CTCTCCCG | TGTGAATACTGG | JH4 | ||||||
| Tumor | VH3 - 23 | TGTGCGAAA | TTGGTG | GGAGGCAGCTGCT | CTCTCCCG | TGTGAATACTGG | JH4 | |||||||
| 3 | BAL | VH* | TGTGCGAGAGA | CTTCTCGGGTCCTCCA | AGTGGGAGCTAC | C | TCTACTACTGG | JH4 | ||||||
| Tumor | VH* | TGTGCGAGAGA | CTTCTCGGGTCCTCCA | AGTGGGAGCTAC | C | TCTACTACTGG | JH4 | |||||||
| 4 | BAL | VH3 - 15 | TGTACCACAGA | TCCGCTTG | ATTACGATTTTTGGAGTGG | CGGT | TACTACAACTCCATGGACGTCTGG | JH6 | ||||||
| Tumor | VH3 - 15 | TGTACCACAGA | TCCGCTTG | ATTACGATTTTTGGAGTGG | CGGT | TACTACAACTCCATGGACGTCTGG | JH6 | |||||||
| 5 | BAL | VH3 - 30 | TGTGCGAGAGA | AGGATATTTTGACTGGTTATT | TGCGCAACTCTTTGGCCCA | TTTGACTGCTGG | JH4 | |||||||
| Tumor | VH3 - 30 | TGTGCGAGAGA | AGGATATTTTGACTGGTTATT | TGCGCAACTCTTTGGCCCA | TTTGACTGCTGG | JH4 | ||||||||
| 6 | BAL | VH* | TGTGCGAGAGA | T | CATCTGCGGGGAC | G | TGTCTTTGATGTCTGG | JH3 | ||||||
| Tumor | VH* | TGTGCGAGAGA | T | CATCTGCGGGGAC | G | TGTCTTTGATGTCTGG | JH3 | |||||||
| 7 | BAL | VH3 - 7 | TGTGCGAGA | GGAA | TTCGGAGTGATGA | CGGTCTTAGTGGAGGGGCAA | TGGACGTCTGG | JH3 | ||||||
| 8 | BAL | VH5 - 51 | TGTGCGAGA | ACGA | AATACGGTTACAACTC | GG | ACGGTATGGACGTCTGG | JH6 | ||||||
| 9 | BAL | VH5 - a03 | TGTGCGAGA | GTGGTACCAGCTGATAT | AGTCGGGAT | CTACTACTACGCTTTGGACGTCTGG | JH6 | |||||||
| 10 | BAL | VH3 - 13 | TGTGCAAGAG | GA | TATTACTATGCTTCGGGGAGTT | TGG | TTGACTACTGG | JH4 | ||||||
. | . | CDR3 Sequences . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | . | VH . | . | n1 . | DH . | n2 . | JH . | . | ||||||
| 1 | BAL | VH3 - 23 | TGTGCGAAAG | CCTTGCGGC | GGATTACGATTTTTGGAGTGGGTAT | CGGGAAGGATGGGGG | CTTTGAGTACTGG | JH4 | ||||||
| Tumor | VH3 - 23 | TGTGCGAAAG | CCTTGCGGC | GGATTACGATTTTTGGAGTGGGTAT | CGGGAAGGATGGGGG | CTTTGAGTACTGG | JH4 | |||||||
| 2 | BAL | VH3 - 23 | TGTGCGAAA | TTGGTG | GGAGGCAGCTGCT | CTCTCCCG | TGTGAATACTGG | JH4 | ||||||
| Tumor | VH3 - 23 | TGTGCGAAA | TTGGTG | GGAGGCAGCTGCT | CTCTCCCG | TGTGAATACTGG | JH4 | |||||||
| 3 | BAL | VH* | TGTGCGAGAGA | CTTCTCGGGTCCTCCA | AGTGGGAGCTAC | C | TCTACTACTGG | JH4 | ||||||
| Tumor | VH* | TGTGCGAGAGA | CTTCTCGGGTCCTCCA | AGTGGGAGCTAC | C | TCTACTACTGG | JH4 | |||||||
| 4 | BAL | VH3 - 15 | TGTACCACAGA | TCCGCTTG | ATTACGATTTTTGGAGTGG | CGGT | TACTACAACTCCATGGACGTCTGG | JH6 | ||||||
| Tumor | VH3 - 15 | TGTACCACAGA | TCCGCTTG | ATTACGATTTTTGGAGTGG | CGGT | TACTACAACTCCATGGACGTCTGG | JH6 | |||||||
| 5 | BAL | VH3 - 30 | TGTGCGAGAGA | AGGATATTTTGACTGGTTATT | TGCGCAACTCTTTGGCCCA | TTTGACTGCTGG | JH4 | |||||||
| Tumor | VH3 - 30 | TGTGCGAGAGA | AGGATATTTTGACTGGTTATT | TGCGCAACTCTTTGGCCCA | TTTGACTGCTGG | JH4 | ||||||||
| 6 | BAL | VH* | TGTGCGAGAGA | T | CATCTGCGGGGAC | G | TGTCTTTGATGTCTGG | JH3 | ||||||
| Tumor | VH* | TGTGCGAGAGA | T | CATCTGCGGGGAC | G | TGTCTTTGATGTCTGG | JH3 | |||||||
| 7 | BAL | VH3 - 7 | TGTGCGAGA | GGAA | TTCGGAGTGATGA | CGGTCTTAGTGGAGGGGCAA | TGGACGTCTGG | JH3 | ||||||
| 8 | BAL | VH5 - 51 | TGTGCGAGA | ACGA | AATACGGTTACAACTC | GG | ACGGTATGGACGTCTGG | JH6 | ||||||
| 9 | BAL | VH5 - a03 | TGTGCGAGA | GTGGTACCAGCTGATAT | AGTCGGGAT | CTACTACTACGCTTTGGACGTCTGG | JH6 | |||||||
| 10 | BAL | VH3 - 13 | TGTGCAAGAG | GA | TATTACTATGCTTCGGGGAGTT | TGG | TTGACTACTGG | JH4 | ||||||
Underlined nucleotides are mutated when compared with the more homologous germline segment.
VH nucleotide sequence was too short for the assessment of the more homologous VH germline segment.
The detection or the nondetection of a dominant peak was identical in biopsies and BAL fluid in 15 (83%) of 18 studied patients. Of the 3 nonconcordant results, one patient had a pulmonary lymphoma with a clonal pattern in the biopsy and an oligoclonal pattern in the BAL fluid. A peak with an identical CDR3 size to the tumoral peak was found in the BAL fluid, but in the absence of a unique expansion, it was not possible to assume that it was the tumoral clone.
A clonal B-cell population in the BAL fluid could reflect a highly restricted circulating B-cell repertoire as seen in chronic lymphocytic leukemias. Therefore, patients with a known chronic lymphocytic leukemia were not included in the study, and clonality in blood was assessed in 41 patients with available blood samples and compared with BAL fluid clonality (Table 5). This series included 29 patients with previously undiagnosed NHL and 12 patients with a previously diagnosed B-cell lymphoproliferative disorder. Of the 41 patients, 6 patients whose diagnosis is given in Table 5 had a circulating dominant clonal B-cell population. Three patients had a dominant B-cell clone also in the BAL fluid. In the 3 cases, the clone CDR3 size was identical in both blood and BAL fluid, but the clonal identity was not ascertained by sequencing. Seven of the 41 patients had a BAL fluid dominant clone in the absence of a dominant clone in blood. Thus, the detection in BAL fluid of a dominant B-cell clone was not merely related to the presence of a dominant clone circulating in blood (P > .05).
Comparison of clonality in blood and BAL fluid in 41 patients
Characteristic . | No. patients . |
|---|---|
| Patients with unpreviously diagnosed B-NHL | 29 |
| Clonal BAL fluid | |
| Blood+ | |
| Al* | 1 |
| Blood− | |
| NHL | 2 |
| Al | 2 |
| Infectious pneumoniae | 1 |
| Nonclonal BAL fluid | |
| Blood+ | |
| Al* | 1 |
| Undertermined | 1 |
| Blood− | |
| Diffuse interstitial pneumoniae | 11 |
| Infectious pneumoniae | 8 |
| Castleman disease | 1 |
| Undetermined | 1 |
| Patients with previously diagnosed B-cell lymphoproliferative disorder | 12 |
| Clonal BAL fluid | |
| Blood+ | |
| Kaposi sarcoma | 1 |
| DLBCL | 1 |
| Blood− | |
| Undetermined | 2 |
| Nonclonal BAL fluid | |
| Blood+ | |
| Cryoglobulinemia | 1 |
| Blood− | |
| NHL | 1 |
| Diffuse interstitial pneumoniae | 3 |
| Infectious pneumoniae | 2 |
| Cancer | 1 |
| Undetermined | 1 |
Characteristic . | No. patients . |
|---|---|
| Patients with unpreviously diagnosed B-NHL | 29 |
| Clonal BAL fluid | |
| Blood+ | |
| Al* | 1 |
| Blood− | |
| NHL | 2 |
| Al | 2 |
| Infectious pneumoniae | 1 |
| Nonclonal BAL fluid | |
| Blood+ | |
| Al* | 1 |
| Undertermined | 1 |
| Blood− | |
| Diffuse interstitial pneumoniae | 11 |
| Infectious pneumoniae | 8 |
| Castleman disease | 1 |
| Undetermined | 1 |
| Patients with previously diagnosed B-cell lymphoproliferative disorder | 12 |
| Clonal BAL fluid | |
| Blood+ | |
| Kaposi sarcoma | 1 |
| DLBCL | 1 |
| Blood− | |
| Undetermined | 2 |
| Nonclonal BAL fluid | |
| Blood+ | |
| Cryoglobulinemia | 1 |
| Blood− | |
| NHL | 1 |
| Diffuse interstitial pneumoniae | 3 |
| Infectious pneumoniae | 2 |
| Cancer | 1 |
| Undetermined | 1 |
Al indicates diffuse interstitial pneumoniae associated with autoimmune disease.
Clonal pattern of bronchoalveolar lymphocytes is associated with the diagnosis of pulmonary B-NHL
Immunoglobulin gene rearrangement analysis was performed prospectively on alveolar lymphocytes recovered by BAL from the 106 patients. In 7 patients, the PCR was not interpretable, because of either the presence of inhibitor of PCR in the sample32 or an insufficient number of B cells to be analyzed. Ninety-nine Genscan PCR results were analyzed (Table 6). The sensitivity and specificity of BAL fluid clonality analysis with regard to the detection of a pulmonary lymphoma were determined from the 70 patients with previously undiagnosed B-NHL, whose diagnoses were ascertained and whose BAL fluid clonality were determined (82 - 6 - 6 = 70 patients) (Table 6). Indeed, in this group, diagnosis was made by using established criteria, including histology when necessary (half of patients), and particularly for all primary pulmonary lymphomas. A clonal pattern could be demonstrated in 18 of 70 BAL fluids. The presence of a clonal pattern in BAL fluid was statistically more frequent in the pulmonary B-NHL group (10 of 12 versus 8 of 58, P < .0001) (Table 6).
PCR analysis of BAL fluid lymphocyte clonality
. | Interpretable PCR, n . | Clonal/interpretable PCR . | . | . | ||
|---|---|---|---|---|---|---|
. | . | Strong peak . | Weak peak . | Total clonal (%) . | ||
| Patients with unpreviously diagnosed B-NHL | ||||||
| Primary pulmonary lymphomas | 12 | 9 | 1 | 10/12 (83) | ||
| Nonlymphomatous pulmonary lesions | 58 | 2 | 6 | 8/58 (14)* | ||
| Undetermined pulmonary diagnosis | 6 | 0 | 1 | 1/6 | ||
| Patients with B-cell lymphoproliferative disorder | ||||||
| Secondary pulmonary B-cell NHL | 8 | 6 | 0 | 6/8 (75) | ||
| Nonlymphomatous pulmonary lesions | 12 | 1 | 1 | 2/12 (17)† | ||
| Undetermined pulmonary diagnosis; n = 3 | 3 | 2 | 0 | 2/3 | ||
. | Interpretable PCR, n . | Clonal/interpretable PCR . | . | . | ||
|---|---|---|---|---|---|---|
. | . | Strong peak . | Weak peak . | Total clonal (%) . | ||
| Patients with unpreviously diagnosed B-NHL | ||||||
| Primary pulmonary lymphomas | 12 | 9 | 1 | 10/12 (83) | ||
| Nonlymphomatous pulmonary lesions | 58 | 2 | 6 | 8/58 (14)* | ||
| Undetermined pulmonary diagnosis | 6 | 0 | 1 | 1/6 | ||
| Patients with B-cell lymphoproliferative disorder | ||||||
| Secondary pulmonary B-cell NHL | 8 | 6 | 0 | 6/8 (75) | ||
| Nonlymphomatous pulmonary lesions | 12 | 1 | 1 | 2/12 (17)† | ||
| Undetermined pulmonary diagnosis; n = 3 | 3 | 2 | 0 | 2/3 | ||
Total clonal = strong peak + weak peak. Values in the total clonal column are total clonal/interpretable PCR.
1 of 15 fibrosis/BOOP/LIP/drug induced, 6 of 13 pneumoniae associated with autoimmune disease, 1 of 18 infectious pneumoniae.
1 of 5 infectious pneumoniae, 1 of 2 miscellaneous.
Then, the peak intensity of strong or weak type (described in “Patients and methods”) was noted, and the clinical relevance of a strong clonal pattern was analyzed (Table 6). The specificity of a strong clonal pattern was 97% (56 of 56 + 2), with high 82% (9 of 9 + 2) and 95% (56 of 56 + 3) positive and negative predictive values, respectively. Therefore, the detection of a strong clonal population in BAL fluid is a good argument for the diagnosis of pulmonary lymphoma, and, conversely, in the absence of either a strong or weak clonal population in BAL fluid, the probability to have a pulmonary lymphoma is very low.
A weak monoclonal B-cell expansion was found with high frequency (38%) in patients with an autoimmune disease involving the lungs (ie, 2 Sjögren syndromes, 2 pulmonary vasculitis, and 1 dermatopolymyositis).
Within the group of 20 patients with a previously diagnosed B-cell lymphoproliferative disorder whose pulmonary diagnosis was ascertained and whose BAL fluid clonality was determined (24 - 3 - 1 = 20 patients), the detection of a clonal population in BAL fluid also was associated with the diagnosis of pulmonary B-NHL (6 of 8 versus 2 of 12; P < .01) (Table 6).
Because Genscan software only analyzes CDR3 length diversity, one cannot exclude that one dominant peak includes several clones which present the same CDR3 size. We could further analyze clonality of 25 of 29 dominant peaks detected in BAL fluid (19 strong dominant peaks and 6 weak dominant peaks). The clonal nature of 19 strong dominant peak was established in 18 of 19 patients either by CDR3 nucleotide sequencing (n = 10; Table 4) or heteroduplex analysis (n = 8, data not shown). In 1 of 19 patient peaks monoclonality could not be ascertained by heteroduplex analysis. In this case, Genscan peak intensity was borderline between strong and weak peak, and the patient presented with infectious pneumonia. In the 6 weak dominant peaks, in only one patient heteroduplex analysis showed a monoclonal band with a faint intensity (data not shown). Interestingly, this patient had a primary pulmonary lymphoma, and, again, Genscan peak intensity was borderline between strong and weak peak.
Discussion
The diagnosis of pulmonary lymphoma may be evoked in 2 clinical settings: primary pulmonary lymphoma or pulmonary localization of an otherwise extended NHL. Whether primary or secondary, the demonstration of pulmonary involvement by lymphoma is difficult, and the presence of a clonal B-cell population in alveolar lymphocytes may be of indirect value for the diagnosis of pulmonary lymphoma.15-17,33 In the first clinical setting, the detection of a B-cell clone in BAL fluid would argue for the initiation of invasive investigations as transbronchial or surgical biopsies for both histologic analysis and immunohistologic staining.3,5,6 In the second clinical setting, in the absence of chronic lymphocytic leukemia, the detection in the BAL fluid of the same B-cell clone as in an extrapulmonary NHL lesion would be sufficient for the diagnosis of pulmonary involvement and would avoid the more invasive investigations. To assess both the feasibility in routine practice of the clonality analysis and the diagnostic value of B-cell clonality in BAL fluid, a prospective study was carried out, including patients in whom pulmonary lymphoma was considered as a putative diagnosis. Interpretable PCR results were obtained in most analyzed BAL fluids (99 [94%] of 106), demonstrating that our PCR analysis may become a routine procedure. When detected in both BAL fluid and tumor biopsy, dominant B-cell clones were identical, as assessed by Genscan analysis and CDR3 nucleotide sequences, confirming that dominant B-cell clone detected in BAL fluid were derived from tumor cells. The positive and negative predictive values of the analysis of BAL fluid B-cell clonality by PCR for diagnosis of pulmonary NHL were remarkably high, 82% and 95%, respectively.
A BAL fluid dominant clonal B-cell population could be detected by PCR in 10 of 12 patients with a histologically diagnosed primary pulmonary NHL and in 4 of 5 patients with a histologically diagnosed secondary pulmonary B-NHL. Thus, in 3 patients such a B-cell clone could not be evidenced in BAL fluid, although the lymphoma involved the lung. One was MALT lymphoma, one diffuse large B cell lymphoma, and one lymphoplasmocytic lymphoma. A first hypothesis to explain this false-negative result with regard to the diagnosis of lymphoma is the failure to amplify the malignant cell immunoglobulin gene rearrangement. Indeed, the incidence of detected clonal immunoglobulin gene rearrangement by PCR in B-NHL varies with the malignant cell differentiation status, particularly their pregerminal or postgerminal center phenotype which correlates with the somatic mutation rate.34 By using 2 PCRs (FR2/JH, FR3/JH) we analyzed lymphomatous biopsies from 11 patients and found a dominant B-cell clone in 10 (90%) of 11 samples, in accordance with previously reported data in MALT, follicular, and diffuse large B-cell lymphomas.21 A second hypothesis is the lack of sensitivity of the technique. Indeed, clonality analysis aims at the detection of a dominant clonal population relative to other B cells in the sample and could fail to detect a minority of malignant B cells.
In the 18 patients in whom biopsy was analyzed, the B-cell clonality in BAL fluid compared to the biopsy with a correlation of 83%. In 2 patients, the presence of a tumoral population in BAL fluid could not be demonstrated despite the characterization of the CDR3 size of dominant B-cell population in the tumoral biopsy and the detection of a peak of the same CDR3 size but nondominant in the BAL fluid. This finding underlines the limit of the technique which characterizes a clone only by its CDR3 size and not by its CDR3 sequence, as denaturing gradient gel electrophoresis (DGGE) does.35 However, the DGGE technique detects intraclonal diversity because of ongoing somatic mutations which characterizes germinal center derived lymphomas and has been reported in pulmonary MALT lymphomas.36 Intraclonal diversity could result in an oligoclonal pattern of the PCR product analysis, thus being useless for the diagnosis of B-cell NHL.
A detectable B-cell clone in BAL fluid does not mean a pulmonary B-NHL. Thus, 8 (14%) of 58 patients with nonlymphomatous pulmonary lesions were found to have a dominant B-cell population in the BAL fluid. Interestingly, in 6 of the 8 patients, the peak in Genscan analysis was of weak intensity, suggesting a minor dominant B-cell clone. This minor dominant B-cell clone was not observed in the 5 cases further analyzed by heteroduplex. The more likely hypothesis is that heteroduplex analysis is less sensitive than Genscan analysis, ie, cannot detect faint clonal population in a high polyclonal background, but one cannot exclude that some of those weak peaks are truly oligoclonal. Moreover, 2 of 8 patients had a peak of strong intensity with 1 of the 2 peaks not detected by heteroduplex analysis. The latter was retrospective of borderline intensity between strong and weak. Thus, we can hypothesize that, if the strong or weak classification is refined by a heteroduplex analysis of borderline cases, the predictive value of strong peak would be enhanced. Of note, none of those 8 patients have developed a lymphoma with a median follow-up of 4 years after inclusion. Longer follow-up is ongoing.
From the 8 patients with a dominant B-cell population in the BAL fluid and nonlymphomatous pulmonary lesions, 5 patients had diffuse interstitial pneumonia associated with an autoimmune disease, and 1 patient had an infectious pneumonia. Chronic antigenic stimulation either driven by infectious pathogens or autoantigens has been shown to result in oligoclonal or clonal expansions of B cells in involved tissue. Helicobacter infection and gastritis37 or hepatitis C virus (HCV) infection and hepatitis38 as infectious diseases, and Sjögren syndrome as autoimmune disease39,40 are well-documented conditions with reported nonmalignant and clonal B-cell expansions. All these diseases are risk factors for developing lymphomas.41 Some data suggest, at least in HCV infection, that malignant cells could derive from the rheumatoid factor-producing cells.42 The hypothesis that Sjögren syndrome may share a common pathogenesis with HCV-related lymphomas (ie, a chronic stimulation of rheumatoid factor producing B cells) is suggested by the demonstration of a nonrandom utilization of heavy and light V gene segments by Sjögren syndrome–associated lymphoma B cells and the demonstration that these lymphoma B cells may display a rheumatoid factor activity.43
In conclusion, our study is the first prospective study using PCR for alveolar lymphocyte clonality analysis in a large population of patients included on the basis of clinical criteria. We show on one hand that the detection of a strong peak in PCR product, ie, a major clonal B-lymphocyte population in the BAL fluid, has good positive and negative predictive values in the diagnosis of pulmonary B-NHL. PCR should be performed to sustain the biopsy indication in case of an isolated pulmonary lesion and possibly to avoid the biopsy in case of pulmonary lesion in an extended B-NHL. On the other hand, a benign minor B-cell clone can be detected notably in autoimmune and chronic infectious diseases, the predictive value of which for developing lymphoma remains to be determined.
Prepublished online as Blood First Edition Paper, January 8, 2004; DOI 10.1182/blood-2003-07-2335.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Chantal Lahet and Christophe Graube for their excellent technical assistance; Drs Stern, Rivaud, Grenet, Bonan, Gonnot of Foch hospital, and Drs Milleron, Souidi L'huillier of Tenon hospitals for their clinical contribution; and Mr Avignan for reviewing the English manuscript. We thank Patrick Maison for his critical review of the statistical analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal