Abstract
The chemokine stromal-derived factor-1 (SDF-1), which is constitutively expressed in most tissues as SDF-1α and SDF-1β resulting from alternative gene splicing, regulates hematopoiesis, lymphocyte homing, B-lineage cell growth, and angiogenesis. Because SDF-1α and SDF-1β are constitutively and ubiquitously expressed, their degradation must serve an important regulatory role. Here we show that SDF-1α and SDF-1β are secreted as full-length molecules. When exposed to human serum, full-length SDF-1α (1-68) undergoes processing first at the COOH terminus to produce SDF-1α 1-67 and then at the NH2 terminus to produce SDF-1α 3-67. By contrast, full-length SDF-1β (1-72) is processed only at the NH2 terminus to produce SDF-1β 3-72. CD26/dipeptidyl peptidase is responsible for serum cleavage of SDF-1α and SDF-1β at the NH2 terminus. Serum processing of SDF-1α at the COOH terminus, which has not been previously reported, reduces the ability of the polypeptide to bind to heparin and to cells and to stimulate B-cell proliferation and chemotaxis. The additional processing at the NH2 terminus renders both forms of SDF-1 unable to bind to heparin and to activate cells. The differential processing of SDF-1α and SDF-1β provides biologic significance to the existence of 2 splice forms of the chemokine and adds a tool to precisely regulate SDF-1's biologic activity by changes in specific activity.
Introduction
The chemokine stromal-derived factor-1 (SDF-1) is a key regulator of physiological cell motility during embryogenesis and after birth, and it has been implicated in pathological cell motility associated with tumor metastasis.1-6 The biologic effects of SDF-1 are mediated by a specific G protein–coupled receptor, CXCR4, which has also been shown to serve as a coreceptor for T-cell tropic HIV-1.7-13 SDF-1 is constitutively expressed in stromal cells, endothelial cells, dendritic cells, and other cells.1,4,14-16 On the basis of sequence analysis, SDF-1 is remarkably conserved among species. A single conservative substitution at position 18 (I → V) distinguishes human and murine SDF-1.17 Two splice forms of SDF-1 have been identified, SDF-1α and SDF-1β, which have identical amino acid sequences except for the presence of 4 additional amino acids at the carboxy terminus of SDF-1β.1,17 The significance of the existence of 2 splice forms of SDF-1 has remained unclear.
Structure-function analysis of SDF-1α (1-67) has identified the NH2-terminal amino acids (residues 1-8) as critical to CXCR4 binding and activation. Modification of the first 2 amino acids (K-1 and P-2) alone resulted in loss of receptor activation, and deletion of the first 8 amino acids resulted in loss of receptor binding activity.18 However, the NH2-terminus alone, which is a highly mobile region of SDF-1α, was found to be insufficient for receptor binding and activation, and an additional site consisting of a RFFESH motif (residues 12-17) was identified as necessary for SDF-1α docking to CXCR4.18,19 Furthermore, the cluster of basic residues K-24, H-25, K-27, and R-41 was proposed to provide surface charge complementarity for the negatively charged extracellular portion of CXCR419-21 and to contribute to a heparan sulfate binding site anchoring SDF-1α (1-67) to cell surface proteoglycans.22
Recently, proteolytic degradation of endogenous SDF-1 in the bone marrow was identified as playing a critical role in mobilization of hematopoietic progenitor cells to the peripheral circulation.23,24 Endogenous SDF-1 provides a retention signal for hematopoietic stem and progenitor cells, which express CXCR4,25 such that its local degradation would release the cells from this site. In vitro, SDF-1α can be enzymatically cleaved by metalloproteinases, CD26/dipeptidyl peptidase IV, serine proteases, and leukocyte elastase to generate distinct N-terminally truncated forms of the molecule.26-29 However, it is unclear whether these enzymes play a role in the physiologic processing of SDF-1 in vivo.
In vitro, SDF-1α can inhibit T-cell tropic HIV-1 infection of target cells acting as a competitor for CXCR4-mediated viral entry.9,10 Recently, we found endogenous SDF-1 to be inactive in normal adult serum, and this could explain why endogenous SDF-1 is generally ineffective as a natural anti–HIV-1 agent.30 Here, we examined SDF-1 inactivation further. These studies reveal that serum enzymes can selectively cleave SDF-1α both at the carboxy and NH2 terminus, and SDF-1β at the NH2 terminus only, generating molecules with differing specific activity. The different sensitivity of SDF-1α and SDF-1β to proteolytic processing provides a mechanism for chemokine functional regulation and reveals a functional difference between the 2 splice forms of the chemokine.
Materials and methods
Reagents
Recombinant SDF-1α and SDF-1β were from R&D Systems (Minneapolis, MN). Synthetic forms of SDF-1α lacking the C-terminal lysine (amino acids 1-67) or the C-terminal lysine (K) plus the 2 NH2-terminal amino acids lysine (K) and proline (P) (amino acids 3-67) were from Upstate Biotechnology (Lake Placid, NY) or from Dr I. Clark-Lewis (University of British Columbia, Vancouver, Canada). SDF-1 protein concentration in phosphate-buffered saline (PBS) was verified by optic density (OD) measurements at 280 nm and the extinction coefficient of 8200. Antigen-affinity purified goat antihuman SDF-1α and SDF-1β (BAF 310), antigen-affinity purified goat antihuman SDF-1β (BAF 311), mouse immunoglobulin G1 (IgG1) control (hybridoma clone 11711.11), and mouse IgG1 monoclonal antihuman/mouse SDF-1 antibodies (MAB 350 and MAB 310) were from R&D Systems; rabbit antihuman SDF-1 antigen-affinity purified antibodies were from PeproTech (Rocky Hill, NJ). The CD26/dipeptidyl peptidase inhibitor AB19231 was a gift from Dr A. M. Lambeir (University of Antwerp, Belgium). Heparin (porcine, intestinal mucosa) was from Sigma Chemical (St Louis, MO).
Cells, flow cytometry, calcium flux, cell proliferation, and chemotaxis
Primary human umbilical vein endothelial cells (HUVECs) were prepared from umbilical cord and propagated through passage 5 as previously described.32 The human Jurkat and BL-41 cell lines were cultured in RPMI 1640 (Gibco-Invitrogen, Grand Island, NY) medium supplemented with 10% fetal bovine serum (Biofluids, Rockville, MD). The murine bone marrow stromal cell line MS-5,33 a gift from Dr A. Berardi, Ospedale Bambin Gesù, Rome, Italy) was cultured in α-minimum essential medium (α-MEM) (Gibco-Invitrogen) supplemented with 5% fetal bovine serum (Biofluids, Rockville, MD) and 2 × 10-2 M 2-mercaptoethanol. Conditioned medium was prepared in serum-free Opti-MEM (Gibco-Invitrogen) over 24 hours of incubation. Flow cytometry was performed as described previously.16 Briefly, HUVECs (passage 4) were incubated with SDF-1α, SDF-1β, synthetic SDF-1 (1-67 or 3-67), or recombinant SDF-1α that had been treated (10 minutes or 20 hours) with 10% human serum. For detection of surface SDF-1, cells were stained with mouse monoclonal anti–SDF-1 antibody (clone MAB 350, R&D Systems; 5 μg/mL for 45 minutes at 4°C) followed by a phycoerythrin (PE)–labeled goat antimouse F(ab′)2 fragment (30 minutes at 4°C; Jackson ImmunoResearch, West Grove, PA). Calcium flux was detected using a ratio fluorescence spectrometer (Photon Technology International, South Brunswick, NJ).34 For proliferation assays, the murine DW34 cells,35 (a gift from Dr P. Kincade, Oklahoma Medical Research Foundation, Oklahoma City), were cultured in triplicate at 2 × 104 cells per well in RPMI 1640 medium containing 10% fetal calf serum and 50 μM 2-mercaptoethanol in 96-well flat-bottom plates for 30 hours at 37°C.35 Cultures were pulsed with 0.5 μCi (5.0 × 104 Bq) thymidine (Perkin-Elmer, Boston, MA) for 6 hours, cells harvested, and radioactivity measured. For chemotaxis assays,36 the human Burkitt lymphoma cell line BL-41 (0.5 × 106 cells per transwell) was incubated (RPMI 1640 medium containing 10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] buffer and 0.5% bovine serum albumin [BSA]) in the upper chamber of transwells (5-μm pore size; Costar, Cambridge, MA) for 4 hours at 37°C.
Immunoprecipitation, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting
Cell lysates were prepared in lysis buffer (10 mM Tris [tris(hydroxymethyl) aminomethane] [pH 7.4], 0.5% Nonidet P-40 [NP-40], 150 mM NaCl, and protease inhibitor cocktail) (Roche Complete; Roche Diagnostic, Indianapolis, IN) and dilution (1:2) in tricine sodium dodecyl sulfate (SDS) sample buffer (Novex, San Diego, CA). Immunoprecipitation, performed as described,30 utilized mouse monoclonal anti–SDF-1 antibody (10 μg/mL, MAB 310; R&D Systems) followed by addition of protein G beads to the antibody/protein mixture. Immunoblotting, performed as described,30 utilized rabbit antihuman SDF-1α antigen-affinity purified polyclonal antibody (PeproTech), goat IgG antihuman SDF-1α antigen-affinity purified antibody (BAF 310; R&D Systems), or goat IgG antihuman SDF-1β antigen-affinity purified antibody (BAF 310; R&D Systems).
SDF-1–heparin-binding assays
Porcine intestinal mucosa heparin (Sigma Chemical) suspended in 0.1 M MES (2-N-morpholinoethanesulfonic acid [pH 5.5]) at 1810 U/mL was reacted at room temperature with 1.25 mM biotin LC hydrazide (Pierce Biotechnology, Rockford, IL) and 1.25 mM EDC (1-ethyl-3-(3-dimethylaminopropryl)carbodiimide hydrochloride; Pierce Biotechnology) for 18 hours. The mixture was extensively dialyzed against PBS to remove unreacted biotin. Biotinylated heparin (1.8 U/mL in PBS) was injected onto the flow cell of the Sensor Chip SA (Biacore, Uppsala, Sweden) that is coated with streptavidin; 375 resonance units (RUs) of biotinylated heparin were immobilized. The flow cell was then conditioned with several injections of 1.5 M NaCl. SDF-1–heparin-binding assays were performed by surface plasmon resonance using a Biacore 3000 system (Biacore). Test samples were diluted in HEPES buffer saline containing 3 mM EDTA (ethylenediaminetetraacetic acid) and 0.005% Surfactant P20 (HBS-EP, Biacore) maintained at 25°C and injected over the heparin-coated or control flow cell surfaces at a flow rate of 50 μL/min. Association and dissociation phases were evaluated over 2 minutes. To evaluate the dissociation phase, the formed complexes were washed with HBS-EP (Biacore) at a flow rate of 50 μL/min over 2 minutes. Sensor chips were regenerated with 3 time pulses of 1.5 M NaCl for 30 seconds. Kinetic constants were obtained from the sensorgrams by fitting the data using BIAevaluation software (Biacore). Dissociation constants (Kd) were calculated from the ratio of dissociation and association rate constants (Kd = koff/kon).
SDF-1 evaluation by HPLC-ESI-MS
Intact and truncated forms of SDF-1α and SDF-1β were analyzed by high-performance liquid chromatography electrospray ionization mass spectrometry (HPLC-ESI-MS). Capillary HPLC was performed using a MAGIC 2002 binary gradient pump (Michrom BioResources, Auburn, CA) operating at 40 μL/min. A Michrom capillary precolumn splitter assembly reduced the flow being delivered to a C-18 capillary column (BetaBasic-18, 100 × 0.32 mm, 3 μm; ThermoHypersil-Keystone Scientific, Bellefonte, PA) to approximately 2 μL/min. Solvent A was HPLC grade water with 0.1% formic acid, and solvent B was HPLC grade acetonitrile with 0.1% formic acid. Five microliters of sample solution was injected into a Michrom Cap-Trap (Michrom BioResources), washed with 20 μL HPLC grade water, and eluted onto the column by the following gradient: 10% B (0.00 minutes) → 10% B (5.00 minutes) → 50% B (50 minutes) → 100% B (75 minutes) → 100% B (100 minutes). The eluent was introduced into a Finnigan LCQ ion trap mass spectrometer (ThermoQuest, San Jose, CA) by a modified nanospray device for mass analysis.37 No sheath or auxiliary gases were used. The temperature of the heated capillary was set at 200°C. The spray voltage was 2.5 kV. The scan range was set from 150 to 2000 m/z for full-scan mode. Centroid data were collected and interpreted using the Xcalibur software package (Thermo Finnigan, San Jose, CA). Masses of the intact and truncated forms of SDF-1α and SDF-1β were determined using the BIOMASS deconvolution algorithm.
Statistical analysis
Data were compared by the Student t test; P values less than .05 were considered significant.
Results
Comparison of SDF-1 produced by MS-5 cells and SDF-1 present in serum
We looked for SDF-1α and SDF-1β in cell lysates and culture supernatants of MS-5 stromal cells. By immunoprecipitation followed by immunoblotting with antibodies that recognize both SDF-1α and SDF-1β or antibodies that preferentially recognize SDF-1β, specific bands corresponding in size to full-length SDF-1α and SDF-1β are identified in MS-5 cell lysates and culture supernatants (Figure 1A), suggesting that MS-5 stromal cells produce full-length SDF-1α and SDF-1β. By the same method, we identified SDF-1–related bands in human sera (Figure 1B). Notably, these bands appeared to be of somewhat lower molecular weight relative to the full-length recombinant SDF-1α and SDF-1β molecules used as controls, suggesting the occurrence of serum cleavage. To test for this possibility, recombinant SDF-1α (2 μg in 40 μL PBS) and SDF-1β (2 μg in 40 μL PBS) were incubated (20 hours at room temperature) either alone or with human serum (4 μL in 40 μL PBS containing SDF-1) and subsequently analyzed by mass spectrometry. SDF-1α incubated for 20 hours in serum revealed a relative molecular mass of 7606 Da (average [av.]), which is 353 lower than that of the control SDF-1α (7959 Da. [av.]; residues 1-68, calculated 7959 [average for 4 oxidized cysteines]) and is consistent with the recovery of SDF-1α encompassing residues 3-67, reflecting the loss of 2 residues (KP, 225 Da) at the NH2 terminus and of 1 residue (K, 128 Da) at the carboxy terminus (Figure 2B). In addition, mass spectrometry analysis of SDF-1β incubated for 20 hours in serum revealed a relative molecular mass of 8297 Da (av.), which is lower than that of the control SDF-1β (mass 8522 Da [av.]; residues 1-72) and is consistent with the recovery of a truncated SDF-1β species encompassing residues 3-72, reflecting the loss of 2 NH2 terminal residues (KP) (Figure 2C).
Detection of SDF-1α and SDF-1β in MS-5 cells and in serum. (A) Culture supernatants from MS-5 cells produced in serum-free medium (8 mL), control serum-free medium (8 mL), cell lysates of MS-5 cells (1 × 106 cell equivalents), recombinant human SDF-1α (20 ng), and SDF-1β (20 ng) were subjected to immunoprecipitation with anti–SDF-1α plus anti–SDF-1β antibodies and immunoblotted with antibodies that recognize both SDF-1α and SDF-1β or antibodies that preferentially recognize SDF-1β. (B) Aliquots of recombinant SDF-1α and SDF-1β and serum from 2 healthy individuals (a and b; precleared with protein G) were subjected to immunoprecipitation with anti–SDF-1α plus anti–SDF-1β antibodies and the immunoprecipitates immunoblotted with antibodies that recognize both SDF-1α and SDF-1β.
Detection of SDF-1α and SDF-1β in MS-5 cells and in serum. (A) Culture supernatants from MS-5 cells produced in serum-free medium (8 mL), control serum-free medium (8 mL), cell lysates of MS-5 cells (1 × 106 cell equivalents), recombinant human SDF-1α (20 ng), and SDF-1β (20 ng) were subjected to immunoprecipitation with anti–SDF-1α plus anti–SDF-1β antibodies and immunoblotted with antibodies that recognize both SDF-1α and SDF-1β or antibodies that preferentially recognize SDF-1β. (B) Aliquots of recombinant SDF-1α and SDF-1β and serum from 2 healthy individuals (a and b; precleared with protein G) were subjected to immunoprecipitation with anti–SDF-1α plus anti–SDF-1β antibodies and the immunoprecipitates immunoblotted with antibodies that recognize both SDF-1α and SDF-1β.
Demonstration of SDF-1α and SDF-1β cleavage by human serum. Recombinant SDF-1α and SDF-1β were incubated for 20 hours at room temperature alone or with 10% normal human serum. (A) Mass spectrometry of recombinant SDF-1α after 20 hours of incubation at room temperature alone or with 10% normal human serum. (B) Mass spectrometry of recombinant SDF-1β after 20 hours of incubation at room temperature alone or with 10% normal human serum. The average molecular mass after deconvolution of the indicated ion peaks (left) is shown on the right. Representative experiments are shown.
Demonstration of SDF-1α and SDF-1β cleavage by human serum. Recombinant SDF-1α and SDF-1β were incubated for 20 hours at room temperature alone or with 10% normal human serum. (A) Mass spectrometry of recombinant SDF-1α after 20 hours of incubation at room temperature alone or with 10% normal human serum. (B) Mass spectrometry of recombinant SDF-1β after 20 hours of incubation at room temperature alone or with 10% normal human serum. The average molecular mass after deconvolution of the indicated ion peaks (left) is shown on the right. Representative experiments are shown.
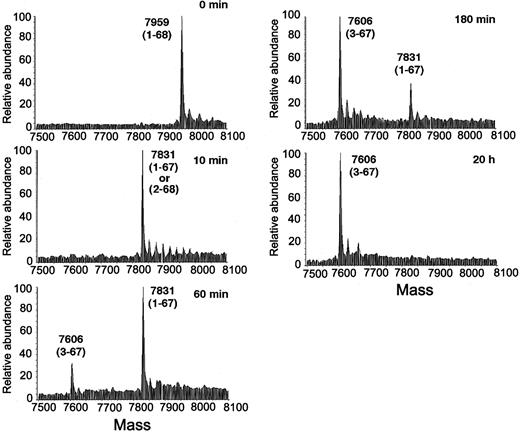
A kinetic analysis of SDF-1α conversion from full-length (1-68) to the cleaved form 3-67 (lacking KP at the NH2 terminus and K at the carboxy terminus) in the presence of serum is shown in Figure 3. At the 10-minute time point, the mass spectrum of SDF-1α shows a mass of 7831 Da (av.), corresponding to the loss of a K at either termini. At the 60-minute time point, in addition to a main SDF-1α–related peak (mass 7831 Da [av.]; loss of 1 K), a smaller peak is noted with a relative mass of 7606 Da (av.), corresponding to the SDF-1α 3-67 species, lacking both KP at the NH2 terminus and K at the carboxy terminus. At the 180-minute and 20-hour time points, SDF-1α has mostly converted to the cleaved form 3-67. These results provide evidence for a 2-step SDF-1α processing by serum resulting in the final cleaved product in which the carboxy-terminal K is rapidly removed followed by the cleavage of the NH2-terminal KP.
Kinetic analysis of SDF-1α cleavage by human serum. Recombinant SDF-1α (1-68) was incubated at room temperature for the indicated times (0 to 20 hours) with 10% human serum and analyzed by mass spectrometry. The average molecular mass after deconvolution of the indicated ion peaks (left) is shown on the right. Representative experiment is shown.
Kinetic analysis of SDF-1α cleavage by human serum. Recombinant SDF-1α (1-68) was incubated at room temperature for the indicated times (0 to 20 hours) with 10% human serum and analyzed by mass spectrometry. The average molecular mass after deconvolution of the indicated ion peaks (left) is shown on the right. Representative experiment is shown.
Characterization of SDF-1 cleavage by serum
CD26/dipeptidyl peptidase IV can cleave dipeptides from the NH2 terminus of polypeptide chains, preferably after a proline or alanine.38 SDF-1α, which possesses the residues KPV- at the NH2 terminus, can be a substrate for CD26/dipeptidyl peptidase IV.26 To test whether CD26/dipeptidyl peptidase IV is responsible for serum cleavage of SDF-1α and SDF-1β, we used AB192, a diaryl phosphonate ester, which is a potent and irreversible specific inhibitor of CD26/dipeptidyl peptidase IV.31 In the presence of 50 μM AB192, most SDF-1α incubated for 20 hours with 10% serum had a mass of 7831 Da (av.), corresponding to SDF-1α 1-67, whereas in the absence of the inhibitor all SDF-1α had a mass of 7606 Da (av.), corresponding to SDF-1 3-67 (Figure 4A). This protection of SDF-1α cleavage by AB192 provides evidence that CD26/dipeptidyl peptidase IV is responsible for serum cleavage of SDF-1α at the NH2 terminus and that the sequence of cleavage events is SDF-1α 1-68 to 1-67 to 3-67.
Characterization of SDF-1α cleavage by serum evaluated by mass spectrometry. (A) Fresh human serum was preincubated for 15 minutes at 37°C with or without the CD26/dipeptidyl peptidase inhibitor AB192 (50 μM), added (10% final dilution) to recombinant SDF-1α, and then incubated at room temperature for 20 hours. (B) Fresh human serum was heated at 56°C for 30 minutes or left at 4°C for 30 minutes, added (10% final dilution) to recombinant SDF-1α, and then incubated at room temperature for 20 hours. (C) Fresh human serum was preincubated at room temperature for 15 minutes with or without 20 mM EDTA, added (10% final dilution) to recombinant SDF-1α, and then incubated at room temperature for 10 minutes.
Characterization of SDF-1α cleavage by serum evaluated by mass spectrometry. (A) Fresh human serum was preincubated for 15 minutes at 37°C with or without the CD26/dipeptidyl peptidase inhibitor AB192 (50 μM), added (10% final dilution) to recombinant SDF-1α, and then incubated at room temperature for 20 hours. (B) Fresh human serum was heated at 56°C for 30 minutes or left at 4°C for 30 minutes, added (10% final dilution) to recombinant SDF-1α, and then incubated at room temperature for 20 hours. (C) Fresh human serum was preincubated at room temperature for 15 minutes with or without 20 mM EDTA, added (10% final dilution) to recombinant SDF-1α, and then incubated at room temperature for 10 minutes.
We tested for the effects of heat on the ability of serum to cleave SDF-1. When SDF-1α was treated for 20 hours with human serum that had been heated (56 °C, 30 minutes), the mass spectra showed the conversion of full-length SDF-1α (mass 7959 Da [av.]; residues 1-68) to a species with relative mass of 7734 Da. (av.), corresponding to SDF-1α 3-68, consistent with the loss of the NH2-terminal residues KP (Figure 4B). Based on these results, we conclude that the serum activity that removes the carboxy-terminal K from SDF-1α is heat sensitive. A comparison of the time course of SDF-1α conversion to the 3-68 amino acids species (in the presence of heat-treated serum) with the time course of SDF-1α conversion to the 3-67 amino acids species (in the presence of fresh serum) revealed minimal differences with respect to the processing at the NH2 terminus (not shown), suggesting that cleavage at the carboxy and NH2 termini is independently regulated.
To test for metal dependency of serum processing of SDF-1 we used EDTA, which was added to fresh serum at 20 mM 15 minutes prior to testing. Serum containing EDTA was then added to recombinant SDF-1α for 10 minutes at room temperature. As shown (Figure 4C), most SDF-1α exposed to serum plus EDTA had a mass of 7959 Da (av.), corresponding to the full-length molecule (1-68). Instead, 10-minute incubation with fresh serum without EDTA reduced SDF-1α to a mass of 7831 Da (av.), corresponding to a loss of 1 K. We therefore conclude that serum truncation of SDF-1α at the carboxy terminus is, at least in part, metal dependent.
Several sera were tested at 10% concentration for their ability to cleave SDF-1α over 20 hours of incubation at room temperature, including 3 normal human sera, 1 serum from an individual who had received granulocyte colony-stimulating factor (G-CSF) for hematopoietic cell mobilization, a bovine serum, a rabbit serum, and a mouse serum. When analyzed by mass spectrometry, all sera were indistinguishable in their ability to process SDF-1α from full-length molecule (1-68) to the cleaved 3-67 species lacking KP at the NH2 terminus and K at the carboxy terminus (not shown). Thus, these sera contain catalytically active CD26/dipeptidyl peptidase IV, which cleaves SDF-1α and SDF-1β at the NH2 terminus to generate chemokines lacking the NH2-terminal peptides KP. Sera also possess the ability to selectively cleave SDF-1α at the carboxy terminus to generate a chemokine lacking the carboxy-terminal K.
Heparin binding by SDF-1 species and effects of heparin on serum SDF-1 processing
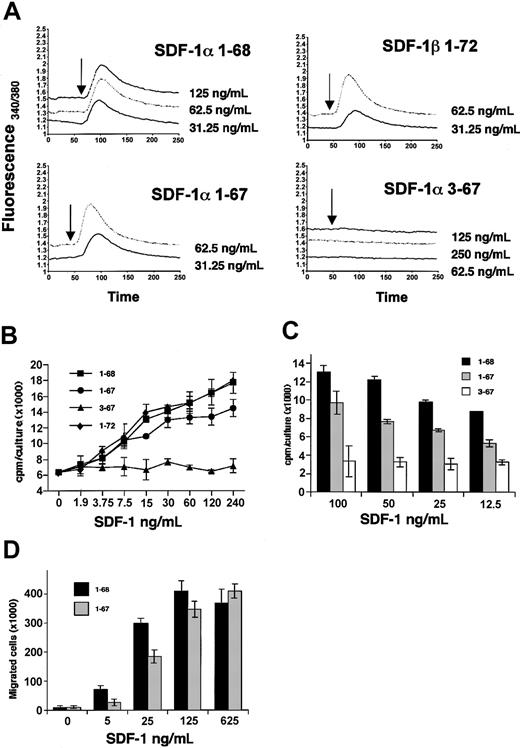
The binding of full-length SDF-1α and SDF-1β to heparin was evaluated by surface plasmon resonance using the Biacore system. In preliminary experiments (not shown), injection of SDF-1α (200 nM) over the heparin-coated sensor chip containing 375 RU heparin gave a signal of 2000 RU, whereas similar injection over the control chip caused minimal signal (not shown). Also, preincubation of SDF-1α with heparin (200 μg/mL) prior to injection over the heparin-activated sensor chip reduced markedly SDF-1α binding, demonstrating the specificity of the SDF-1–heparin interaction. Using this system, full-length SDF-1α and SDF-1β and the SDF-1α 1-67 fragment displayed significant dose-dependent binding to immobilized heparin, whereas SDF-1α 3-67 displayed little binding (Figure 5A). Kinetic constants derived from the sensograms revealed that the dissociation equilibrium constants (Kd = koff/kon) for SDF-1β (1-72) and for SDF-1α (1-68) were similar at 17 and 24 nM, respectively. However, the Kd for SDF-1α 1-67 was measured at 83 nM, which is more than 3-fold higher than that displayed by full-length SDF-1α (1-68), suggesting that the carboxy-terminal K contributes, in part, to heparin binding. Because the Kd for SDF-1α 3-67 was measured at 6.4 μM, this form of SDF-1α has lost most of the heparin-binding capacity.
SDF-1 affinity for heparin and cell surface proteoglycans. (A) Recombinant full-length SDF-1β (1-72) and SDF-1α (1-68) and synthetic SDF-1α 1-67 and 3-67 were injected at various concentrations (200, 100, 50, 25, and 12.5 nM) over a Biacore sensor chip containing streptavidin plus biotinylated heparin. The signal (measured in resonance units [RUs]) was recorded over a 120-second association phase and 120-second dissociation phase. The dissociation equilibrium constants (Kd) are shown for each set of data. Representative results from 3 independent experiments performed are shown. (B) SDF-1α binding to HUVECs evaluated by fluorescence-activated cell sorter (FACS) analysis. Recombinant full-length SDF-1α (1-68) and synthetic SDF-1α 1-67 were added at various concentrations (2000, 1000, and 500 ng) to HUVECs (1 × 106 cells). After incubation (4°C, 30 minutes), the cells were stained for surface SDF-1. A representative experiment of 3 performed is shown. Filled histograms reflect background fluorescence. (C) Recombinant SDF-1α (50 ng) was incubated for 10 minutes in buffer alone or with heparin (50 or 1000 μg/mL) with or without addition of fresh human serum. Lane 1: SDF-1α alone; lane 2: SDF-1α plus 10% human serum; lane 3: SDF-1α plus heparin 50 μg/mL; lane 4: SDF-1α plus heparin 1000 μg/mL; lane 5: SDF-1α plus heparin 50 μg/mL plus 10% human serum; lane 6: SDF-1α plus heparin 1000 μg/mL plus 10% human serum. Samples were immunoblotted with rabbit (top) or goat (bottom) antibodies against SDF-1α and SDF-1β.
SDF-1 affinity for heparin and cell surface proteoglycans. (A) Recombinant full-length SDF-1β (1-72) and SDF-1α (1-68) and synthetic SDF-1α 1-67 and 3-67 were injected at various concentrations (200, 100, 50, 25, and 12.5 nM) over a Biacore sensor chip containing streptavidin plus biotinylated heparin. The signal (measured in resonance units [RUs]) was recorded over a 120-second association phase and 120-second dissociation phase. The dissociation equilibrium constants (Kd) are shown for each set of data. Representative results from 3 independent experiments performed are shown. (B) SDF-1α binding to HUVECs evaluated by fluorescence-activated cell sorter (FACS) analysis. Recombinant full-length SDF-1α (1-68) and synthetic SDF-1α 1-67 were added at various concentrations (2000, 1000, and 500 ng) to HUVECs (1 × 106 cells). After incubation (4°C, 30 minutes), the cells were stained for surface SDF-1. A representative experiment of 3 performed is shown. Filled histograms reflect background fluorescence. (C) Recombinant SDF-1α (50 ng) was incubated for 10 minutes in buffer alone or with heparin (50 or 1000 μg/mL) with or without addition of fresh human serum. Lane 1: SDF-1α alone; lane 2: SDF-1α plus 10% human serum; lane 3: SDF-1α plus heparin 50 μg/mL; lane 4: SDF-1α plus heparin 1000 μg/mL; lane 5: SDF-1α plus heparin 50 μg/mL plus 10% human serum; lane 6: SDF-1α plus heparin 1000 μg/mL plus 10% human serum. Samples were immunoblotted with rabbit (top) or goat (bottom) antibodies against SDF-1α and SDF-1β.
Heparan sulfate proteoglycans, which are structurally related to heparin, are found ubiquitously on cell surfaces.39 We therefore examined whether the different affinities for heparin exhibited by SDF-1 species might correlate with differences in binding to cell surface proteoglycans. Primary human umbilical vein endothelial cells (HUVECs) naturally express low-level SDF-1 on the cell surface detected by specific antibodies (Figure 5B). Incubation (1 hour, 4°C) with full-length SDF-1α (1-68) resulted in a dose-dependent increase of surface SDF-1. This binding of SDF-1α to the cell surface is mostly unrelated to receptor engagement because CXCR4 is saturated by SDF-1α at low nanomolar concentrations.18 Similar results were derived from full-length SDF-1β (not shown). Synthetic SDF-1α 1-67, or SDF-1α 1-67 generated by serum cleavage (not shown), bound less efficiently than the full-length molecule to HUVECs, such that approximately twice as much protein was required to achieve similar cell surface chemokine levels (Figure 5B). However, synthetic SDF-1 3-67 (Figure 5B) or SDF-1 3-67 generated by serum cleavage (not shown) bound poorly to HUVECs even at the highest concentration tested. Thus, the different species of SDF-1 differ in their ability to bind to cell surface.
Because primary endothelial cells naturally express low-level surface SDF-1, which appears to be full length based on its motility on SDS-PAGE and is functional as a regulator of endothelial cell morphogenesis and angiogenesis,16 we examined whether SDF-1 binding to cell surface proteoglycans might affect SDF-1 cleavage by serum. We took advantage of the observation that the rabbit anti–SDF-1α and anti–SDF-1β antibodies (PeproTech) recognize full-length (1-68) SDF-1α but not the truncated SDF-1α species 1-67, whereas the goat anti–SDF-1α and anti–SDF-1β antibodies (R&D Systems) recognize both SDF-1α species equally well.30 Using these antibodies for detection, we examined serum cleavage of SDF-1α in the presence or absence of heparin. SDF-1α (50 ng) in PBS alone (10 μL) or PBS with heparin (1000 or 50 μg/mL) was exposed for 10 minutes to human serum (10%). As expected (Figure 5C), 10 minutes of exposure to fresh serum resulted in cleavage of the carboxy-terminal K from SDF-1α as evidenced by low-level recognition by the rabbit antibody and the slightly increased mobility of the specific band visualized on reprobing with the goat anti–SDF-1 antibodies (Figure 5C, lane 2). In the presence of heparin at the higher (1000 μg/mL) concentration (Figure 5C, lane 6), little SDF-1 cleavage was detected as evidenced by recognition with the rabbit antibodies and relative protein mobility (Figure 5C, compare lanes 2 and 6). Thus, SDF-1 can bind to heparin and presumably heparan sulfate proteoglycans, which can protect the chemokine from carboxy-terminal serum cleavage.
Receptor activation and biologic function of SDF-1 species
Binding of chemokines to their receptors leads to a transient rise in Ca2+ ions. Recombinant full-length SDF-1α and SDF-1β similarly stimulated a dose-dependent rise in Ca2+ ions in the T-cell line Jurkat, which expresses CXCR4 (Figure 6A). The synthetic 1-67 SDF-1α was indistinguishable from the full-length molecule in its ability to cause a dose-dependent rise in Ca2+ ions in the target Jurkat cells, whereas the synthetic 3-67 SDF-1α failed to induce a rise in Ca2+ ions in the target Jurkat cells even at the highest concentration (250 ng/mL). This confirms the critical importance of the NH2-terminal residues KP for SDF-1α receptor activation18 and suggests that the loss of the carboxy-terminal K does not impair SDF-1α's ability to activate the CXCR4 receptor.
Receptor engagement, stimulation of cell proliferation, and chemotaxis by SDF-1 species. (A) Calcium fluxes in fura-2–loaded Jurkat cells in response to varying concentrations of recombinant SDF-1α (1-68) and SDF-1β (1-72) and synthetic SDF-1α (1-67) and SDF-1α 3-67. The results reflect fluorescence measurements as a ratio of excitation at 340 and 380 nm. Arrows indicate chemokine addition. (B) DW34 pre–B-cell proliferation in response to varying concentrations (1.9 to 240 ng/mL) of recombinant full-length SDF-1β (1-72), SDF-1α (1-68), SDF-1α 1-67 (generated by 10 minutes of incubation with 10% fresh human serum at room temperature), and SDF-1α 3-67 (generated by 20 hours of incubation with fresh serum). The results reflect the mean counts per minute (cpm) per culture (± SEM) of triplicate determinations; a representative experiment of 5 performed is shown. (C) DW34 pre–B-cell proliferation in response to varying concentrations (12.5 to 100 ng/mL) of recombinant full-length SDF-1α (1-68), synthetic SDF-1α 1-67, and synthetic SDF-1α 3-67. The results reflect the mean counts per minute per culture (± SEM) of triplicate determinations; a representative experiment is shown. (D) BL-41 B-cell chemotaxic response to recombinant full-length SDF-1α (1-68) and synthetic SDF-1α (1-67) at varying concentrations (5 to 625 ng/mL). The results reflect the mean (± SEM) number of cells that have migrated to the lower chamber in 5 experiments performed.
Receptor engagement, stimulation of cell proliferation, and chemotaxis by SDF-1 species. (A) Calcium fluxes in fura-2–loaded Jurkat cells in response to varying concentrations of recombinant SDF-1α (1-68) and SDF-1β (1-72) and synthetic SDF-1α (1-67) and SDF-1α 3-67. The results reflect fluorescence measurements as a ratio of excitation at 340 and 380 nm. Arrows indicate chemokine addition. (B) DW34 pre–B-cell proliferation in response to varying concentrations (1.9 to 240 ng/mL) of recombinant full-length SDF-1β (1-72), SDF-1α (1-68), SDF-1α 1-67 (generated by 10 minutes of incubation with 10% fresh human serum at room temperature), and SDF-1α 3-67 (generated by 20 hours of incubation with fresh serum). The results reflect the mean counts per minute (cpm) per culture (± SEM) of triplicate determinations; a representative experiment of 5 performed is shown. (C) DW34 pre–B-cell proliferation in response to varying concentrations (12.5 to 100 ng/mL) of recombinant full-length SDF-1α (1-68), synthetic SDF-1α 1-67, and synthetic SDF-1α 3-67. The results reflect the mean counts per minute per culture (± SEM) of triplicate determinations; a representative experiment is shown. (D) BL-41 B-cell chemotaxic response to recombinant full-length SDF-1α (1-68) and synthetic SDF-1α (1-67) at varying concentrations (5 to 625 ng/mL). The results reflect the mean (± SEM) number of cells that have migrated to the lower chamber in 5 experiments performed.
The pre–B-cell clone DW34 responds to SDF-1 with increased proliferation.2 We used DW34 cell proliferation as an in vitro biologic assay for SDF-1 (Figure 6B). Full-length recombinant SDF-1α and SDF-1β similarly induced a dose-dependent rise in DW34 cell proliferation (up to 240 ng/mL; higher concentrations were less stimulatory). The SDF-1α species 1-67 (generated by 10 minutes of exposure to 10% fresh human serum; final human serum concentration 0.1%) also induced a dose-dependent rise in cell proliferation (up to 240 ng/mL; higher concentrations were less stimulatory). However, approximately twice as much SDF-1α 1-67 was required to achieve levels of stimulation comparable to those induced by full-length SDF-1α (P < .01). Control SDF-1α 1-68 incubated for 10 minutes with 10% heat-inactivated serum (which does not cleave the molecule at this time point) was indistinguishable from the full-length SDF-1α, providing evidence that decreased proliferation by the serum-processed 1-67 SDF-1α was likely attributable to the cleavage. SDF-1α 3-67 (generated by 20 hours of incubation with fresh human serum) retained minimal ability to stimulate DW34 cell proliferation. The significant loss of SDF-1α biologic activity associated with serum processing was confirmed (Figure 6C) by comparing DW34 cell stimulation by full-length recombinant SDF-1α (1-68) and by the synthetic SDF-1α species 1-67 (P < .01) and 3-67 (P < .01). In additional testing, we compared the chemotactic activity of recombinant SDF-1α 1-68 and synthetic SDF-1α 1-67. The human Burkitt lymphoma cell line BL-41, which expresses CXCR4 at high levels,36 displayed a dose-dependent chemotactic response to both SDF-1α species, but the response to SDF-1α 1-67 was significantly reduced (P < .01) compared with SDF-1α 1-68 at suboptimal concentrations of 5 and 25 ng/mL (Figure 6D). Thus, 2 different biologic assays detect a loss of activity associated with COOH-terminal processing of SDF-1α.
Discussion
We show that SDF-1α and SDF-1β are expressed and secreted by cells as apparently intact polypeptides but are present in the circulation as processed forms as a result of proteolytic processing. When exposed to serum, recombinant human (rHu) SDF-1α (1-68) undergoes processing first at the COOH terminus to produce SDF-1α 1-67 and then at the NH2 terminus to produce SDF-1α 3-67, whereas rHu SDF-1β 1-72 is processed only at the NH2 terminus to produce SDF-1β 3-72. In functional assays, proteolytic removal of the COOH-terminal K from SDF-1α reduces the polypeptide's ability to bind to heparin and to cells and to stimulate pre–B-cell proliferation and B-cell chemotaxis. The additional processing at the NH2 terminus reduces markedly SDF-1's ability to bind to heparin and to activate cells. The secretion of full-length SDF-1α and SDF-1β by cells and the subsequent processing of SDF-1α and SDF-1β by serum to generate SDF-1 species of differing specific activities has not been previously reported. In addition, the existence of a serum protease, which cleaves SDF-1α at COOH terminus and reduces its biologic activities, has not been previously suspected. Unlike other chemokines whose expression is induced by specific signals, SDF-1 is constitutively expressed. Based on the new information presented here, regulated degradation is likely to play a critical role in the control of SDF-1 function.
Previous studies have recognized the potential importance of NH2-terminal proteolytic processing as a modification of functional importance for SDF-1α. Synthetic forms of SDF-1α lacking the NH2-terminal residues KP had no ability to activate the CXCR4 receptor.18 Several metalloproteinases were reported to cleave 4 or 5 NH2-terminal amino acids from SDF-1.27,28 Leukocyte elastase, a protease that removes 3 amino-terminal residues from SDF-1α,29 was proposed to contribute to the release of hematopoietic stem cells from the bone marrow to the peripheral circulation by inactivating SDF-1.23 CD26/dipeptidyl peptidase IV was reported to remove the amino-terminal 2 amino acids from SDF-1α to generate an inactive chemokine.26 However, the physiological significance of CD26/dipeptidyl peptidase processing of SDF-1α had not been previously demonstrated. Based upon the nature of the SDF-1 cleavage products, blocking experiments with a specific inhibitor, and the fact that CD26/dipeptidyl peptidase is present in serum in catalytically active soluble form40 and at sufficiently high concentrations,26 we conclude that this enzyme is responsible for the NH2-terminal processing and inactivation of SDF-1α and SDF-1β in serum. Because CD26/dipeptidyl peptidase is present on cells of many lineages, it could also regulate SDF-1 biologic activity as a cell-bound enzyme.41
Prior to the current studies, there was little information on COOH-terminal cleavage of SDF-1α. A form of SDF-1α consisting of residues 1-67, lacking the COOH-terminal lysine, was purified from the culture supernatant of MS-5 stromal cells,4 but the origin and significance of this isoform were not explored. Because this cleaved form of SDF-1α was biologically active, subsequent studies have utilized the SDF-1α 1-67 variant. Even the crystal and nuclear magnetic resonance (NMR) spectroscopy structure of SDF-1α was derived from this variant form of SDF-1α.18,19 Proteolytic processing at the carboxy terminus, although uncommon among CXC chemokines, is not unique to SDF-1α.34,42-45 Noteworthy, the pattern of SDF-1α cleavage at the COOH terminus and metal dependency of the reaction bear similarity to the characteristics of Armillaria mellea protease, a metalloenzyme that can work close to the termini of polypeptide chains and displays primary specificity toward peptide bonds where a lysine residue contributes an α amino group.46,47
Studies of the structural basis for SDF-1α biologic activity have failed to reveal a role for the COOH domain. However, it is worth noting that such studies have utilized the SDF-1α 1-67 variant, which lacks the carboxy-terminal lysine. Interestingly, it was observed that SDF-1β, which is identical to SDF-1α, except for 4 additional amino acid residues present at the carboxy terminus, was about 2-fold more potent than SDF-1α 1-67.18 This difference was not explained. We confirm here this difference in activity and further show that it is attributable to the lack of the carboxy-terminal lysine in SDF-1α, which reduces the affinity of SDF-1α for heparin. Earlier experiments showed that the combined substitution of the basic amino acids K24, H25, and K27 by serine generated a variant SDF-1α molecule that had lost heparin-binding capacity.22 The same cluster of basic amino acids was proposed to contribute to electrostatic interactions with the negatively charged extracellular domain of CXCR4.19 Based on our results, we suggest that the carboxy-terminal lysine (K68), perhaps in conjunction with adjacent lysines at positions 64, 56, and 54, contributes to heparan sulfate binding.
The binding of SDF-1 to glycosaminoglycans, both on the cell surface and bound to the extracellular matrix, is believed to be important for chemokine function, presumably because it provides a means for achieving enhanced concentrations of immobilized SDF-1 available for binding to CXCR4.18,19,22 Recently, cell surface–bound SDF-1 was found to mediate lymphocyte attachment and transmigration under physiologic flow conditions.48 Here, we further show that heparin binding protects SDF-1 from cleavage at the carboxy terminus and thus serves to preserve SDF-1α activity in local sites.
Because the COOH-terminal cleavage reduces the heparin-binding affinity and the biologic activity of SDF-1α, such processing would be expected to have a regulatory role in vivo. In particular, this would add another tool to precisely regulate the biologic activity of the chemokine by effecting changes in the specific activity of the ligand.
SDF-1 has emerged as a critical regulator for normal development of the nervous, hematopoietic, and cardiovascular systems. After birth, the chemokine is constitutively expressed in virtually all tissues and regulates hematopoiesis, lymphocyte homing, and angiogenesis. It is not surprising that in the absence of transcriptional regulation, evolution has devised a complex and hierarchic pathway of degradation.
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-08-2857.
M.S. and F.Y. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Drs Josh Farber, Lei Yao, Yoshiyasu Aoki, Giovanna Zappala, Anne-Marie Lambeir, Ian Clark-Lewis, Phil Owen, and Elias Lolis for their help on various aspects of this work.




![Figure 5. SDF-1 affinity for heparin and cell surface proteoglycans. (A) Recombinant full-length SDF-1β (1-72) and SDF-1α (1-68) and synthetic SDF-1α 1-67 and 3-67 were injected at various concentrations (200, 100, 50, 25, and 12.5 nM) over a Biacore sensor chip containing streptavidin plus biotinylated heparin. The signal (measured in resonance units [RUs]) was recorded over a 120-second association phase and 120-second dissociation phase. The dissociation equilibrium constants (Kd) are shown for each set of data. Representative results from 3 independent experiments performed are shown. (B) SDF-1α binding to HUVECs evaluated by fluorescence-activated cell sorter (FACS) analysis. Recombinant full-length SDF-1α (1-68) and synthetic SDF-1α 1-67 were added at various concentrations (2000, 1000, and 500 ng) to HUVECs (1 × 106 cells). After incubation (4°C, 30 minutes), the cells were stained for surface SDF-1. A representative experiment of 3 performed is shown. Filled histograms reflect background fluorescence. (C) Recombinant SDF-1α (50 ng) was incubated for 10 minutes in buffer alone or with heparin (50 or 1000 μg/mL) with or without addition of fresh human serum. Lane 1: SDF-1α alone; lane 2: SDF-1α plus 10% human serum; lane 3: SDF-1α plus heparin 50 μg/mL; lane 4: SDF-1α plus heparin 1000 μg/mL; lane 5: SDF-1α plus heparin 50 μg/mL plus 10% human serum; lane 6: SDF-1α plus heparin 1000 μg/mL plus 10% human serum. Samples were immunoblotted with rabbit (top) or goat (bottom) antibodies against SDF-1α and SDF-1β.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/7/10.1182_blood-2003-08-2857/6/m_zh80070459310005.jpeg?Expires=1769096137&Signature=oSL-AcdvJ7oE9c~APVUWdjIpBu7UCu-2xvnhjfmYzJUaFjc9Sw5mYucAJdy-i2OC1ewGicJ1HHmEJEGVg7zmm8N4YTlPBlKdixUlUx-5hjxAEV1XFXCwHGW6MYp2Uil3RNZ6CqXgu-ag-oHArprv3oMmisQTreNK3KX6TqQlDjiEySSZx8g2~mZf25FOXo-ocUpItmu-3oS7ks5XK43~4EFzWWWbY2-qHIrP43JzSMV8HNRKhmILtrFxw9z-ONSx6b74tOe1uz8LB~LwwCY2mqKAH9kgVFKNscJebEcQWosvOYZZ2GJxi2QcBdTM41M~PH6FokhvSbGyujrryXXLYg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal