Abstract
We have addressed whether aberrant ongoing hypermutation can be detected in the proto-oncogenes PIM1, c-MYC, RhoH/TTF, PAX5, and the tumor-suppressor gene CD95 in primary central nervous system lymphomas (PCNSLs) derived from immunocompetent HIV-negative patients. Nine of 10 PCNSLs analyzed harbored somatic mutations in the PIM1, c-MYC, RhoH/TTF, and PAX5 genes, but not in the CD95 gene, with 8 tumors carrying alterations in at least 2 of these genes. Furthermore, ongoing aberrant mutation was evidenced in a subset of PCNSLs (2 of 3). Although most of the mutations corresponded to base pair substitutions, deletions were also present. The mean mutation frequency was approximately 60-fold lower for these genes compared with the values obtained for immunoglobulin genes in PCNSL. They were increased 2- to 5-fold compared with extracerebral diffuse large B-cell lymphoma (DLBCL). In summary, our data demonstrate aberrant somatic hypermutations at high frequency in the PIM1, PAX5, RhoH/TTF, and c-MYC genes in most PCNSLs. These findings may indicate a pathogenic role for aberrant somatic hypermutation in PCNSL development. In contrast, although mutations were detected in exon 9 of the CD95 gene, the lack of mutations in the 5′ region provides no evidence for the CD95 gene as a target for aberrant somatic mutation.
Introduction
Primary central nervous system lymphomas (PCNSLs) are classified as diffuse large B-cell lymphomas (DLBCLs) according to the World Health Organization (WHO) system.1 However, they differ from extracerebral DLBCLs in their significantly poorer prognosis.2 This clinically relevant observation has raised the question whether PCNSLs comprise a distinct disease entity. The pathogenesis of PCNSL is largely unknown. Recent studies have identified characteristic recurrent translocations mainly involving the immunoglobulin H (IgH) and the BCL6 gene loci, which are considered to be pathogenetically relevant.3
Histogenetically, PCNSLs are derived from germinal center (GC) B cells, as demonstrated by their expression of the bcl6 protein and a still active process of hypermutation.4-6 Interestingly, they carry an extremely high load of somatic mutations, exceeding that of all other lymphoma entities.4 This propensity prompted us to investigate whether the process of somatic hypermutation in growthregulatory genes plays a role in the development of PCNSL.
Aberrant hypermutation has recently been described for several lymphoma entities. In extracerebral DLBCL, aberrant somatic mutations affected multiple gene loci, including the protooncogenes PIM1, c-MYC, RhoH/TTF, and PAX5, which were somatically mutated in more than 50% of the tumors analyzed.7 Aberrant hypermutation targeting the 5′ region was restricted to malignant B cells and could not be detected in normal, nonmalignant GC B cells.
The gene PIM1, which encodes a serine/threonine protein kinase involved in cell proliferation and survival, has occasionally been observed in chromosomal translocations in lymphomas.8,9 c-MYC, which encodes a transcription factor controlling cell growth, proliferation, and apoptosis, is characteristically mutated in endemic Burkitt lymphoma carrying t(8;14) translocations.10 The RhoH/TTF gene encodes a guanosine triphosphate (GTP)–binding protein of the Ras superfamily. This proto-oncogene may be involved in different lymphoma entities.11 PAX5 represents a transcription factor essential for B-cell lineage commitment and differentiation.12 The tumor-suppressor gene CD95, with a critical role in the regulation of cell survival and apoptosis, is expressed at high levels in normal GC B cells. Expression of this gene increases their susceptibility to apoptosis. Interestingly, somatic mutations of the CD95 gene have been described in 21% and 60% of nodal and extranodal DLBCLs,13 respectively.
Considering that PCNSLs exceed other extracerebral DLBCLs with respect to their mean mutation frequency of immunoglobulin genes, it is tempting to speculate that hypermutation aberrantly targeting genes with oncogenic potential may be of functional relevance for their development. The aim of the present study was to analyze the proto-oncogenes PIM1, c-MYC, RhoH/TTF, and PAX5 and the tumor-suppressor gene CD95 for the presence of somatic mutations in a series of PCNSLs derived from immunocompetent, HIV-negative patients. Interestingly, 90% (9 of 10) of the PCNSLs harbored somatic mutations in at least one of these genes, supporting the hypothesis that aberrant somatic mutations in growth-associated genes may provide an alternative pathway for PCNSL development.
Materials and methods
Material, diagnoses, and DNA extraction
DNA was extracted from frozen samples containing at least 80% of tumor cells of 10 histopathologically classified primary DLBCLs of the central nervous system with the NucleoSpin Tissue Kit (BC Clontech, Heidelberg, Germany). DNA was dissolved in 100 μL TE buffer (10 mmol/L Tris, 1 mmol/L ethylene-diamine-tetraacetic acid; pH 7.6). Two microliters to 5 μL of this stock DNA solution, corresponding to 100 ng of DNA, was used in each amplification reaction.
IgH gene rearrangements and sequence analysis of V region genes
Tumors were investigated for their rearrangements of IgH genes by polymerase chain reaction (PCR) and were directly sequenced, as described previously.4
Sequencing analysis of CD95, PAX5, RhoH/TTF, c-MYC, and PIM1
Mutational analysis of the CD95, PAX5, RhoH/TTF, c-MYC, and PIM1 genes was restricted to regions previously shown to contain more than 90% of mutations in systemic DLBCLs.7 Oligonucleotides used in the PCR and further primers used for sequencing are summarized in Table 1. All PCR products were directly sequenced from both sides using the BigDye Kit (Applied Biosystems, Weiterstadt, Germany) with an ABI377 automated sequencer (Applied Biosystems).
Oligonucleotides used in mutation analysis
Gene (accession no.) . | Primer sequence . | Use . | PCR product, bp . |
|---|---|---|---|
| CD95 5′ UTR (al157394) | AAGAGTGACACACAGGTGTTC | P + S | 478 |
| AAGGCCCAAGAAAAGCAAGTC | P + S | ||
| CD95 exon 8 (al157394) | CCTTCTTAATCACTTAATCTAGC | P + S | 336 |
| CTGCCTGATAAATGCTTATGCTG | P + S | ||
| CD95 exon 9 (m67454) | TGGGAATTTCATTTAGAAAAACA | P + S | 413 |
| TACTCAGAACTGAATTTGTTGT | P + S | ||
| PAX5 (af386791) | AGGGACCTCAGAAGCATCGAGGCC | P + S | 932 |
| TGAAAAAGGCGCCATCGGAGTAG | P + S | ||
| CACTGTAAGCACGACCCG | S | ||
| CGGGTCGTGCTTACAGTG | S | ||
| RhoH/TTF (af386789) | GCTTTTACTCTAGGCCAAACATCG | P + S | 875 |
| CTTCTACCGACACTTCGCATTCTT | P + S | ||
| CTGAGGTGGTTTGATTTGG | S | ||
| CCAAATCAAACCACCTCAG | S | ||
| c-MYC exon 1 (x00364) | CACCGGCCCTTTATAATGCG | P + S | 1302 |
| ACGATTCCAGGAGAATCGGA | P + S | ||
| CTTGCCGCATCCACGAAAC | S | ||
| GGAGAGGAGAAGGCAGAG | S | ||
| CCCAGAGAGCAATTAACAC | S | ||
| GTGTTGTAAGTTCCAGTGC | S | ||
| c-MYC intron 1 (x00364) | TCTCTAGAGGTGTTAGGACG | P + S | 865 |
| AGAGCTATCCCCTAAAGCGG | P + S | ||
| GCTGGCAAAAGGAGTGTTG | S | ||
| CTCCCAACCTTCCCTCTC | S | ||
| c-MYC exon 2 (x00364) | CCGCTGGTTCACTAAGTGCG | P + S | 1167 |
| GGATGGGAGGAAACGCTAAAG | P + S | ||
| CAGCGAGGATATCTGGAAG | S | ||
| CTTGTACCTGCAGGATCTG | S | ||
| CTTCCAGATATCCTCGCTG | S | ||
| CAGATCCTGCAGGTACAAG | S | ||
| PIM1 (af386792) | TTCTCCGGCGTCATTAGGC | P + S | 1025 |
| CGTTTGTTAGGTTAAGCCGC | P + S | ||
| GCCAGCTGAACCTGTAATG | S | ||
| CATTACAGGTTCAGCTGGC | S |
Gene (accession no.) . | Primer sequence . | Use . | PCR product, bp . |
|---|---|---|---|
| CD95 5′ UTR (al157394) | AAGAGTGACACACAGGTGTTC | P + S | 478 |
| AAGGCCCAAGAAAAGCAAGTC | P + S | ||
| CD95 exon 8 (al157394) | CCTTCTTAATCACTTAATCTAGC | P + S | 336 |
| CTGCCTGATAAATGCTTATGCTG | P + S | ||
| CD95 exon 9 (m67454) | TGGGAATTTCATTTAGAAAAACA | P + S | 413 |
| TACTCAGAACTGAATTTGTTGT | P + S | ||
| PAX5 (af386791) | AGGGACCTCAGAAGCATCGAGGCC | P + S | 932 |
| TGAAAAAGGCGCCATCGGAGTAG | P + S | ||
| CACTGTAAGCACGACCCG | S | ||
| CGGGTCGTGCTTACAGTG | S | ||
| RhoH/TTF (af386789) | GCTTTTACTCTAGGCCAAACATCG | P + S | 875 |
| CTTCTACCGACACTTCGCATTCTT | P + S | ||
| CTGAGGTGGTTTGATTTGG | S | ||
| CCAAATCAAACCACCTCAG | S | ||
| c-MYC exon 1 (x00364) | CACCGGCCCTTTATAATGCG | P + S | 1302 |
| ACGATTCCAGGAGAATCGGA | P + S | ||
| CTTGCCGCATCCACGAAAC | S | ||
| GGAGAGGAGAAGGCAGAG | S | ||
| CCCAGAGAGCAATTAACAC | S | ||
| GTGTTGTAAGTTCCAGTGC | S | ||
| c-MYC intron 1 (x00364) | TCTCTAGAGGTGTTAGGACG | P + S | 865 |
| AGAGCTATCCCCTAAAGCGG | P + S | ||
| GCTGGCAAAAGGAGTGTTG | S | ||
| CTCCCAACCTTCCCTCTC | S | ||
| c-MYC exon 2 (x00364) | CCGCTGGTTCACTAAGTGCG | P + S | 1167 |
| GGATGGGAGGAAACGCTAAAG | P + S | ||
| CAGCGAGGATATCTGGAAG | S | ||
| CTTGTACCTGCAGGATCTG | S | ||
| CTTCCAGATATCCTCGCTG | S | ||
| CAGATCCTGCAGGTACAAG | S | ||
| PIM1 (af386792) | TTCTCCGGCGTCATTAGGC | P + S | 1025 |
| CGTTTGTTAGGTTAAGCCGC | P + S | ||
| GCCAGCTGAACCTGTAATG | S | ||
| CATTACAGGTTCAGCTGGC | S |
P indicates PCR; S, sequencing.
All PCR reactions were performed in duplicate in 2 independent reactions. Sequences were aligned to the corresponding germline sequences (accession numbers are given in Table 1). Double peaks in the sequencing reaction indicated most mutations. All mutations corresponding to published allele sequences were excluded from further analysis. Only mutated sequences were taken into consideration for calculation of the mean mutation frequencies.
In addition, PCR products of PAX5 and RhoH/TTF of 3 patients with PCNSL (patients 2, 6, and 10) were cloned using the TOPO TA Cloning Kit (Invitrogen, Karlsruhe, Germany). From each patient, 20 clones were sequenced from both sides. To exclude possible Taq errors, only mutations that were detected in at least 2 clones derived from the same patient were considered. Sequences, which seemed to have been caused by PCR artifacts, were also excluded. Informed consent from the patients was provided according to the Declaration of Helsinki.
Results
In our series of 10 patients with PCNSL, 9 (90%) harbored somatic mutations in the tumor-suppressor gene CD95 and in the protooncogenes c-MYC, PAX5, RhoH/TTF, and PIM1. Eight tumors (80%) displayed somatic mutations in at least 2 of these genes, and 1 tumor had mutations detectable in 1 proto-oncogene only. Results are summarized in Table 2.
Mutation status of the IgH, CD95, PAX5, RhoH/TTF, c-MYC, and PIM1 genes
. | . | CD95 . | . | . | . | . | . | . | . | . | . | c-MYC . | . | . | . | . | . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | 5′ UTR . | . | Exon 8 . | . | Exon 9 . | . | PAX5 . | . | RhoH/TTF . | . | Exon 1 . | . | Intron 1 . | . | Exon 2 . | . | PIM1 . | . | ||||||||||
| Patient . | IGH (%) . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | ||||||||||
| 1 | V3-7 (8.9) | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | ||||||||||
| 2 | V4-34 (16.6) | 0 | - | 0 | - | 0 | - | 19 | 776C>T, 848C>T, 882G>C, 1001C>T, 1017G>C, 1025G>A, 1066C>A, 1070G>A, 1074G>A, 1077C>A, 1194C>G, 1230G>C, 1349G>T, 1354G>A, 1435G>C, 1446G>A, 1449C>G, Δ(1467-1501), 1536G>T | 7 | 413G>T, 494T>C, 539C>T, 694G>C, 838C>T, 839C>G, 885C>T | 2 | 3020G>T, 3461G>C | 2 | 3627C>A, 3741G>A | 0 | - | 5 | 1909G>A: Ala→Thr, 1932G>A: Val→Val, 1969C>T: Leu→Phe, 2172C>A, 2227G>A | ||||||||||
| 3 | V3-74 (14.1) | 0 | - | 0 | - | 1 | 941T>G: Val→Val | 8 | 848C>T, 943G>A, 1046C>T, 1048C>T, 1082C>T, 1117G>A, 1382C>T, 1384C>T | 2 | 694G>A, 1098G>A | 3 | 3266G>A, 3481C>T, 3516G>A | 0 | - | 0 | - | 0 | - | ||||||||||
| 4 | - | 0 | - | 0 | - | 0 | - | 0 | - | 1 | 486T>G | 1 | 2506C>G: Ser→Cys | 0 | - | 0 | - | 0 | - | ||||||||||
| 5 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 1 | 3052C>G | 0 | - | 0 | - | 1 | 2486G>A | ||||||||||
| 6 | V1-2 (23.3) | 0 | - | 0 | - | 4 | 862A>G: Asn→Ser, 880T>A: Leu→STOP, 888T>C + 890T>C: Tyr→His | 3 | 1113C>T, 1209T>C, 1295C>T | 1 | 420T>G | 1 | 2397G>A: Glu→Lys | 0 | - | 0 | - | 0 | - | ||||||||||
| 7 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 2 | 1862T>C: Asp→Asp, 1875C>A: Phe→Leu | ||||||||||
| 8 | V3-7 (12.6) | 0 | - | 0 | - | 0 | - | 2 | 743C>A, 1017G>A | 2 | 987T>G, 1057C>T | 0 | - | 2 | 4151T>A, 4224C>T | 0 | - | 3 | 2022G>C: Glu→Asp, 2029C>G: Leu→Val, 2734G>C | ||||||||||
| 9 | V4-34 (12.2) | 0 | - | 0 | - | 0 | - | 4 | 910G>C, 1071C>T, 1193G>A, 1334C>A | 5 | 470G>C, 479G>A, 649C>T, 900G>A, 959G>A | 0 | - | 1 | 4099C>T | 0 | - | 0 | - | ||||||||||
| 10 | V3-7 (16.0) | 0 | - | 0 | - | 0 | - | 29 | 736G>A, 745G>A, Δ(846-1083), 1099G>A, 1100C>T, 1178G>A: Met*→Ile, 1182A>G: Ile→Val, 1185C>T: His→Tyr, 1189G>A, 1193G>T, 1200C>T, 1214G>T, 1259G>A, 1265G>A, 1283G>A, 1333G>C, 1334C>G, 1338C>G, 1347G>C, 1349G>A, 1353G>C, Δ(1383-1543), 1549A>C, 1558G>A, 1559G>C, 1561G>A, 1572G>A, 1573A>T, 1584G>A | 7 | 533T>G, 539G>T, 743G>A, 759C>T, 962G>A, 1056G>A, 1092G>A | 2 | 2588G>A: Trp→STOP, 2821C>A: Ser→Tyr | 1 | 3757C>A | 0 | - | 8 | 1882G>C: Glu→Gln, 1920C>G: Phe→Leu, 2023C>T: Leu→Phe, 2168C>G, 2322G>C, 2554A>C, 2636G>A, 2719C>T | ||||||||||
. | . | CD95 . | . | . | . | . | . | . | . | . | . | c-MYC . | . | . | . | . | . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | 5′ UTR . | . | Exon 8 . | . | Exon 9 . | . | PAX5 . | . | RhoH/TTF . | . | Exon 1 . | . | Intron 1 . | . | Exon 2 . | . | PIM1 . | . | ||||||||||
| Patient . | IGH (%) . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | No. mutations . | Mutation type and position . | ||||||||||
| 1 | V3-7 (8.9) | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | ||||||||||
| 2 | V4-34 (16.6) | 0 | - | 0 | - | 0 | - | 19 | 776C>T, 848C>T, 882G>C, 1001C>T, 1017G>C, 1025G>A, 1066C>A, 1070G>A, 1074G>A, 1077C>A, 1194C>G, 1230G>C, 1349G>T, 1354G>A, 1435G>C, 1446G>A, 1449C>G, Δ(1467-1501), 1536G>T | 7 | 413G>T, 494T>C, 539C>T, 694G>C, 838C>T, 839C>G, 885C>T | 2 | 3020G>T, 3461G>C | 2 | 3627C>A, 3741G>A | 0 | - | 5 | 1909G>A: Ala→Thr, 1932G>A: Val→Val, 1969C>T: Leu→Phe, 2172C>A, 2227G>A | ||||||||||
| 3 | V3-74 (14.1) | 0 | - | 0 | - | 1 | 941T>G: Val→Val | 8 | 848C>T, 943G>A, 1046C>T, 1048C>T, 1082C>T, 1117G>A, 1382C>T, 1384C>T | 2 | 694G>A, 1098G>A | 3 | 3266G>A, 3481C>T, 3516G>A | 0 | - | 0 | - | 0 | - | ||||||||||
| 4 | - | 0 | - | 0 | - | 0 | - | 0 | - | 1 | 486T>G | 1 | 2506C>G: Ser→Cys | 0 | - | 0 | - | 0 | - | ||||||||||
| 5 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 1 | 3052C>G | 0 | - | 0 | - | 1 | 2486G>A | ||||||||||
| 6 | V1-2 (23.3) | 0 | - | 0 | - | 4 | 862A>G: Asn→Ser, 880T>A: Leu→STOP, 888T>C + 890T>C: Tyr→His | 3 | 1113C>T, 1209T>C, 1295C>T | 1 | 420T>G | 1 | 2397G>A: Glu→Lys | 0 | - | 0 | - | 0 | - | ||||||||||
| 7 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 0 | - | 2 | 1862T>C: Asp→Asp, 1875C>A: Phe→Leu | ||||||||||
| 8 | V3-7 (12.6) | 0 | - | 0 | - | 0 | - | 2 | 743C>A, 1017G>A | 2 | 987T>G, 1057C>T | 0 | - | 2 | 4151T>A, 4224C>T | 0 | - | 3 | 2022G>C: Glu→Asp, 2029C>G: Leu→Val, 2734G>C | ||||||||||
| 9 | V4-34 (12.2) | 0 | - | 0 | - | 0 | - | 4 | 910G>C, 1071C>T, 1193G>A, 1334C>A | 5 | 470G>C, 479G>A, 649C>T, 900G>A, 959G>A | 0 | - | 1 | 4099C>T | 0 | - | 0 | - | ||||||||||
| 10 | V3-7 (16.0) | 0 | - | 0 | - | 0 | - | 29 | 736G>A, 745G>A, Δ(846-1083), 1099G>A, 1100C>T, 1178G>A: Met*→Ile, 1182A>G: Ile→Val, 1185C>T: His→Tyr, 1189G>A, 1193G>T, 1200C>T, 1214G>T, 1259G>A, 1265G>A, 1283G>A, 1333G>C, 1334C>G, 1338C>G, 1347G>C, 1349G>A, 1353G>C, Δ(1383-1543), 1549A>C, 1558G>A, 1559G>C, 1561G>A, 1572G>A, 1573A>T, 1584G>A | 7 | 533T>G, 539G>T, 743G>A, 759C>T, 962G>A, 1056G>A, 1092G>A | 2 | 2588G>A: Trp→STOP, 2821C>A: Ser→Tyr | 1 | 3757C>A | 0 | - | 8 | 1882G>C: Glu→Gln, 1920C>G: Phe→Leu, 2023C>T: Leu→Phe, 2168C>G, 2322G>C, 2554A>C, 2636G>A, 2719C>T | ||||||||||
The indicated codon for patient 10 in PAX5 corresponds to the starting point of translation.
The CD95 gene was analyzed in 3 different regions: the 5′ untranslated region, exon 8, and exon 9. Two patients with PCNSL carried somatic mutations, which were confined to exon 9 in both. In 1 of these tumors (patient 3), a single base pair substitution corresponded to a silent mutation, whereas the second tumor (patient 6) harbored 3 point mutations resulting in an amino acid exchange from asparagine to serine and from tyrosine to histidine, respectively. A mutation in patient 6 caused a stop codon. In contrast, somatic mutations were absent from the 5′ untranslated region and from exon 8 in all these patients and in 3 other PCNSL patients also studied (data not shown). The mean mutation frequency for CD95 exon 9 was 0.30% (range, 0.12%-0.48%).
Mutations of the PAX5 gene were detected in 6 PCNSL patients, with a mean mutation frequency of 0.58% (range, 0.11%-1.56%). Most mutations were point mutations. In patients 2 and 10, 1 and 2 deletions were detected, respectively. In patient 10, 3 mutations corresponded to missense mutations, but nonsense mutations were absent.
Seven (70%) of 10 of our PCNSL patients harbored mutations in the RhoH/TTF gene, all of which were identified as base pair substitutions. Calculation of the mutation frequency revealed a mean of 0.20% (range, 0.06%-0.4%).
Within the c-MYC gene, exon 1, intron 1, and exon 2 were studied. Exon 1 and intron 1, but not exon 2, contained somatic mutations. Overall, 8 (80%) of 10 PCNSL patients carried a somatically mutated c-MYC gene. In 6 (60%) of 10 and 4 (40%) of 10 tumors, somatic mutations were identified in exon 1 and intron 1, respectively. All mutations corresponded to base pair exchanges; 3 resulted in an amino acid substitution, and another mutation introduced a stop codon (Table 2). The mean mutation frequency was calculated as 0.06% (range, 0.04%-0.12%) and 0.09% (range, 0.06%-0.12%) for exon 1 and intron 1, respectively.
The PIM1 gene showed mutations in 5 (50%) of 10 of the tumors, most of which resulted in an amino acid exchange. Deletions or insertions were not detectable. Overall, 9 missense mutations were detected, and nonsense mutations were absent. The mean mutation frequency was 0.19% (range, 0.05%-0.39%).
Patient 10 carried an unusually high number of mutations in all genes except the CD95 gene. Twenty-nine mutations were present in PAX5, including 2 deletions and 27 point mutations resulting in an amino acid exchange in 3 of them; the other point mutations were not located in protein coding regions. Furthermore, this tumor harbored 7 mutations in the RhoH/TTF gene and 8 mutations in the PIM1 gene. All mutations located in protein coding regions led to amino acid exchanges. Interestingly, 2 mutations affected exon 1 of the c-MYC gene, yielding a stop codon and an amino acid substitution, respectively.
Overall analysis of the mutation pattern and frequency for all genes analyzed revealed a maximum of 29 mutations (including 2 deletions) per gene, which was identified for PAX5. For all genes included in this analysis, transitions were more common than transversions (Table 3). To determine whether aberrant hypermutation was occurring, subcloning experiments were performed for the PAX5 and the RhoH/TTF genes in 3 patients with PCNSL.
Mutation patterns of the PAX5, RhoH/TTF, c-MYC, and PIM1 genes
Gene . | All mutations . | del/ins . | A . | T . | G . | C . | TS . | TV . | RGYW WRCY . |
|---|---|---|---|---|---|---|---|---|---|
| PAX5 (%) | 65 (100) | 3 (5) | 3 (5) | 1 (2) | 35 (53) | 23 (35) | 39 (60) | 23 (35) | 22 (34) |
| RhoH/TTF (%) | 25 (100) | 0 (0) | 0 (0) | 4 (16) | 14 (56) | 7 (28) | 16 (64) | 9 (36) | 14 (56) |
| c-MYC (%) | 16 (100) | 0 (0) | 0 (0) | 1 (6) | 7 (44) | 8 (50) | 8 (50) | 8 (50) | 4 (25) |
| PIM1 (%) | 19 (100) | 0 (0) | 1 (5) | 1 (5) | 9 (47) | 8 (43) | 9 (47) | 10 (53) | 7 (37) |
| Σ (%) | 125 (100) | 3 (2) | 4 (3) | 7 (6) | 65 (52) | 46 (37) | 72 (58) | 50 (40) | 47 (38) |
Gene . | All mutations . | del/ins . | A . | T . | G . | C . | TS . | TV . | RGYW WRCY . |
|---|---|---|---|---|---|---|---|---|---|
| PAX5 (%) | 65 (100) | 3 (5) | 3 (5) | 1 (2) | 35 (53) | 23 (35) | 39 (60) | 23 (35) | 22 (34) |
| RhoH/TTF (%) | 25 (100) | 0 (0) | 0 (0) | 4 (16) | 14 (56) | 7 (28) | 16 (64) | 9 (36) | 14 (56) |
| c-MYC (%) | 16 (100) | 0 (0) | 0 (0) | 1 (6) | 7 (44) | 8 (50) | 8 (50) | 8 (50) | 4 (25) |
| PIM1 (%) | 19 (100) | 0 (0) | 1 (5) | 1 (5) | 9 (47) | 8 (43) | 9 (47) | 10 (53) | 7 (37) |
| Σ (%) | 125 (100) | 3 (2) | 4 (3) | 7 (6) | 65 (52) | 46 (37) | 72 (58) | 50 (40) | 47 (38) |
del/ins indicates number of deletions and insertions; A, mutations occurring at adenosine position; T, analogous to A; G, analogous to A; C, analogous to A; RGYW/WRCY, number of mutations embedded in the mutational hot spot motive RGYW (R = A, G; Y = C, T; W = A, T); TS, transitions; TV, transversions; and Σ, the sum of the mutations in the respective column.
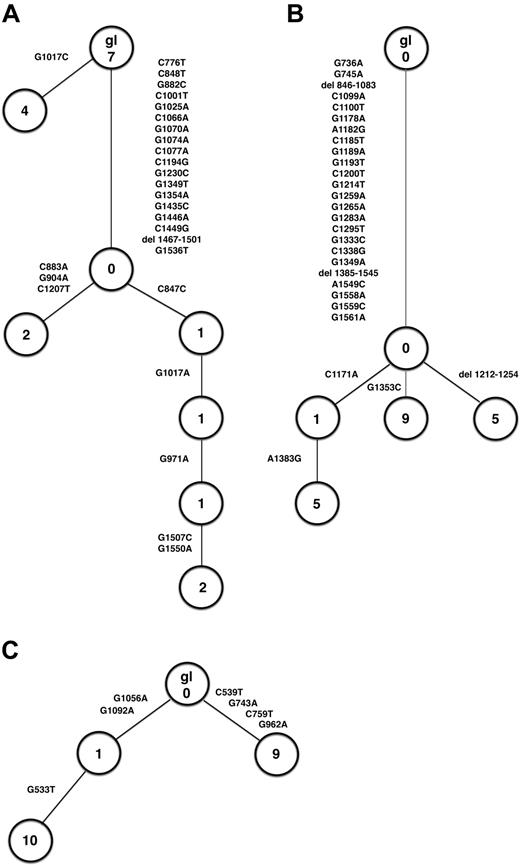
These studies provided evidence for ongoing mutation of the PAX5 gene in patients 2 and 10 but not in patient 6. In patient 2, compared with the respective germline sequence, 4 sequences were identical differing in one mutation from the germline sequence. The second allele gave rise to 5 subclones that shared 18 mutations and also introduced 1, 2, 3, or 5 more mutations, respectively. These additional mutations could be aligned as a treelike structure, thus, demonstrating ongoing mutation in the PAX5 gene (Figure 1A). This observation was confirmed by data derived from patient 10 (Figure 1B). Subcloning was performed using the tumor-derived PCR product, which consistently harbored a deletion. Four subclones were identified; all shared 24 point mutations and 2 deletions compared with the germline sequence. In addition, 1 subclone, which was represented 5 times, harbored another deletion, and a second subclone, which was represented 9 times, introduced another point mutation. The third subclone was represented by 1 sequence and harbored 1 additional point mutation. The fourth subclone, derived from subclone 3, was characterized by an additional point mutation (Figure 1B).
Treelike structure of sequences derived from cloned PCR products. (A) PAX5, patient 2. (B) PAX5, patient 10. (C) RhoH/TTF, patient 10. gl indicates germline sequence.
Treelike structure of sequences derived from cloned PCR products. (A) PAX5, patient 2. (B) PAX5, patient 10. (C) RhoH/TTF, patient 10. gl indicates germline sequence.
With respect to the RhoH/TTF gene, we obtained evidence for ongoing mutations in patient 10 but not in patient 2 or 6. In patient 10, 2 major clones differed from the germline sequence by the introduction of 4 and 2 point mutations, represented 9 and 11 times, respectively. Ten sequences were derived from the latter subclone, which harbored 1 additional point mutation (Figure 1C).
To confirm whether these sequence differences between the cloned PCR products were indeed indicative of intraclonal diversity or whether they simply reflected Taq DNA polymerase errors, we calculated the expected errors for our PCR conditions. Based on a Taq DNA polymerase error rate of 10–5/bp and cycle,14,15 we expected 1 error/2857 bp and 1 error/3333 bp for PAX5 (35 cycles) and RhoH/TTF (30 cycles), respectively. Considering the number of base pairs of the individual sequences and the number of sequences analyzed (18, 20, and 20 for PAX5, patients 2 and 10, and for RhoH/TTF, patient 10, respectively), one would have expected the introduction of 6, 6, and 5 mutations because of Taq DNA polymerase errors for PAX5, patients 2 and 10, and for RhoH/TTF, patient 10, respectively. With 18, 27, and 7 point mutations in patient 2 and patient 10 for the PAX5 gene and in patient 10 for the RhoH/TTF gene (Table 2), respectively, the number of point mutations is clearly above this threshold. In this regard it is important to note that only those mutations that were detected in at least 2 individual sequences of each patient were considered in the analysis for ongoing mutation. All mutations recognized only once (24, 5, and 14 for PAX5, patients 2 and 10, and for RhoH/TTF, patient 10, respectively) and, thus, at least partly reflecting Taq DNA polymerase errors were excluded to ensure that only point mutations, which can be reliably attributed to the process of aberrant somatic mutation because of their repeated detection in independent clones, were considered for analysis. Taken together, our data indicate ongoing aberrant somatic hypermutation in PCNSL.
Rearranged IgH genes could be identified in 7 PCNSL patients (70%) in this series. Three tumors lacked dominant amplificates, probably because highly mutated rearranged gene segments are sometimes not recognized by primers. All tumors with detectable monoclonally rearranged IgH genes harbored somatic mutations (Table 2). The mean mutation frequency was 14.81% (range, 8.9%-23.3%; Table 2).
Discussion
The present study demonstrates that PCNSLs are targeted by aberrant somatic hypermutations with involvement of 4 potent proto-oncogenes—PAX5, PIM1, c-MYC, and RhoH/TTF—although not the 5′ untranslated region of the CD95 gene. All these genes play an important role in B-cell development and differentiation and in the regulation of proliferation and apoptosis.
In general, the mutation pattern with a predominance of base pair exchanges, more frequent transitions than transversions, and an elevated G+C ratio over A+T substitutions exhibits characteristic features of normal somatic hypermutation. Furthermore, there was evidence for ongoing active somatic hypermutation at least in a subset of PCNSL. Nevertheless, these data do not exclude a possible contribution of other mechanisms to the introduction of mutations. The mean mutation frequency of the aberrantly mutated genes was significantly (approximately 60-fold) lower than values reported for IgH genes in PCNSL (Table 2).4 Interestingly, comparison of the mean mutation frequencies calculated for the individual genes analyzed in PCNSLs with data obtained for extracerebral DLBCLs7 revealed that PCNSLs generally exceeded values for extracerebral DLBCLs by 2- to 5-fold. This is in line with this study and our earlier observation that PCNSLs are characterized by extremely high mutation frequencies for the IgH and the IgL genes, with values of 13.2% and 8.3%, respectively. These values exceed those obtained for all other lymphoma entities analyzed so far.4 Taken together, our data suggest that the neoplastic cell population or its precursors are characterized by a prolonged or even ongoing germinal center reaction. This concept is further corroborated by the observation of ongoing aberrant hypermutation in at least in a fraction of PCNSLs.
Remarkably, patient 2, who had a high number of somatic mutations in all 4 proto-oncogenes, also had a high mean mutation frequency of 16.6% for the IgH gene and IgH and a BCL6 translocations (patient 8, as reported by Montesinos-Rongen et al3 ). This observation supports the hypothesis that an abnormally active hypermutation favors the occurrence of translocations as a result of a misdirected somatic hypermutation. Thus, our data lend support to the concept that active somatic hypermutation is of crucial importance for PCNSL development by introducing aberrant mutations and by causing translocations.
Because the genes targeted by aberrant somatic hypermutation are proto-oncogenes and tumor-suppressor genes, respectively, introducing somatic mutations in these genes may have important functional consequences. The signal transducers RhoH/TTF and PIM1, which harbored a remarkable number of 25 and 19 mutations in 7 and 5 PCNSLs, respectively, have been identified as translocation partners of BCL6 in different lymphoma types.11,16 Mutations of the RhoH/TTF gene, which encodes a small GTP-binding protein of the RAS superfamily involved in signal transduction, were spread across the entire analyzed region without further clustering. In contrast, mutations in the PIM1 gene clustered in the last part of the region analyzed in the vicinity of a putative phosphorylation site, represented by the nucleotides at positions 2081 to 2083,17 which, however, was not directly targeted in our PCNSL series. Remarkably, mutations of the PIM1 gene frequently resulted in an amino acid exchange, thereby possibly altering the structure and, subsequently, the function of the PIM1 protein, which plays an important role in cell proliferation and survival. PAX5, which encodes a B-cell–specific transcription factor required for B-cell lineage commitment, differentiation, and isotype switching, also harbored mutations in regions involved in the initiation of translation (patient 10).
In contrast to these proto-oncogenes, somatic mutations of the c-MYC gene occurred at relatively low frequencies (0.06% and 0.09% for exon 1 and intron 1, respectively). In patient 10, such a point mutation in exon 1 generated a stop codon, leading to a truncated protein; the functional consequences remain to be elucidated. In this regard, one may speculate that during tumor development c-MYC expression played a role, but that at the time of stop codon generation the tumor cells had become independent of c-MYC–mediated effects.
In the CD95 gene, mutations were confined to the death domain encoding exon 9, a region far downstream from the 5′ region, which is characteristically targeted by the hypermutation machinery. Thus, 2 different mechanisms may underlie the introduction of mutations in the CD95 gene and the other genes analyzed. Exon 9 mutations were present in 2 of 10 patients in this series; in patient 3 only a silent point mutation, which might also have represented a rare polymorphism, made the overall incidence of CD95 mutations in PCNSL low. Nevertheless, the base pair exchange in patient 6 generated a stop codon at position 880. This resulted in the loss of a functional death domain in this tumor, which may render the tumor cells resistant to CD95/CD95L-mediated apoptosis, as has been described in in vitro experiments for CD95-mutant B cells.18
CD95 mutations have also been described for other lymphoma entities.19 Interestingly, they appear particularly frequent in lymphomas with extranodal manifestation.19 Furthermore, the observation of autoreactive diseases suggested a link between CD95 mutation, lymphomagenesis, and autoimmunity, although this is still controversially discussed because CD95 gene mutations appeared to play little, if any, role in the generation of the pool of plasmablasts in patients with systemic lupus erythematosus.13,20 However, although CD95 mutations alone are unlikely to commit a GC B cell to malignant transformation, they may represent an early event in lymphomagenesis by favoring the persistence of mutant GC B cells, which may acquire additional transforming modifications, ultimately resulting in malignant lymphoma formation. In this context it is interesting that PCNSLs preferentially rearranged the V4-34 gene segment, which has been implicated in autoimmune disorders, including cold agglutinin disease.21
From these mutation analyses, direct functional consequences of potential pathogenetic relevance for lymphomagenesis can be anticipated from amino acid exchanges, the introduction of stop codons (patient 6, CD95, exon 9; patient 10, c-MYC, exon 1) and the mutations of start codons (patient 10, PAX5). Furthermore, it is conceivable that mutations may influence the expression and regulation of these genes in a mechanism analogous to observations obtained for the BCL6 gene.22
In patient 1, the only PCNSL of our series without evidence for aberrant somatic hypermutation, one may wonder whether the tumor cells have experienced a germinal center reaction at all. This was demonstrated by the presence of mutations in the IgH gene; however, the mutation frequency of 8.9% was rather low. Fluorescence in situ hybridization (FISH) analysis revealed a polyploidy of the tumor cells (patient 9, as reported by Montesinos-Rongen et al3 ).
Comparison of our data derived from immunocompetent, HIV-negative patients with a study of 4 AIDS-associated PCNSL, which were also targeted by aberrant somatic hypermutations,17 may point to differences in the genes affected. Although somatic hypermutations involved the RhoH/TTF gene and exon 2 of the c-MYC gene in 1 of 4 AIDS-associated PCNSLs, respectively, somatic mutations were absent from the PIM1 and PAX5 genes.17 Thus, taking into account that the number of tumors investigated is still low, these divergent observations may be consistent with the concept that pathogenic pathways differ in HIV-associated PCNSLs as opposed to PCNSLs of immunocompetent patients.
Taken together, the data of the present study indicate aberrant somatic hypermutations at relatively high frequency in the proto-oncogenes PIM1, PAX5, RhoH/TTF, and c-MYC and in the tumor-suppressor gene CD95 in PCNSL. These changes may be of functional relevance in the development of PCNSL.
Prepublished online as Blood First Edition Paper, October 30, 2003; DOI 10.1182/blood-2003-05-1465.
Supported by the Deutsche Krebshilfe/Dr Mildred-Scheel-Stiftung für Krebsforschung (grant no. 10-1641-De 1).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Ralf Küppers for helpful discussion and critical reading of the manuscript. We also thank Marek Franitza and Alexandra Kuklik for expert technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal