Abstract
Differential expression of Hox genes is associated with normal hematopoiesis, whereas inappropriate maintenance of Hox gene expression, particularly Hoxa7 and Hoxa9, is a feature of leukemias harboring mixed-lineage leukemia (MLL) mutations. To understand the pathogenic roles of Hox genes in MLL leukemias, we assessed the impact of Hoxa7 or Hoxa9 nullizygosity on hematopoietic progenitor compartments and their susceptibility to MLL-induced leukemias. Selective reductions in the absolute numbers of committed progenitors, but not of hematopoietic stem cells, distinguished Hoxa7- and Hoxa9-deficient mice. Megakaryocytic/erythroid progenitor (MEP) reductions in Hoxa7-/-mice correlated with reticulocytosis and thrombocytopenia without anemia. Conversely, Hoxa9-/- mice displayed marked lymphopenia and substantial reductions of common lymphoid progenitors (CLPs) and lymphoid precursors, in addition to significant reductions of common myeloid progenitors (CMPs) and granulocyte/monocyte progenitors (GMPs). In retroviral transduction/transplantation assays, Hoxa7- and Hoxa9-deficient progenitors remained susceptible to transformation by MLL-GAS7, which activates MLL through a dimerization-dependent mechanism. However, Hoxa7-/- or Hoxa9-/-progenitors were less efficient in generating transformed blast colony-forming units (CFUs) in vitro and induced leukemias with longer disease latencies, reduced penetrance, and less mature phenotypes. Thus, Hoxa7 and Hoxa9 contribute to hematopoietic progenitor homeostasis but are not necessary for MLL-GAS7–mediated leukemogenesis, yet they appear to affect disease latency, penetrance, and phenotypes consistent with their critical roles as downstream targets of MLL fusion proteins. (Blood. 2004;103:3192-3199)
Introduction
The mixed-lineage leukemia (MLL) gene is a frequent target of chromosomal translocations that result in its structural rearrangement in 10% of adult and 80% of infant acute leukemias. MLL rearrangements induce its promiscuous fusion to more than 30 different partner proteins, resulting in leukemias of different lineages.1 Others2 and we3 have shown that heterologous fusion with transcriptional effector domains constitutes a common mechanism for oncogenic activation of MLL by a subset of its fusion partners. Recently, an alternative pathway was reported showing MLL is oncogenically activated by dimerization domains contributed by another subset of its fusion partners that do not display inherent transactivation properties.4 Hematopoietic cells transformed by either category of MLL fusion protein consistently express a subset of Hox genes, which are candidate downstream effectors of MLL oncogenic activity.4,5 These findings are also consistent with studies of infant leukemias showing that expression of specific Hox genes (in particular HoxA9 and HoxA7) and the Hox cofactor Meis1 are up-regulated in MLL leukemias.6-9
Hox genes are implicated in normal and abnormal hematopoiesis.10,11 Differential expression of Hox genes is closely associated with specific lineages and stages of hematopoietic differentiation. Experimentally, hyperexpression of HoxB4 leads to expansion of the hematopoietic stem cell (HSC) pool without inducing leukemia in mice,12 whereas forced expression of HoxA1013 or HoxB314 induces hematologic malignancies, albeit with long disease latency. Conversely coexpression of the Hox-cofactor Meis1 with Hoxa7 or Hoxa9 efficiently induces short-latency acute myeloid leukemia (AML) in mice.15 Overexpression of HoxA9 in human AML has also been reported to be an independent marker associated with poor prognostic outcomes.16 Moreover, subsets of Hox genes (HoxA9, HoxA11, HoxC13, and HoxD13) are directly involved in the formation of chimeric fusion proteins with Nup98 as a result of chromosomal translocations in human AML.17-21
Although the aberrant expression of Hox genes has been directly linked to certain leukemias, their roles in MLL-mediated leukemogenesis are not well defined. Expression of HoxA7 did not correlate with MLL rearrangements in an expression profile study of human leukemias and cell lines.22 Although expression profile studies of infant acute lymphoblastic leukemia (ALL) revealed preferential expression of HoxA9 in MLL leukemias that exhibited myeloid and lymphoid phenotypes, it was also overexpressed in most AMLs lacking MLL rearrangements.16 Targeted down-regulation of MLL-AF9 resulted in apoptosis of a human leukemia cell line but did not down-regulate the expression of HoxA9.23 Inducible expression of MLL-AF9 in the 32Dcl3 myeloid progenitor cell line inhibited, rather than activated, the expression of certain Hox genes, including HoxA7, that are otherwise up-regulated by granulocyte colony-stimulating factor(G-CSF)–mediated differentiation.24 However, most of these in vitro studies were performed using cell lines that likely harbor multiple genetic mutations, some of which may affect their Hox dependence. The current study was undertaken using a genetic approach with Hox-deficient primary hematopoietic progenitors, together with retroviral transduction/transplantation assays, to molecularly dissect the functional requirements of individual Hox genes for in vitro and in vivo MLL–mediated leukemogenic transformation.
Materials and methods
DNA constructs and mice
Cell sorting and immunophenotypic analysis by FACS
All experiments were performed using bone marrow and thymus from 4- to 8-week-old mice with age- and sex-matched controls. Immunophenotypic analysis was performed using fluorescence-activated cell sorter (FACS) analysis with fluorochrome-conjugated monoclonal antibodies to Sca-1 (E13-161-7 clone), c-Kit (2B8 clone), Flt3 (A2F10.1), CD34 (RAM34), CD43 (S7 clone), Mac-1 (M1/70 clone), Gr-1 (RB6-8C5 clone), CD19 (1D3 clone), B220 (RA3-6B2 clone), CD24/HSA (30-F1 clone), BP-1 (6C3 clone), CD3ϵ (145-2C11 clone), CD4 (GK1.5 clone), CD8a (53-6.7 clone), anti–interleukin-2 receptor α (anti–IL-2Rα)/CD25 (A2F10.1 clone), CD44 (IM781 clone), CD16/CD32 (FcγRII/III) (24G2 clone), T-cell receptor β (TCRβ; H57-597 clone), Thy1.1 (19XE5 clone), Thy1.2 (53-2.1), Ter-119 (TER-119), CD45.1 (A20.1.7), and CD45.2 (AL1-4A2). Antibodies were purchased from either PharMingen (San Diego, CA) or eBioscience (San Diego, CA). Staining was generally performed on ice for 15 to 30 minutes. Cells were washed twice in staining medium and resuspended in 1 μg/mL propidium iodine (PI) before analysis using a Moflops (modified triple laser Cytomation/Becton Dickinson hybrid FACS) or a Vantage (modified triple laser [488-nm argon laser, 599-nm dye laser, and UV laser]; Becton Dickinson Immunocytometry System, Mountain View, CA). Dead cells were gated out using high PI staining and forward light scatter. To analyze and sort HSCs, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), granulocyte/monocyte progenitors (GMPs), and megakaryocytic/erythroid progenitor (MEPs), bone marrow cells were stained with specific conjugated and unconjugated antibodies, as previously described,27-30 before analysis or double sorting using the FACS Vantage. Briefly, phenotypic populations were defined as HSC (Lin-/lo/Thy1.1lo/c-Kithi/Sca-1hi), CLP (Lin-/c-Kitlo/Sca-1lo/IL-7Rlo/Thy1.1-), CMP (c-Kithi/Sca-1-/CD34+/CD16/32lo), GMP (Lin-/c-Kithi/Sca-1-/CD34+/CD16/32hi), and MEP (Lin-/c-Kithi/Sca-1-/CD34-/CD16/32lo). B-lymphoid progenitor analysis was performed as previously described.31 For T-lymphoid progenitor studies, pro-T1 and pro-T2 populations were gated on c-Kithi/Lin-/lo/CD44+/CD25- and c-Kithi/Lin-/lo/CD44+/CD25+, respectively.32,33
In vitro and in vivo transformation assays
In vitro hematopoietic progenitor transformation assays (HPTAs) were performed as previously described.34 Enriched hematopoietic progenitors were prepared from the bone marrows of 4- to 10-week-old C57BL/6 mice by positive selection for c-Kit expression using magnetic activated cell sorting (MACS) before transduction by spinoculation with viral supernatants collected 3 days after transfection of Phoenix cells. For in vitro assays, transduced cells were plated in 1% methylcellulose (StemCell Technologies, Vancouver, BC, Canada) supplemented with either myeloid or lymphoid cytokines in the presence or absence of 1 mg/mL G418. Myeloid-conditioned methylcellulose contained Iscove modified Dulbecco medium (IMDM)–based Methocult (Methocult M3231; StemCell Technologies) supplemented with 20 ng/mL stem cell factor (SCF) and 10 ng/mL each of IL-3, IL-6, and granulocyte-macrophage colony-stimulating factor (GM-CSF; R&D Systems, Minneapolis, MN). Single-cell suspensions (104 cells) of G418-resistant colonies were then replated in methylcellulose medium supplemented with growth factors without G418. Plating was repeated every 7 days. In each round of replating, single-cell suspensions were also expanded in RPMI liquid medium containing 20% fetal calf serum (FCS) plus 20% WEHI-conditioned medium. For tumorigenicity assays, 106 immortalized cells were injected into the retro-orbital venous sinuses of 6- to 10-week-old syngeneic C57BL/6 mice that had received sublethal doses (5.25 Gy) of total body γ irradiation (cesium Cs 135 [135Cs]). For in vivo assays, 2 × 105 transduced cells were transplanted together with 2 × 105 total bone marrow rescue cells into the retro-orbital venous sinuses of 6- to 10-week-old syngeneic, lethally irradiated (10.5 Gy total body γ irradiation [135Cs]) C57BL/6 mice.35 Mice were maintained on antibiotic water to avoid infection and were monitored for the development of leukemia by complete blood count, blood smear, and FACS analysis. Tissues were fixed in buffered formalin, sectioned, and stained with hematoxylin and eosin (H&E) for histologic analysis.
TCR gene rearrangement analyses
Genomic DNAs were extracted from tumor tissues and digested with the EcoRI restriction enzyme. Digested DNAs were separated in 0.8% agarose, transferred to nylon membranes, and hybridized with radiolabeled DNA probes. The probe used for mouse TCR Jβ2 gene segments has been described previously.36
Reverse transcription–polymerase chain reaction analyses
Cells harvested from cultures or mouse tissues were lysed in Trizol (Life Technologies) for RNA extraction. cDNAs were synthesized using oligo-dT primers and Superscript II (Life Technologies). Polymerase chain reaction (PCR) was performed using primers specific for Hoxa7, Hoxa9, MLLGAS7, E2A-HLF, or β-actin.4 Details regarding primer sequences and PCR conditions are available on request.
Results
Hoxa7- and Hoxa9-deficient mice display selective reductions in hematopoietic progenitor populations
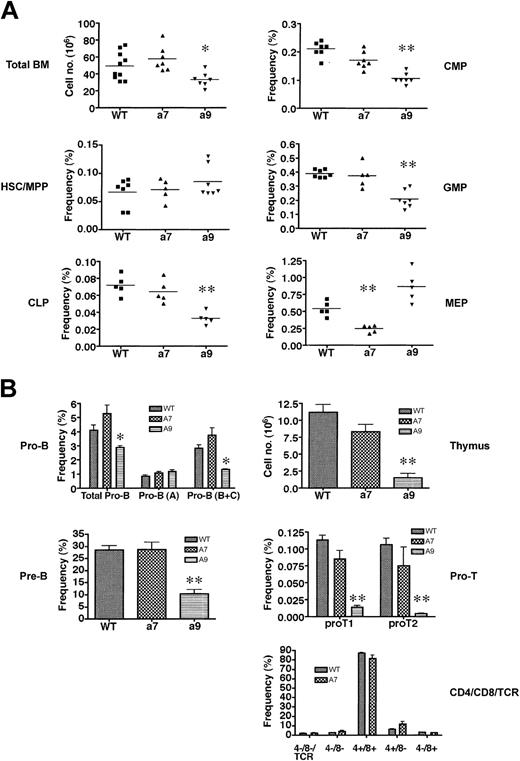
To assess the contributions of Hoxa7 and Hoxa9 in hematopoiesis, we determined the impact of their absence on the hematopoietic compartments of Hoxa7 and Hoxa9 null mice using FACS analysis of surface antigens that define specific progenitor populations. Total numbers of bone marrow cells were unaffected by the absence of Hoxa7, but they reduced to 30% of wild-type levels in Hoxa9-/- mice (Figure 1A).
Hoxa7-andHoxa9-deficient mice exhibit quantitative reductions inthe frequency of phenotypically defined hematopoietic progenitors. (A) Analysis of adult bone marrow for phenotypically defined populations of HSCs/MPPs and early progenitors (CLPs, CMPs, GMPs, MEPs). Horizontal lines indicate the mean frequencies for specific progenitors. *P < .01; **P < .001. (B) Analysis of B-cell (left panel) and T-cell (right panel) progenitors from bone marrow and thymus, respectively. *P < .01; **P < .001. Error bars indicate SD.
Hoxa7-andHoxa9-deficient mice exhibit quantitative reductions inthe frequency of phenotypically defined hematopoietic progenitors. (A) Analysis of adult bone marrow for phenotypically defined populations of HSCs/MPPs and early progenitors (CLPs, CMPs, GMPs, MEPs). Horizontal lines indicate the mean frequencies for specific progenitors. *P < .01; **P < .001. (B) Analysis of B-cell (left panel) and T-cell (right panel) progenitors from bone marrow and thymus, respectively. *P < .01; **P < .001. Error bars indicate SD.
The reduced bone marrow cellularity in Hoxa9-/- mice was not accounted for by reductions in HSCs and multipotent progenitors (MPPs) because their numbers (Table 1) and overall frequencies (Figure 1A) in Hoxa9- and Hoxa7-deficient mice were comparable to those in wild-type controls.
Absolute cell numbers in different hematopoietic compartments of wild-type, Hoxa7−/−, and Hoxa9−/− mice
. | Wild-type . | Hoxa7−/− . | Hoxa9−/− . |
|---|---|---|---|
| HSC and MPP count, × 103 | 32 ± 13 | 36 ± 11 | 34 ± 21 |
| CLP count, ×103 | 35 ± 11 | 37 ± 10 | 8 ± 2.4† |
| CMP count, ×103 | 108 ± 21 | 93 ± 23 | 33 ± 12† |
| GMP count, ×103 | 190 ± 33 | 210 ± 45 | 69 ± 31† |
| MEP count, ×103 | 256 ± 55 | 140 ± 42* | 280 ± 135 |
| Total Pro-B count, ×106 | 1.9 ± 0.39 | 2.5 ± 0.58 | 1.2 ± 0.45* |
| Pro-B (Fr. A) count, ×103 | 480 ± 170 | 640 ± 185 | 435 ± 135 |
| Pro-B (Fr. B-C) count, ×106 | 1.4 ± 0.31 | 1.8 ± 0.45 | 0.71 ± 0.21† |
| Pre-B count, ×106 | 13.8 ± 1.5 | 15.3 ± 2.9 | 3.4 ± 2.1† |
| Pro-T1 count, ×103 | 11.3 ± 0.33 | 8.3 ± 1.7 | 0.72 ± 0.13† |
| Pro-T2 count, ×103 | 10.9 ± 0.31 | 7.2 ± 2.3 | 0.31 ± 0.064† |
| CD4−/CD8−/TCRβ− count, ×106 | 0.21 ± 0.032 | 0.29 ± 0.065 | ND |
| CD4−/CD8− count, ×106 | 0.3 ± 0.045 | 0.4 ± 0.12 | ND |
| CD4+/CD8+ count, ×106 | 9.8 ± 1.2 | 6.8 ± 2.3 | ND |
| CD4+/CD8− count, ×106 | 0.73 ± 0.12 | 0.8 ± 0.34 | ND |
| CD4−/CD8+ count, ×106 | 0.36 ± 0.08 | 0.26 ± 0.036 | ND |
. | Wild-type . | Hoxa7−/− . | Hoxa9−/− . |
|---|---|---|---|
| HSC and MPP count, × 103 | 32 ± 13 | 36 ± 11 | 34 ± 21 |
| CLP count, ×103 | 35 ± 11 | 37 ± 10 | 8 ± 2.4† |
| CMP count, ×103 | 108 ± 21 | 93 ± 23 | 33 ± 12† |
| GMP count, ×103 | 190 ± 33 | 210 ± 45 | 69 ± 31† |
| MEP count, ×103 | 256 ± 55 | 140 ± 42* | 280 ± 135 |
| Total Pro-B count, ×106 | 1.9 ± 0.39 | 2.5 ± 0.58 | 1.2 ± 0.45* |
| Pro-B (Fr. A) count, ×103 | 480 ± 170 | 640 ± 185 | 435 ± 135 |
| Pro-B (Fr. B-C) count, ×106 | 1.4 ± 0.31 | 1.8 ± 0.45 | 0.71 ± 0.21† |
| Pre-B count, ×106 | 13.8 ± 1.5 | 15.3 ± 2.9 | 3.4 ± 2.1† |
| Pro-T1 count, ×103 | 11.3 ± 0.33 | 8.3 ± 1.7 | 0.72 ± 0.13† |
| Pro-T2 count, ×103 | 10.9 ± 0.31 | 7.2 ± 2.3 | 0.31 ± 0.064† |
| CD4−/CD8−/TCRβ− count, ×106 | 0.21 ± 0.032 | 0.29 ± 0.065 | ND |
| CD4−/CD8− count, ×106 | 0.3 ± 0.045 | 0.4 ± 0.12 | ND |
| CD4+/CD8+ count, ×106 | 9.8 ± 1.2 | 6.8 ± 2.3 | ND |
| CD4+/CD8− count, ×106 | 0.73 ± 0.12 | 0.8 ± 0.34 | ND |
| CD4−/CD8+ count, ×106 | 0.36 ± 0.08 | 0.26 ± 0.036 | ND |
Values are mean ± SD.
Fr. A indicates Hardy Fraction A; Fr. B-C, Hardy Fraction B-C; and ND, not determined.
*P<.01; †P<.001.
However, further analysis of the progenitor compartments revealed significant reductions in absolute numbers of common lymphoid progenitors (CLPs) and common myeloid progenitors (CMPs) in Hoxa9-/- mice (80% and 70%, respectively) (Table 1). Reductions (65%) were also observed in the downstream granulocyte/monocyte progenitors (GMPs), but the megakaryocyte/erythroid progenitors (MEPs), which are also descended from CMPs, appeared to be unaffected by the absence of Hoxa9 (Figure 1A; Table 1). Conversely, the absolute numbers (Table 1) and frequencies (Figure 1A) of CLPs, CMPs, and GMPs appeared to be unaffected in Hoxa7-/- mice, whereas the MEP population was reduced to 40% of wild-type levels. Consistent with their severe CLP reductions, Hoxa9-/- mice exhibited marked reductions in B-cell precursors and thymocytes (Figure 1B; Table 1). Most significantly affected were the pro-T1 and pro-T2 populations in the thymus, which were reduced by more than 15- and 35-fold, respectively (Table 1). Hoxa9-/- bone marrow also displayed significant reductions in the pro-B– and pre-B–cell compartments, which were reduced more than 40% and 70%, respectively (Figure 1B; Table 1). In contrast to Hoxa9-/- mice, Hoxa7-/- mice displayed normal numbers and distributions of B-cell progenitors in the bone marrow and T-cell progenitors in the thymus (Figure 1B; Table 1). Hoxa9-/- mice displayed significant reductions in total peripheral white blood cell (WBC) counts affecting the myeloid and lymphoid lineages (Table 2).
Cell numbers in peripheral blood of wild-type, Hoxa7−/−, and Hoxa9−/− mice
. | Wild-type . | Hoxa7−/− . | Hoxa9−/− . |
|---|---|---|---|
| Complete WBC count, × 109/L | 8.9 ± 0.96 | 8.2 ± 0.42 | 4.6 ± 0.99† |
| RBC count, × 1012/L | 8.7 ± 0.45 | 8.6 ± 0.20 | 9.9 ± 0.29 |
| Absolute neutrophil count, × 109/L | 1.969 ± 0.629 | 0.950 ± 0.110* | 0.845 ± 0.163* |
| Absolute lymphocyte count, × 109/L | 7.013 ± 0.497 | 7.163 ± 0.386 | 3.537 ± 0.835† |
| Platelet count, × 109/L | 1095 ± 55 | 766 ± 27† | 807 ± 50* |
| Reticulocyte count, % | 4.77 ± 0.56 | 9.1 ± 0.82* | 3.3 ± 0.20 |
. | Wild-type . | Hoxa7−/− . | Hoxa9−/− . |
|---|---|---|---|
| Complete WBC count, × 109/L | 8.9 ± 0.96 | 8.2 ± 0.42 | 4.6 ± 0.99† |
| RBC count, × 1012/L | 8.7 ± 0.45 | 8.6 ± 0.20 | 9.9 ± 0.29 |
| Absolute neutrophil count, × 109/L | 1.969 ± 0.629 | 0.950 ± 0.110* | 0.845 ± 0.163* |
| Absolute lymphocyte count, × 109/L | 7.013 ± 0.497 | 7.163 ± 0.386 | 3.537 ± 0.835† |
| Platelet count, × 109/L | 1095 ± 55 | 766 ± 27† | 807 ± 50* |
| Reticulocyte count, % | 4.77 ± 0.56 | 9.1 ± 0.82* | 3.3 ± 0.20 |
Values are mean ± SD.
RBC indicates red blood cell.
P<.05.
P<.01.
In contrast, Hoxa7-/- mice had normal WBC counts, though the reduced platelet and elevated reticulocyte numbers might reflect defects in the MEP compartment. Interestingly, reductions in the absolute neutrophil count were also observed in Hoxa7-/- mice (Table 2), suggesting potential functional defects in myeloid progenitors downstream of GMP. These results, in conjunction with previous hematologic findings,37,38 indicate that Hoxa7 and Hoxa9 contribute to the development of select hematopoietic progenitor populations, but neither is essential for hematopoiesis.
MLL-GAS7 is capable of transforming Hoxa7- or Hoxa9-deficient hematopoietic progenitors in vitro
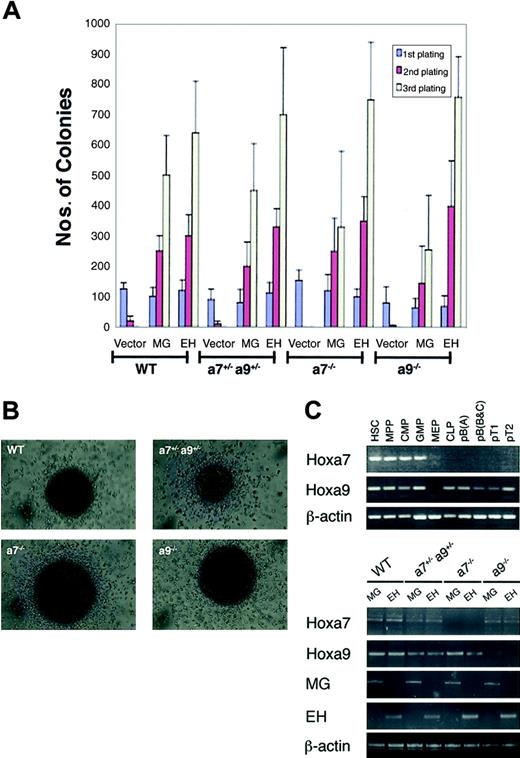
To investigate the functional requirements for Hoxa7 and Hoxa9 in MLL-mediated leukemogenesis, hematopoietic progenitor transformation assays (HPTAs) were performed using progenitors harvested from Hoxa7-/-, Hoxa9-/-, Hoxa7+/-Hoxa9+/-, or wild-type mice. Progenitors were transduced with retroviruses encoding MLL-GAS7 or, as a positive control, E2A-HLF, which has previously been shown to mediate Hoxa7- and Hoxa9-independent transformation.39 As expected, E2A-HLF enhanced the replating capacity of hematopoietic progenitors from all mutant and wild-type genotypes (Figure 2A). Conversely, cells transduced with vector alone lost their proliferative capacity in the second round of plating. Similar to E2A-HLF, MLL-GAS7 induced continued replating of hematopoietic progenitors regardless of their genotype (Figure 2A). However, the numbers of colonies induced by MLL-GAS7 in Hoxa7-/- and Hoxa9-/- progenitors were reduced compared with wild-type progenitors, though they displayed similar colony morphologies and cellular phenotypes (positive for c-Kit, Mac-1, Gr-1) (Figure 2A-B and data not shown).
In vitro transformation ofHox-deficient hematopoietic progenitors byMLL-GAS7. (A) HPTAs using various MSCV retroviruses encoding MLL-GAS7 (MG), E2A-HLF (HF), or vector alone were performed on progenitors harvested from wild-type, Hoxa7-/-, Hoxa9-/-, and Hoxa7+/-Hoxa9+/- mice. Bar charts represent the average number of colonies obtained in each round of methylcellulose plating in 3 different independent assays. Error bars indicate SD. (B) Blast-type morphology of colonies induced by MLL-GAS7 in progenitors derived from different Hox-deficient genetic backgrounds. (C) RT-PCR analysis of Hoxa7 and Hoxa9 expression in normal and transformed hematopoietic cells. (Top) Differential expression of Hox genes in highly purified populations of hematopoietic stem cells and progenitors. (Bottom) Appropriate expression of Hox and fusion transcripts in transformed cells. Transcripts are listed to the left; progenitor genotypes and transduced genes are indicated above the lanes.
In vitro transformation ofHox-deficient hematopoietic progenitors byMLL-GAS7. (A) HPTAs using various MSCV retroviruses encoding MLL-GAS7 (MG), E2A-HLF (HF), or vector alone were performed on progenitors harvested from wild-type, Hoxa7-/-, Hoxa9-/-, and Hoxa7+/-Hoxa9+/- mice. Bar charts represent the average number of colonies obtained in each round of methylcellulose plating in 3 different independent assays. Error bars indicate SD. (B) Blast-type morphology of colonies induced by MLL-GAS7 in progenitors derived from different Hox-deficient genetic backgrounds. (C) RT-PCR analysis of Hoxa7 and Hoxa9 expression in normal and transformed hematopoietic cells. (Top) Differential expression of Hox genes in highly purified populations of hematopoietic stem cells and progenitors. (Bottom) Appropriate expression of Hox and fusion transcripts in transformed cells. Transcripts are listed to the left; progenitor genotypes and transduced genes are indicated above the lanes.
RT-PCR analysis demonstrated that though Hoxa7 and Hoxa9 were expressed in wild-type HSCs, CMPs, and GMPs, only Hoxa9 was expressed in lymphoid progenitors, including CLP, pro-B, and pro-T cells (Figure 2C). These results using highly purified subpopulations of hematopoietic progenitors are consistent with, but significantly extend, previous studies, which showed preferential Hoxa9 expression in Sca+/Lin- cells potentially enriched in HSCs but not in Sca-/Lin+ cells.40 Interestingly, neither Hox gene was expressed in wild-type MEPs though a very weak Hoxa7 signal was detected in 1 of 3 MEP samples when using 38 PCR cycles (Figure 2C and data not shown). High-level expression of Hoxa7 and Hoxa9 was detected in MLL-GAS7-transformed cells originating from wild-type bone marrow, and somewhat lower levels were expressed in Hoxa7+/-Hoxa9+/- transformed cells (Figure 2C). Transformed Hox null progenitors lacked expression of the respective Hoxa7 or Hoxa9 genes. Thus, early hematopoietic progenitors, which are likely targets for transformation by MLL oncogenes,35 express Hoxa7 and Hoxa9, which are maintained in wild-type cells transformed by MLL-GAS7.
MLL-GAS7-transformed Hoxa7-/- progenitors induce AML with longer disease latency and reduced penetrance
To assess their leukemogenic potentials, cells transformed in vitro by MLL-GAS7 (wild-type or Hoxa7-/-) were expanded in liquid cultures and injected into sublethally irradiated syngeneic mice. All mice injected with wild-type cells transformed by MLL-GAS7 succumbed to leukemia within 6 months. Conversely, only half the cohort injected with Hoxa7-/- cells transformed by MLL-GAS7 developed leukemia in the same time frame (Figure 3A; Table 3) with a longer mean latency (138 vs 110 days). Immunophenotypic and histologic analyses demonstrated that the leukemias from both genotypes had features of AML, with extensive extramedullary involvement of the liver. However, leukemias arising from Hoxa7-/- cells typically expressed higher levels of c-Kit–and exhibited less mature cellular morphologies (Figure 3B-C).
Hoxa7-/- cells transformed by MLL-GAS7 induce AML in mice.(A) Survival curves are shown for cohorts (n = 10) of mice injected with MLL-GAS7-transformed cells derived from wild-type (red) and Hoxa7-/- (blue) bone marrow or mock injected controls (black). (B) Phenotypic analysis of leukemic blasts resulting from MLL-GAS7 transduction of wild-type and Hoxa7-/- progenitors. Blue profiles represent FACS staining obtained with antibodies specific for the indicated cell surface antigens. Red profiles represent unstained controls or staining obtained with isotype control antibodies. (C) Histologic analysis of tissues from mice injected with MLL-GAS7-transformed wild-type or Hoxa7-/- cells. Original magnifications for peripheral blood and livers × 1200 and × 40, respectively.
Hoxa7-/- cells transformed by MLL-GAS7 induce AML in mice.(A) Survival curves are shown for cohorts (n = 10) of mice injected with MLL-GAS7-transformed cells derived from wild-type (red) and Hoxa7-/- (blue) bone marrow or mock injected controls (black). (B) Phenotypic analysis of leukemic blasts resulting from MLL-GAS7 transduction of wild-type and Hoxa7-/- progenitors. Blue profiles represent FACS staining obtained with antibodies specific for the indicated cell surface antigens. Red profiles represent unstained controls or staining obtained with isotype control antibodies. (C) Histologic analysis of tissues from mice injected with MLL-GAS7-transformed wild-type or Hoxa7-/- cells. Original magnifications for peripheral blood and livers × 1200 and × 40, respectively.
Features of AML induced by normal or Hoxa7−/− progenitors transformed by MLL-GAS7
. | Control . | Wild-type . | Hoxa7−/− . |
|---|---|---|---|
| Latency, d | NA | 110 ± 33 | 138 ± 60 |
| WBC, ×109/L | 7.5 ± 1.5 | 87 ± 62 | 60 ± 43 |
| Blasts in PB, % | 0 ± 0 | 43 ± 19 | 25 ± 12 |
| Spleen weight, g | 0.22 ± 0.08 | 1.3 ± 0.31 | 0.9 ± 0.42 |
| Liver weight, g | 0.51 ± 0.12 | 1.64 ± 0.51 | 1.35 ± 0.48 |
. | Control . | Wild-type . | Hoxa7−/− . |
|---|---|---|---|
| Latency, d | NA | 110 ± 33 | 138 ± 60 |
| WBC, ×109/L | 7.5 ± 1.5 | 87 ± 62 | 60 ± 43 |
| Blasts in PB, % | 0 ± 0 | 43 ± 19 | 25 ± 12 |
| Spleen weight, g | 0.22 ± 0.08 | 1.3 ± 0.31 | 0.9 ± 0.42 |
| Liver weight, g | 0.51 ± 0.12 | 1.64 ± 0.51 | 1.35 ± 0.48 |
Values shown are the mean ± SD from wild-type (n = 10) and Hoxa7−/− (n = 6) mice at the time of killing. PB indicates peripheral blood; NA, not applicable.
Hoxa7 and Hoxa9 are not required for the induction of leukemias by MLL-GAS7
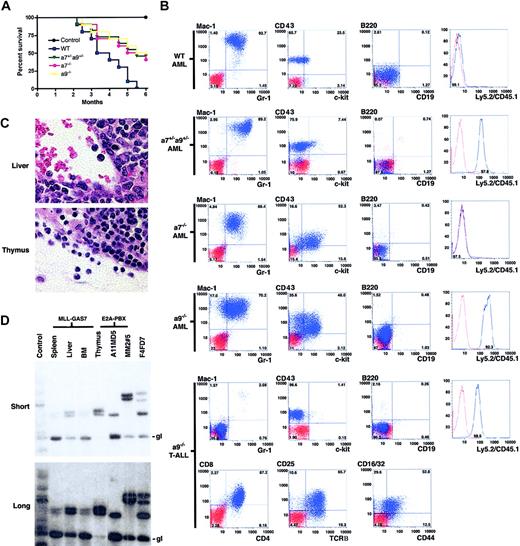
Because in vitro culture and expansion of transformed cells can allow rapid acquisition of secondary mutations that influence the in vivo oncogenic readout, MLL-GAS7-transduced progenitors were directly transplanted into lethally irradiated syngeneic mice to assess the requirements of individual Hox genes in MLL-mediated leukemogenesis. Consistent with previous studies, wild-type progenitors transduced with MLL-GAS7 induced acute leukemias in all recipients within 6 months (Figure 4A). Conversely, only half the cohort that underwent transplantation with MLL-GAS7-transduced cells derived from Hoxa7-/- or Hoxa9-/- bone marrow developed leukemias in the same time frame. Mice that underwent transplantation with MLL-GAS7-transduced cells derived from Hoxa7+/-Hoxa9+/- bone marrow also developed leukemias with disease latencies similar to those of Hox null cells. Although the results suggest a possible genetic interplay between these 2 Hoxa genes, further studies are necessary to validate this possibility. Donor origins of the leukemic cells were confirmed by their expression of Ly5 (CD45) congenic markers in which wild-type and Hoxa7-/- cells were Ly5.1, Hoxa9-/- cells were Ly5.2, and Hoxa7+/-Hoxa9+/- cells were Ly5.1/Ly5.2 (Figure 4B). Leukemias derived from the different genetic backgrounds expressed similar cell surface markers (positive for Mac-1/Gr-1/CD43/c-Kit; negative for lymphoid markers B220, CD19, CD3, CD4, and CD8), except that the leukemic cells derived from Hoxa7-/- or Hoxa9-/- progenitors exhibited higher levels of c-Kit expression compared with those derived from wild-type bone marrow (Figure 4B and data not shown).
Absence of Hoxa7 or Hoxa9 influences the latency, penetrance, and phenotype of MLL-GAS7 AML. (A) Survival curves are shown for cohorts (n = 10) of mice that received transplanted MLL-GAS7-transduced c-Kit+ bone marrow cells derived from wild-type (blue), Hoxa7+/-Hoxa9+/- (green), Hoxa7-/- (pink), or Hoxa9-/- (yellow) mice, or nontransduced wild-type bone marrow cells (black). (B) Typical phenotypic analysis of leukemic blasts from mice that underwent transplantation with transduced wild-type (n = 6), Hoxa7+/-Hoxa9+/- (n = 4), Hoxa7-/- (n = 4), and Hoxa9-/- (n = 4) progenitors. Blue profiles represent FACS staining obtained with antibodies specific for the indicated cell surface antigens. Red profiles represent unstained controls or staining obtained with isotype control antibodies. (C) Histologic analysis of tissues from a mouse that developed T-ALL after transplantation with MLL-GAS7-transduced Hoxa9-/- cells. Original magnification × 1200. (D) TCR gene rearrangement studies of tissues from mice with T-ALL. Thymocytes from normal and E2A-PBX1 transgenic mice were used as negative and positive controls, respectively. Numbers above the positive control lanes refer to sample numbers of the E2A-PBX1 transgenic mice.36 Short and long exposures are shown in upper and lower panels, respectively. gl indicates germline configuration of the TCRβ gene.
Absence of Hoxa7 or Hoxa9 influences the latency, penetrance, and phenotype of MLL-GAS7 AML. (A) Survival curves are shown for cohorts (n = 10) of mice that received transplanted MLL-GAS7-transduced c-Kit+ bone marrow cells derived from wild-type (blue), Hoxa7+/-Hoxa9+/- (green), Hoxa7-/- (pink), or Hoxa9-/- (yellow) mice, or nontransduced wild-type bone marrow cells (black). (B) Typical phenotypic analysis of leukemic blasts from mice that underwent transplantation with transduced wild-type (n = 6), Hoxa7+/-Hoxa9+/- (n = 4), Hoxa7-/- (n = 4), and Hoxa9-/- (n = 4) progenitors. Blue profiles represent FACS staining obtained with antibodies specific for the indicated cell surface antigens. Red profiles represent unstained controls or staining obtained with isotype control antibodies. (C) Histologic analysis of tissues from a mouse that developed T-ALL after transplantation with MLL-GAS7-transduced Hoxa9-/- cells. Original magnification × 1200. (D) TCR gene rearrangement studies of tissues from mice with T-ALL. Thymocytes from normal and E2A-PBX1 transgenic mice were used as negative and positive controls, respectively. Numbers above the positive control lanes refer to sample numbers of the E2A-PBX1 transgenic mice.36 Short and long exposures are shown in upper and lower panels, respectively. gl indicates germline configuration of the TCRβ gene.
Although most of the mice that received transplanted Hoxa9-/--transduced cells died of AML, one developed T-cell ALL (T-ALL). The leukemic cells from this mouse were negative for all myeloid and B-lymphoid antigens tested (Mac-1/Gr-1/B220/CD19) but were positive for various T-lymphoid markers (CD4/CD8/CD25/CD44/TCRβ), suggesting a CD4/CD8 double-positive type of T-ALL (Figure 4B). The morphology of the blasts in the thymus and liver was also consistent with lymphoid origin (Figure 4C). Southern blot analysis showed identical TCRβ gene rearrangements in DNA isolated from tissues involved by leukemia (bone marrow, thymus, liver, and spleen) (Figure 4C), confirming the clonal origin of the leukemia cells at all sites. Taken together, these studies indicate that Hoxa7 and Hoxa9 are not necessary for the induction of leukemias by MLL-GAS7, but they influence disease latency, penetrance, and phenotype.
Discussion
Colinear and differential expression of Hox genes is required for the proper development of hematopoietic cells.10,11 Our current studies, which focused on early hematopoietic stem/progenitor cell compartments, identified significant reductions in phenotypically defined progenitor populations in 2 different lines of Hoxa-deficient mice. Both Hoxa7- and Hoxa9-deficient mice have apparent normal numbers of HSCs but display distinct defects in various hematopoietic progenitor compartments, suggesting that each of these Hox genes is required for different developmental stages of normal hematopoiesis. Hoxa9-/- mice have combined defects in multiple hematopoietic lineages, starting from very early lymphoid and myeloid progenitors (eg, CLPs, CMPs, and GMPs) to more committed lymphoid precursors (pro- and pre-B cells, pro-T cells). Our observations are consistent with previous studies of Hoxa9-deficient mice that revealed functional deficits and hematologic abnormalities in the myeloid and lymphoid lineages.37,38 Conversely, Hoxa7-/- mice have normal numbers of most early progenitors (CMPs, CLPs, GMPs) except MEPs and do not have obvious defects in the committed lymphoid compartments analyzed. These results suggest that Hoxa7 has a more specific role in megakaryocyte and erythrocyte development, whereas Hoxa9 is required for myeloid and lymphoid development initiated from CMPs/CLPs or even earlier. Although our studies did not address the possible functional interplay among Hox genes in the same paralogous group (eg, Hoxb7) that may complement the phenotypes observed in these Hox-deficient mice (Hoxa7-/-),25,26 identification of distinct progenitor reductions in Hoxa7-/- and Hoxa9-/- mice suggests that neighboring Hox genes within the same cluster may have specialized but not overlapping functions in normal hematopoiesis.
Both Hoxa9 and Hoxa7 have been directly implicated in myeloid leukemogenesis, but their roles in MLL-mediated leukemogenesis have not been extensively studied. Using a somatic model of leukemogenesis based on retroviral transduction/transplantation of c-Kit–enriched hematopoietic progenitors, we have demonstrated that neither of these Hoxa genes is absolutely required for MLL-GAS7–mediated oncogenesis. MLL-GAS7 is capable of transforming Hox-deficient progenitors in vitro and inducing leukemias in vivo. However, Hox-deficient cells transformed by MLL-GAS7 are less efficient in forming blast CFUs in vitro and are partially compromised in their ability to induce leukemia in vivo. Thus, these Hox genes appear to serve rate-limiting roles in MLL-mediated transformation likely as downstream transcriptional targets of the MLL oncogenic fusion proteins, consistent with direct binding of the Hoxa9 promoter by wild-type MLL and its chimeric fusion proteins.41,42
The extent of dependence on Hoxa9, however, differs from a recent study of MLL-ENL, which displayed a more severe compromise of in vitro transformation and a complete inability to induce leukemias in Hoxa9-/- progenitors.39 Quantitative differences in Hox dependence may be attributable to different compositions of stem/progenitor populations used for transduction/transplantation experiments. The current study enriched for stem/progenitor cells by positive selection for c-Kit, which is broadly expressed on hematopoietic stem and progenitor cells. Conversely, the previous MLL-ENL study negatively selected against cells expressing lineage-associated antigens after treatment with 5-fluorouracil (5-FU), which up-regulates the expression of certain lineage markers, such as Mac-1, on hematopoietic stem/progenitor cells, known target cells for MLL-mediated transformation.35 Thus, inadvertent depletion of these cellular subpopulations before transduction may influence the transformation readouts. This hypothesis is consistent with our observations that MLL-ENL enhances the in vitro self-renewal of Hoxa7-/- or Hoxa9-/- hematopoietic progenitors/stem cells purified by positive selection for c-Kit expression (C.W.S. and M.L.C., unpublished observations, January 2003). Alternatively, differences in the transformation mechanisms of MLL oncoproteins, which are determined by the specific fusion partner, may influence Hox dependence. ENL is a component of the EBAF chromatin–remodeling complex43 and, in fusion with MLL, may tether EBAF to MLL target genes. Conversely, the oncogenic properties of MLL-GAS7 are critically dependent on the coiled-coil domain of GAS7, which lacks inherent transcriptional effector properties but facilitates oligomerization of the amino portion of MLL. Fusion proteins such as MLL-ENL are presumed to bind DNA as monomers, whereas the multiple DNA-binding domains of forced MLL oligomers may allow interactions with a broader spectrum of downstream target genes. If this is the case, MLL-GAS7 may have greater flexibility for finding alternative targets if one of the Hox genes is absent. Further studies are needed to distinguish between these and other potential factors that influence Hox-dependent initiation of acute myeloid leukemias by the vast array of MLL fusion proteins.
T-ALL associated with MLL mutations is uncommon and has not been successfully modeled by retroviral transduction/transplantation approaches. T-ALL was observed in MLL-LacZ knock-in mice, which contained LacZ artificially fused with MLL.44 However, β-galactosidase is not a bona fide fusion partner for MLL in human leukemias, and the T-ALLs reported were of mixed B- and T-cell lineage carrying both TCR and IG gene rearrangements, which differ from the human T-ALLs associated with MLL rearrangement.6 Conversely, though only one case has been described, the T-ALL induced by MLL-GAS7 in Hoxa9-/- progenitors shares several features with the corresponding human T-ALL. Each expresses early and late T-cell surface markers and displays rearranged TCR genes. Neither expresses B lymphoid or myeloid markers, in contrast to infant B-ALL.6 The lack of other lineage markers in the leukemic T cells suggests that they are arrested at a late stage of T-cell development (after the pro-T2 stage).28,32 Although the mechanistic role of Hoxa9 in T-ALL is unclear, recent studies reveal that the sequential loss of 3′ region Hoxa cluster genes, including Hoxa9, is closely associated with thymocyte development.45 Hoxa9 expression disappeared at the CD4/CD8 double-positive stage, which is also the stage at which MLL-GAS7-transduced Hoxa9-/- cells are arrested. These results further suggest that Hoxa9 may play a role in determining the phenotypes of MLL-mediated leukemias.
Taken together, our results indicate that Hoxa7 and Hoxa9 are not essential for MLL-mediated leukemogenesis, but they appear to influence the latency, penetrance, and phenotype of disease, consistent with their serving important roles as downstream mediators in the MLL leukemogenic pathway. Given the efficient induction of a leukemic phenotype in Hox-deficient cells by an MLL oncoprotein, our results suggest that targeting an individual Hox gene may not be sufficient to inhibit MLL-mediated leukemogenesis. Future experiments delineating the roles of multiple Hox genes and common Hox-cofactors (eg, Meis1) in MLL-mediated leukemogenesis will provide further insight into these issues.
Prepublished online as Blood First Edition Paper, December 30, 2003; DOI 10.1182/blood-2003-10-3722.
Supported by the National Institutes of Health (grant CA55209) and the Children's Health Initiative. C.W.S. is a special fellow of the Leukemia and Lymphoma Society. H.K. is a fellow of the Ernst Schering Research Foundation. P.W. is a scholar of the Croucher Foundation.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Cita Nicolas, Maria Ambrus, and Dustin M. Ray for excellent technical support and Caroline Tudor and Erica Tse for professional artwork.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal