Abstract
The killing of natural killer (NK) cells is regulated by activating and inhibitory NK receptors that recognize mainly class I major histocompatibility complex (MHC) proteins. In transporter associated with antigen processing (TAP2)–deficient patients, killing of autologous cells by NK cells is therefore expected. However, none of the TAP2-deficient patients studied so far have suffered from immediate NK-mediated autoimmune manifestations. We have previously demonstrated the existence of a novel class I MHC–independent inhibitory mechanism of NK cell cytotoxicity mediated by the homophilic carcinoembryonic antigen–related cell adhesion molecule 1 (CEACAM1) interactions. Here, we identified 3 new siblings suffering from TAP2 deficiency. NK cells derived from these patients express unusually high levels of the various killer cell inhibitory receptors (KIRs) and the CEACAM1 protein. Importantly, the patients' NK cells use the CEACAM1 protein to inhibit the killing of tumor and autologous cells. Finally, we show that the function of the main NK lysis receptor, NKp46, is impaired in these patients. These results indicate that NK cells in TAP2-deficient patients have developed unique mechanisms to reduce NK killing activity and to compensate for the lack of class I MHC–mediated inhibition. These mechanisms prevent the attack of self-cells by the autologous NK cells and explain why TAP2-deficient patients do not suffer from autoimmune manifestations in early stages of life.
Introduction
The killing activity of target cells by human natural killer (NK) cells is mediated via a panel of lysis receptors composed of CD16, NKp30, NKp44, NKp46, and NKG2D.1-6 These receptors recognize viral ligands such as hemagglutinin,7,8 stress-induced ligands such as MHC class I chain–related antigen A (MICA) and MICB,1 or other as-yet-undefined, cellular ligands. Normal cells are protected from lysis by NK cells mainly owing to the interactions between class I MHC proteins and the appropriate inhibitory NK receptors.9-12
In addition, inhibition of NK cytotoxicity can be mediated via other receptors.13,14 We have recently identified a novel class I MHC–independent inhibitory mechanism of human NK cytotoxicity, mediated via the carcinoembryonic antigen–related cell adhesion molecule 1 (CEACAM1) homophilic interactions.15-17 Furthermore, the CEACAM1 protein plays a pivotal role in the inhibition of killing, proliferation, and cytokine secretion of interleukin 2 (IL-2)–activated decidual NK, T, and NKT cells, respectively.18
Once class I MHC proteins are removed from the cell surface, these cells become susceptible to NK cell attack.19 It was, therefore, surprising to learn that patients with transporter associated with antigen processing (TAP2) deficiency do not frequently suffer from autoimmune manifestations at early stages of their life.20 It was recently suggested that activated NK cells derived from such patients either express an unknown inhibitory mechanism or are missing an unidentified lysis receptor.20 Earlier reports concerning MHC class I–deficient mice suggested that NK tolerance toward self-cells might be controlled by similar mechanisms.21
Here, we show that the expression of the NKp46 receptor is severely impaired in a newly identified TAP2-deficient family and that the vast majority of activated NK cells derived from these patients use the CEACAM1 protein interactions to avoid tumor and autologous cell killing.
Patients, materials, and methods
Patients
Patients A, B, and C are siblings (19, 14, and 9 years old, respectively). All patients share the same human leukocyte antigen (HLA) haplotype (A*03, B*07, Bw6, Cw*07, DRB1*15, DRB5, DQB1*06). They presented with severe diffuse bronchiectasis, sinusitis, and serous otitis media with no history of severe viral infections. The Institutional Review Board of Schneider Children's Medical Center of Israel approved these studies, and informed consent was provided according to the Declaration of Helsinki.
Generation of NK clones and phytohemagglutinin (PHA)–induced T-cell blasts
Antibodies and immunoglobulin-fused proteins
The following monoclonal antibodies were used: monoclonal antibody (mAb) W6/32 and HP-1F7 directed against class I MHC molecules; anti–β2-microglobulin mAb BBM-1, anti-CEACAM1 mAb 5F4,23 anti–HLA-A3 GAP A3 mAb, anti-CD16 mAb B73.1.1, and anti-CD94 mAb HP-3D9 (Dako, Hamburg, Germany); the rabbit polyclonal anti-CEACAM1, CEACAM5, and CEACAM6 antibodies (Dako) that block the CEACAM1 interactions15,18 ; anti–killer cell inhibitory receptor 2DL1 (anti-KIR2DL1) mAb EB6 (ImmunoTech, Westbrook, ME); anti-KIR2DL2 mAb GL183 (ImmunoTech); anti–leukocyte immunoglobulin-like receptor 1 (anti-LIR1) mAb HP-F1 (a kind gift from Dr López-Botet; Immunologica, Barcelona, Spain); anti–HLA-DQ mAb G46-6 (Pharmingen, San Diego, CA); anti–HLA-DR mAb TU36 (Pharmingen); and the anti-NKG2D mAb (R&D Systems, Minneapolis, MN). The specificity of all anti-CEACAM antibodies was confirmed previously.18 The anti-NKp46 mAb 461-G1 (immunoglobulin G1 [IgG1]) was generated by immunizing mice with the NKp46-Ig fusion protein. The specificity of this mAb was determined by fluorescence-activated cell sorter (FACS) analysis on NKp46 transfectants and on NK cells (freshly isolated and IL-2 activated) (Table 3). The generation and production of fusion proteins KIR2DL2-Ig, CD99-Ig, NKp46-Ig, NKp30-Ig, and NKp44-Ig were previously described.3,7,8
Impaired expression and function of NKp46 on activated NK clones
. | Patient A . | . | Patient B . | . | Patient C . | . | Healthy sister . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | NKp46, MFI . | .221 cells killed, % . | NKp46, MFI . | .221 cells killed, % . | NKp46, MFI . | .221 cells killed, % . | NKp46, MFI . | .221 cells killed, % . | ||||
| NKp46- | 2.2 ± 0.9 (30/30) | 6.7 ± 2.9 (30/30) | 1.7 ± 0.6 (19/49) | 7.2 ± 3.7 (19/49) | 1.8 ± 0.7 (56/70) | 4.8 ± 2.1 (56/70) | N/O | N/O | ||||
| NKp46+ | N/O | N/O | 16.5 ± 6.3 (29/49) | 25 ± 5.9 (29/49) | 13.5 ± 6 (14/70) | 20.9 ± 6 (14/70) | 16.9 ± 5 (40/40) | 26.5 ± 5.3 (40/40) | ||||
. | Patient A . | . | Patient B . | . | Patient C . | . | Healthy sister . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | NKp46, MFI . | .221 cells killed, % . | NKp46, MFI . | .221 cells killed, % . | NKp46, MFI . | .221 cells killed, % . | NKp46, MFI . | .221 cells killed, % . | ||||
| NKp46- | 2.2 ± 0.9 (30/30) | 6.7 ± 2.9 (30/30) | 1.7 ± 0.6 (19/49) | 7.2 ± 3.7 (19/49) | 1.8 ± 0.7 (56/70) | 4.8 ± 2.1 (56/70) | N/O | N/O | ||||
| NKp46+ | N/O | N/O | 16.5 ± 6.3 (29/49) | 25 ± 5.9 (29/49) | 13.5 ± 6 (14/70) | 20.9 ± 6 (14/70) | 16.9 ± 5 (40/40) | 26.5 ± 5.3 (40/40) | ||||
Activated NK clones were prepared from each individual as indicated. Clones were stained for NKp46 expression by means of the 461-G1 mAb in the form of F(ab′)2 and concomitantly tested for cytotoxic activity against .221 cells. NKp46 expression is presented as the mean MFI of different NK clones ± SD. Cytotoxic activity is presented as the mean percentage of .221 cells killed by NK clones. The data represent mean percentage of cells killed ± SD. The number of NK clones included in the analysis out of total NK clones tested in each group are indicated in parentheses.
N/O indicates not observed.
Restoration of class I MHC expression by transient B-cell line fusion
Epstein-Barr virus (EBV)–transformed B-cell lines derived from the patients were mixed either with the 721.174 (a TAP– cell line),24 with the 721.45 (a TAP+ cell line with hemizygous MHC class I haplotype of HLA-A2, HLA-B5, and HLA-Cw1),25 or with EBV-transformed B-cell lines derived from other TAP1- or TAP2-deficient patients. Cells to be fused were mixed together at a 1:1 ratio at a final concentration of 10 × 106/mL and incubated for 1 hour in RPMI containing 10% fetal calf serum (FCS) and 25 μg/mL PHA-P (Sigma, St Louis, MO). Cells were pelleted and resuspended in phosphate-buffered saline (PBS) containing 50% polyethylene glycol (PEG) 1500 (Sigma), and 5% dimethyl sulfoxide (DMSO) (Sigma). After incubation at 37°C for 1 minute, cells were washed, resuspended in PBS, and incubated for a further 30 minutes at 37°C. After overnight incubation in RPMI containing 10% FCS, total fusion products were stained with GAP A3 monoclonal antibody. Cell mixtures without addition of PEG were used as negative control to rule out humoral effect.
FACS staining, generation of F(ab′)2 fragments
FACS multistaining was performed as previously described.15,18 Conjugated antibodies included Kat4c–fluorescein isothiocyanate (Kat4c-FITC) (IgG1; Dako), anti-CD56–phycoerythrin (PE) (IgG1; Dako), anti-CD3– CyChrome (IgG1; Pharmingen), anti-CD16–biotin (IgG1; Serotec, Raleigh, NC), and anti-NKG2D–PE (IgG1; R&D Systems). Controls were nonbinding isotype-matched fluorochrome-matched mAbs. Detection of immunoglobulin-fusion proteins was performed by means of the PE-conjugated secondary goat antihuman IgG antibodies with minimal cross-reaction (Jackson ImmunoResearch Laboratories, Bar Harbor, ME), as previously described.15,18 All FACS data in all of the figures and tables presented in this article were obtained with antibodies in the form of F(ab′)2. Digestion and purification of the F(ab′)2 fragments were performed with the ImmunoPure F(ab′)2 preparation kit (Pierce, Rockford, IL) according to the manufacturer's instructions.
Cytotoxicity assays
The cytotoxic activity of NK cells against the various targets was assayed in 5-hour 35S-release assays, as described previously.26 In experiments in which antibodies were included, the final mAb concentration was 20 μg/mL. In all assays performed, the spontaneous release did not exceed 25% of the maximal labeling.
Results
Identification of a new family of TAP2-deficient patients
A genetic defect in the class I MHC expression was identified in an Arab-Israeli family. Patients A, B, and C (19-year-old female, 14- and 9-year-old males, respectively) displayed clinical manifestations similar to those displayed by other TAP2-deficient patients.27 The other 5 sisters as well as the parents, who are first cousins, showed no clinical symptoms. NK clones were purified from all of the patients as well from a healthy sister. Forty NK clones from each individual were analyzed by FACS for presence of class I MHC, class II MHC, and β2-microglobulin by use of the W6/32 mAb, the TU36 mAb, and the BBM-1 mAb, respectively. A dramatic decrease in expression of class I MHC proteins and of β2-microglobulin (approximately 20-fold) was observed on the surface of NK clones derived from the patients as compared with those derived from the healthy sister (Table 1). Analysis of class II MHC protein expression revealed only 3-fold (patient A) or 2-fold (patients B and C) reduction compared with the healthy sister (Table 1). Similar results were obtained when different types of cells, such as T cells and monocytes, were used (data not shown).
Expression of MHC proteins on NK clones
Antibody . | Patient A . | Patient B . | Patient C . | Healthy sister . |
|---|---|---|---|---|
| Secondary F(ab′)2 background | 3.2 ± 0.7 | 2.2 ± 0.3 | 2.8 ± 0.4 | 3.4 ± 0.8 |
| W6/32 F(ab′)2 | 36.7 ± 8.2 | 23 ± 6.5 | 30.7 ± 8.0 | 629 ± 183 |
| BBM-1 F(ab′)2 | 7.7 ± 2.5 | 5.3 ± 1.7 | 7.1 ± 1.6 | 130 ± 30 |
| MHC class II F(ab′)2 | 33.1 ± 3.6 | 54.3 ± 9.6 | 54.6 ± 7.7 | 94.7 ± 9.1 |
Antibody . | Patient A . | Patient B . | Patient C . | Healthy sister . |
|---|---|---|---|---|
| Secondary F(ab′)2 background | 3.2 ± 0.7 | 2.2 ± 0.3 | 2.8 ± 0.4 | 3.4 ± 0.8 |
| W6/32 F(ab′)2 | 36.7 ± 8.2 | 23 ± 6.5 | 30.7 ± 8.0 | 629 ± 183 |
| BBM-1 F(ab′)2 | 7.7 ± 2.5 | 5.3 ± 1.7 | 7.1 ± 1.6 | 130 ± 30 |
| MHC class II F(ab′)2 | 33.1 ± 3.6 | 54.3 ± 9.6 | 54.6 ± 7.7 | 94.7 ± 9.1 |
Forty NK clones were isolated from each of the 3 patients and from the healthy sister. Clones were stained by various mAbs as indicated and analyzed by FACS. All antibodies used were in the form of F(ab′)2 fragments. Background was the secondary mAb in the form of F(ab′)2. Data are presented as the median fluorescence intensity (MFI) of 40 clones ± standard deviation (SD). All 160 clones were stained at the same time.
To identify the genetic defect responsible for the impaired expression of class I MHC proteins observed in these patients, an EBV-transformed B-cell line was made from each patient (EBV-A, EBV-B, and EBV-C) and from their healthy mother (EBV-mother). The EBV-transformed B-cell lines were further analyzed for the expression of class I MHC, class II MHC, β2-microglobulin, and HLA-A3 by using the W6/32 mAb, the TU36 and G46-6 mAbs, BBM-1 mAb, and the GAP A3 mAb, respectively. In agreement with the results described in the preceding paragraph, a dramatic down-regulation (approximately 20-fold) of class I MHC proteins and of β2-microglobulin surface expression was observed on EBV-A, EBV-B, and EBV-C as compared with EBV-mother (Table 2). There was no significant difference in the expression of class II MHC proteins, such as HLA-DQ and HLA-DR (Table 2). Notably, the HLA-A3 protein was completely absent from the surface of EBV-A, EBV-B, and EBV-C, but not from EBV-mother (all patients and their mother express HLA-A3; see “Patients, materials, and methods”). There is, however, a slight expression of class I MHC detected by W6/32, indicating that class I MHC proteins other than HLA-A3 are still expressed in low levels on the patient EBV cells (Table 2).
Expression of MHC proteins on EBV-transformed B-cell lines
Cells . | Background . | W6/32 . | BBM-1 . | HLA-DQ . | HLA-DR . | HLA-A3 . |
|---|---|---|---|---|---|---|
| EBV-A | 2.5 ± 0.5 | 122.5 ± 4.9 | 12.5 ± 2.1 | 22.5 ± 2.1 | 91.5 ± 23.3 | 2.5 ± 0.5 |
| EBV-B | 3 ± 0.5 | 106.5 ± 2.1 | 8 ± 4.2 | 24 ± 2.8 | 99.5 ± 14.8 | 2.8 ± 0.6 |
| EBV-C | 2 ± 0.4 | 76 ± 1.4 | 7.35 ± 2.9 | 29.5 ± 4.9 | 114.5 ± 17.7 | 3.0 ± 0.5 |
| EBV-mother | 3.5 ± 0.5 | 1300 ± 141 | 155 ± 35 | 35 ± 7.1 | 122.5 ± 10.6 | 110 ± 15 |
Cells . | Background . | W6/32 . | BBM-1 . | HLA-DQ . | HLA-DR . | HLA-A3 . |
|---|---|---|---|---|---|---|
| EBV-A | 2.5 ± 0.5 | 122.5 ± 4.9 | 12.5 ± 2.1 | 22.5 ± 2.1 | 91.5 ± 23.3 | 2.5 ± 0.5 |
| EBV-B | 3 ± 0.5 | 106.5 ± 2.1 | 8 ± 4.2 | 24 ± 2.8 | 99.5 ± 14.8 | 2.8 ± 0.6 |
| EBV-C | 2 ± 0.4 | 76 ± 1.4 | 7.35 ± 2.9 | 29.5 ± 4.9 | 114.5 ± 17.7 | 3.0 ± 0.5 |
| EBV-mother | 3.5 ± 0.5 | 1300 ± 141 | 155 ± 35 | 35 ± 7.1 | 122.5 ± 10.6 | 110 ± 15 |
EBV-transformed B-cell lines were made from the indicated individuals. Cell lines were stained by various mAbs in the form of F(ab′)2 as indicated and analyzed by FACS. Background was the secondary mAb in the form of F(ab′)2. Data are presented as the average MFI of 3 independent experiments ± SD.
Since HLA-A3 was not detected on the patients' EBV cells, we next used an assay that is based on the restoration of HLA-A3 expression following correction of the class I MHC biosynthetic pathway via PEG-mediated fusion of EBV-A, EBV-B, or EBV-C with various cell lines. Importantly, the 721.45 cells that were used in some of the fusion experiments (see next paragraph), are hemizygous for class I MHC expression (HLA-A2, HLA-B5, and HLA-Cw1). Therefore, specific monitoring is required to discriminate between the class I MHC reconstitution (monitored by GAP A3) and the endogenous expression of the class I MHC proteins on 721.45 cells.
Cells were fused, and the total cell mixture was analyzed with the use of the GAP A3 mAb. As not all mixed cells were fused, the fused cells were identified by the presence of HLA-A3 protein. Fusion of EBV-A, EBV-B, or EBV-C either with the 721.45 cell line that expresses both TAP1 and TAP2 subunits or with cells deficient in TAP1, resulted in the emergence of an HLA-A3+ population (Figure 1). In contrast, fusion of EBV-A, EBV-B, or EBV-C with 721.174 cell line (.174), deficient for TAP1 and TAP2 or with B cells derived from other TAP2-deficient patients, failed to restore HLA-A3 expression (Figure 1). The fact that mixture of EBV-A, EBV-B, or EBV-EBV-C with the various cell lines without addition of PEG failed to reconstitute HLA-A3 expression (data not shown) and in addition the fact that some of the fusions performed did not restore HLA-A3 expression (Figure 1) rule out the possibility of humoral effect. These results demonstrate that the genetic defect is in the TAP2 protein.
The reduction in class I MHC expression is due to TAP2 deficiency. Fusion of EBV-A, EBV-B, and EBV-C with various B-cell lines defective either for the TAP1 and TAP2 subunits (.174) or none of them (.45). Total mixture of cells was analyzed by FACS. Fused cells were identified by HLA-A3 expression. Staining with HLA-A3 is on the y-axis, and forward scatter is on the x-axis. One representative experiment is shown of 3 performed.
The reduction in class I MHC expression is due to TAP2 deficiency. Fusion of EBV-A, EBV-B, and EBV-C with various B-cell lines defective either for the TAP1 and TAP2 subunits (.174) or none of them (.45). Total mixture of cells was analyzed by FACS. Fused cells were identified by HLA-A3 expression. Staining with HLA-A3 is on the y-axis, and forward scatter is on the x-axis. One representative experiment is shown of 3 performed.
Impaired expression and function of NKp46
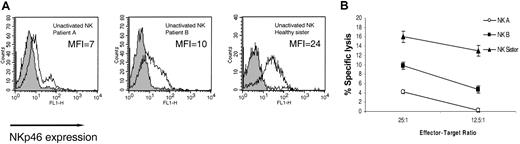
NKp46 is considered to be the main NK killing receptor for NK cells and is uniquely expressed on all NK cells.28 Expression of NKp46 on freshly isolated NK cells was monitored by using an anti-NKp46 mAb (461-G1). In congruence with previous reports,28 relatively low levels of NKp46 were observed on the bulk NK cells isolated from the healthy sister (MFI = 24) (Figure 2A). Sadly, at this stage blood samples could no longer be obtained from patient C owing to the severity of his clinical conditions. Strikingly, the NKp46 receptor could hardly be detected on the surface of more than 85% of the NK cells isolated from patient A (MFI = 7) and on more than 60% of the NK cells isolated from patient B (MFI = 10) (Figure 2A). The function of the NKp46 receptor was assayed concomitantly against 721.221 cells, in which the lysis is controlled by the NKp46 receptor.5 In accordance with the staining results (Figure 2A), very low killing was observed with bulk freshly isolated NK cells derived from patient A; relatively moderate killing was observed with NK cells from patient B; and relatively efficient killing was observed with NK cells from the healthy sister (Figure 2B).
Impaired expression and function of NKp46 on freshly isolated NK cells. (A) NKp46 expression on freshly isolated bulk NK cells. Staining was detected by mAb 461-G1 in the form of F(ab′)2, and the MFI staining is indicated in each histogram. One representative experiment is shown of 3 performed. (B) Killing of .221 cells by freshly isolated NK cells derived from indicated donors. The mean results of 3 independent experiments are shown. The data represent means of the percentage of killing ± SDs.
Impaired expression and function of NKp46 on freshly isolated NK cells. (A) NKp46 expression on freshly isolated bulk NK cells. Staining was detected by mAb 461-G1 in the form of F(ab′)2, and the MFI staining is indicated in each histogram. One representative experiment is shown of 3 performed. (B) Killing of .221 cells by freshly isolated NK cells derived from indicated donors. The mean results of 3 independent experiments are shown. The data represent means of the percentage of killing ± SDs.
We next generated IL-2–activated NK clones from the patients and from the healthy sister. In agreement with previous observations,5 no major difference in the NKp46 expression was observed between freshly isolated and IL-2–activated NK cells (Figure 2A; Table 3). Activated NK clones were analyzed for NKp46 expression and for cytotoxicity against .221 target cells. An impressive reduction in the NKp46 expression was observed in NK clones derived from all of the patients as compared with the healthy sister. All 30 NK clones (100%) derived from patient A did not express the NKp46 protein; 19 (39%) of 49 clones and 56 (80%) of 70 clones derived from patient B and patient C, respectively, were also NKp46– (Table 3). Importantly, the absence of the NKp46 receptor on these NK clones was correlated with poor cytolytic activity against .221 cells (Table 3). NKp46– clones were not observed in the healthy sister (Table 3). On the other hand, 29 (61%) of 49 NK clones derived from patient B, 14 (20%) of 70 from patient C, and 30 (100%) of 30 from the healthy sister were NKp46+ (Table 3). The level of the NKp46 expression was similar in all positive clones (Table 3). Accordingly, the NKp46+ NK clones displayed efficient cytotoxic activity against .221 target cells (Table 3). Efficient killing of other cells types such as EBV-A, EBV-B, EBV-C, 293T, or RPMI 8866 was also observed when assayed against the NKp46+ clones (data not shown).
Activated NK clones derived from the TAP2-deficient patients express unusually high levels of CEACAM1- and class I MHC–recognizing receptors
The results described in the preceding section demonstrate that the killing of targets by NK cells derived from the TAP2-deficient patients can be reduced by the diminished NKp46 expression. However, 61% and 20% of the clones in patients B and C, respectively, still expressed NKp46 (Table 3). We therefore hypothesized the existence of a class I MHC–independent inhibitory mechanism in our patients that controls NK autoreactivity and examined whether CEACAM1 interactions are involved in controlling the killing activity of activated NK cells in TAP2-deficient patients.
Peripheral blood lymphocytes (PBLs) were isolated from all 3 patients and the healthy sister and analyzed by multistaining for the expression of the CD3, CD16, CD56, and CEACAM1. All patients had normal values of lymphocytes in their peripheral blood and a normal lymphocyte distribution, including T, NK, and NKT cells (data not shown). Thus, the low levels of class I MHC proteins (Tables 1, 2) were probably sufficient to select for the development of normal numbers of lymphocytes. PBLs from all 4 donors were also tested for the expression of the CEACAM1 protein. In agreement with previous reports,15,18,23,29 little or no expression of the CEACAM1 protein was observed among all fresh PBLs (data not shown).
Activated NK clones were generated from the 3 patients and from the healthy sister. Sixty NK clones from each individual were assessed for CEACAM1 expression by using the 5F4 mAb. The NK clones from each individual were subgrouped according to the CEACAM1 expression level (negative, low, or high). Low levels of CEACAM1 expression (MFI around 8) had already been observed on NK clones and proved sufficient to confer protection.15,18 However, never before had we observed high expression level of CEACAM1 (MFI around 30) on the surface of NK clones. As previously reported,15,18 53 (88%) of 60 NK clones obtained from the healthy sister were negative for CEACAM1 (Table 4). In contrast, virtually all of the NK clones (98%) obtained from patient A expressed the CEACAM1 protein in unusually high levels. Of the 60 NK clones obtained from patient B, 43 (71%) expressed the CEACAM1 protein in low or high levels (43% and 28%, respectively; Table 4) whereas 44 (73%) of 60 NK cells derived from patient C expressed CEACAM1 in low or high levels (23% and 50%, respectively; Table 4).
Analysis of various NK receptors in accordance with CEACAM1 expression
CEACAM1 expression and receptor staining intensity . | Background . | CD16 . | KIR2DL1 . | KIR2DL2 . | CD94 . | LIR1 . |
|---|---|---|---|---|---|---|
| Patient A | ||||||
| Negative CEACMA1 expressiona | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 2.5 (1/1) | 2.5 (1/1) | N/O | 3 (1/1) |
| Dim | 2 ± 0.5 | 28 (1/1) | N/O | N/O | 48 (1/1) | N/O |
| Bright | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Low CEACMA1 expressionb | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Dim | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Bright | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| High CEACMA1 expressionc | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 5 ± 1 (9/59) | 3 ± 2 (22/59) | 4 ± 1 (20/59) | 4 ± 0 (3/59) | 3 ± 1 (19/59) |
| Dim | 2 ± 0.5 | 15 ± 4 (50/59) | 21 ± 5 (13/59) | 18 ± 12 (18/59) | 19 ± 5 (26/59) | 18 ± 9 (33/59) |
| Bright | 2 ± 0.5 | N/O | 151 ± 39 (24/59) | 198 ± 79 (21/59) | 98 ± 34 (30/59) | 104 ± 13 (7/59) |
| Sum of positive clones (%) | NA | 50/60 (83) | 37/60 (62) | 39/60 (65) | 57/60 (95) | 40/60 (67) |
| Patient B | ||||||
| Negative CEACMA1 expressiond | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 3 ± 0 (2/17) | 2 ± 0.5 (7/17) | N/O | 4 ± 1 (6/17) |
| Dim | 2 ± 0.5 | 15 ± 5 (4/17) | 30 ± 16 (6/17) | 9 ± 2 (2/17) | 24 ± 10 (7/17) | 15 ± 3 (9/17) |
| Bright | 2 ± 0.5 | 161 ± 70 (13/17) | 122 ± 37 (9/17) | 222 ± 77 (8/17) | 95 ± 23 (10/17) | 88 ± 15 (2/17) |
| Low CEACMA1 expressione | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 2 ± 0.4 (9/26) | 3 ± 1 (11/26) | 4 (1/26) | 3 ± 1 (8/26) |
| Dim | 2 ± 0.5 | 20 ± 8 (12/26) | 29 ± 10 (4/26) | 12 ± 3 (3/26) | 18 ± 6 (9/26) | 11 ± 3 (14/26) |
| Bright | 2 ± 0.5 | 142 ± 70 (14/26) | 149 ± 67 (13/26) | 274 ± 140 (12/26) | 90 ± 21 (16/26) | 100 (4/26) |
| High CEACMA1 expressionf | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 3 (1/17) | 2 ± 1 (3/17) | 2 ± 0 (3/17) | 4 (1/17) | 4 ± 1 (5/17) |
| Dim | 2 ± 0.5 | 38 ± 16 (6/17) | 31 ± 15 (4/17) | N/O | 28 ± 13 (8/17) | 10 ± 2 (9/17) |
| Bright | 2 ± 0.5 | 115 ± 47 (10/17) | 168 ± 55 (10/17) | 209 ± 54 (14/17) | 82 ± 15 (8/17) | 93 ± 25 (3/17) |
| Sum of positive clones (%) | NA | 59/60 (98) | 46/60 (77) | 39/60 (65) | 58/60 (97) | 41/60 (68) |
| Patient C | ||||||
| Negative CEACMA1 expressiong | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 2 (1/16) | 3 ± 1 (10/16) | 3 ± 1 (3/16) | 3 ± 1 (2/16) | 3 ± 1 (7/16) |
| Dim | 2 ± 0.5 | 16 ± 10 (13/16) | 43 ± 13 (3/16) | 73 ± 22 (3/16) | 35 ± 9 (14/16) | 9 ± 3 (9/16) |
| Bright | 2 ± 0.5 | 72 ± 4 (2/16) | 149 ± 73 (3/16) | 184 ± 65 (10/16) | N/O | N/O |
| Low CEACMA1 expressionh | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 4 ± 0 (2/14) | 5 ± 0.4 (3/14) | 4 ± 1 (3/14) | 3 ± 2 (1/14) | 3 ± 1 (6/14) |
| Dim | 2 ± 0.5 | 18 ± 9 (8/14) | 44 ± 16 (6/14) | 82 ± 9 (7/14) | 40 ± 13 (13/14) | 9 ± 2 (8/14) |
| Bright | 2 ± 0.5 | 88 ± 32 (4/14) | 146 ± 43 (5/14) | 321 ± 23 (4/14) | N/O | N/O |
| High CEACMA1 expressioni | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 4 ± 2 (5/30) | 4 ± 2 (4/30) | 4 ± 1 (3/30) | 3 ± 1 (3/30) | 3 ± 2 (11/30) |
| Dim | 2 ± 0.5 | 20 ± 11 (23/30) | 27 ± 10 (1/30) | 63 ± 18 (4/30) | 37 ± 15 (27/30) | 10 ± 3 (19/30) |
| Bright | 2 ± 0.5 | 88 ± 10 (2/30) | 188 ± 81 (25/30) | 267 ± 102 (23/30) | N/O | N/O |
| Sum of positive clones (%) | NA | 52/60 (87) | 43/60 (72) | 51/60 (85) | 55/60 (92) | 36/60 (60) |
| Healthy sister | ||||||
| Negative CEACMA1 expressionj | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 3 ± 2 (38/53) | 3 ± 1 (33/53) | N/O | 3 ± 1 (46/53) |
| Dim | 2 ± 0.5 | 45 ± 6 (53/53) | 50 ± 23 (15/53) | 27 ± 14 (5/53) | 26 ± 12 (36/53) | 10 ± 3 (7/53) |
| Bright | 2 ± 0.5 | N/O | N/O | 208 ± 97 (15/53) | 80 ± 25 (17/53) | N/O |
| Low CEACMA1 expressionk | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 4 ± 1 (6/7) | 3 ± 2 (7/7) | 5 ± 2 (6/7) | N/O | 3 ± 1 (5/7) |
| Dim | 2 ± 0.5 | 40 (1/7) | N/O | N/O | 16 ± 3 (4/7) | 10 ± 2 (2/7) |
| Bright | 2 ± 0.5 | N/O | N/O | 108 (1/7) | 71 ± 8 (3/7) | N/O |
| High CEACMA1 expressionb | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Dim | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Bright | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Sum of positive clones (%) | NA | 54/60 (90) | 15/60 (25) | 21/60 (35) | 60/60 (100) | 9/60 (15) |
CEACAM1 expression and receptor staining intensity . | Background . | CD16 . | KIR2DL1 . | KIR2DL2 . | CD94 . | LIR1 . |
|---|---|---|---|---|---|---|
| Patient A | ||||||
| Negative CEACMA1 expressiona | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 2.5 (1/1) | 2.5 (1/1) | N/O | 3 (1/1) |
| Dim | 2 ± 0.5 | 28 (1/1) | N/O | N/O | 48 (1/1) | N/O |
| Bright | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Low CEACMA1 expressionb | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Dim | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Bright | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| High CEACMA1 expressionc | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 5 ± 1 (9/59) | 3 ± 2 (22/59) | 4 ± 1 (20/59) | 4 ± 0 (3/59) | 3 ± 1 (19/59) |
| Dim | 2 ± 0.5 | 15 ± 4 (50/59) | 21 ± 5 (13/59) | 18 ± 12 (18/59) | 19 ± 5 (26/59) | 18 ± 9 (33/59) |
| Bright | 2 ± 0.5 | N/O | 151 ± 39 (24/59) | 198 ± 79 (21/59) | 98 ± 34 (30/59) | 104 ± 13 (7/59) |
| Sum of positive clones (%) | NA | 50/60 (83) | 37/60 (62) | 39/60 (65) | 57/60 (95) | 40/60 (67) |
| Patient B | ||||||
| Negative CEACMA1 expressiond | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 3 ± 0 (2/17) | 2 ± 0.5 (7/17) | N/O | 4 ± 1 (6/17) |
| Dim | 2 ± 0.5 | 15 ± 5 (4/17) | 30 ± 16 (6/17) | 9 ± 2 (2/17) | 24 ± 10 (7/17) | 15 ± 3 (9/17) |
| Bright | 2 ± 0.5 | 161 ± 70 (13/17) | 122 ± 37 (9/17) | 222 ± 77 (8/17) | 95 ± 23 (10/17) | 88 ± 15 (2/17) |
| Low CEACMA1 expressione | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 2 ± 0.4 (9/26) | 3 ± 1 (11/26) | 4 (1/26) | 3 ± 1 (8/26) |
| Dim | 2 ± 0.5 | 20 ± 8 (12/26) | 29 ± 10 (4/26) | 12 ± 3 (3/26) | 18 ± 6 (9/26) | 11 ± 3 (14/26) |
| Bright | 2 ± 0.5 | 142 ± 70 (14/26) | 149 ± 67 (13/26) | 274 ± 140 (12/26) | 90 ± 21 (16/26) | 100 (4/26) |
| High CEACMA1 expressionf | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 3 (1/17) | 2 ± 1 (3/17) | 2 ± 0 (3/17) | 4 (1/17) | 4 ± 1 (5/17) |
| Dim | 2 ± 0.5 | 38 ± 16 (6/17) | 31 ± 15 (4/17) | N/O | 28 ± 13 (8/17) | 10 ± 2 (9/17) |
| Bright | 2 ± 0.5 | 115 ± 47 (10/17) | 168 ± 55 (10/17) | 209 ± 54 (14/17) | 82 ± 15 (8/17) | 93 ± 25 (3/17) |
| Sum of positive clones (%) | NA | 59/60 (98) | 46/60 (77) | 39/60 (65) | 58/60 (97) | 41/60 (68) |
| Patient C | ||||||
| Negative CEACMA1 expressiong | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 2 (1/16) | 3 ± 1 (10/16) | 3 ± 1 (3/16) | 3 ± 1 (2/16) | 3 ± 1 (7/16) |
| Dim | 2 ± 0.5 | 16 ± 10 (13/16) | 43 ± 13 (3/16) | 73 ± 22 (3/16) | 35 ± 9 (14/16) | 9 ± 3 (9/16) |
| Bright | 2 ± 0.5 | 72 ± 4 (2/16) | 149 ± 73 (3/16) | 184 ± 65 (10/16) | N/O | N/O |
| Low CEACMA1 expressionh | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 4 ± 0 (2/14) | 5 ± 0.4 (3/14) | 4 ± 1 (3/14) | 3 ± 2 (1/14) | 3 ± 1 (6/14) |
| Dim | 2 ± 0.5 | 18 ± 9 (8/14) | 44 ± 16 (6/14) | 82 ± 9 (7/14) | 40 ± 13 (13/14) | 9 ± 2 (8/14) |
| Bright | 2 ± 0.5 | 88 ± 32 (4/14) | 146 ± 43 (5/14) | 321 ± 23 (4/14) | N/O | N/O |
| High CEACMA1 expressioni | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 4 ± 2 (5/30) | 4 ± 2 (4/30) | 4 ± 1 (3/30) | 3 ± 1 (3/30) | 3 ± 2 (11/30) |
| Dim | 2 ± 0.5 | 20 ± 11 (23/30) | 27 ± 10 (1/30) | 63 ± 18 (4/30) | 37 ± 15 (27/30) | 10 ± 3 (19/30) |
| Bright | 2 ± 0.5 | 88 ± 10 (2/30) | 188 ± 81 (25/30) | 267 ± 102 (23/30) | N/O | N/O |
| Sum of positive clones (%) | NA | 52/60 (87) | 43/60 (72) | 51/60 (85) | 55/60 (92) | 36/60 (60) |
| Healthy sister | ||||||
| Negative CEACMA1 expressionj | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | 3 ± 2 (38/53) | 3 ± 1 (33/53) | N/O | 3 ± 1 (46/53) |
| Dim | 2 ± 0.5 | 45 ± 6 (53/53) | 50 ± 23 (15/53) | 27 ± 14 (5/53) | 26 ± 12 (36/53) | 10 ± 3 (7/53) |
| Bright | 2 ± 0.5 | N/O | N/O | 208 ± 97 (15/53) | 80 ± 25 (17/53) | N/O |
| Low CEACMA1 expressionk | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | 4 ± 1 (6/7) | 3 ± 2 (7/7) | 5 ± 2 (6/7) | N/O | 3 ± 1 (5/7) |
| Dim | 2 ± 0.5 | 40 (1/7) | N/O | N/O | 16 ± 3 (4/7) | 10 ± 2 (2/7) |
| Bright | 2 ± 0.5 | N/O | N/O | 108 (1/7) | 71 ± 8 (3/7) | N/O |
| High CEACMA1 expressionb | ||||||
| Receptor staining intensity | ||||||
| Negative | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Dim | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Bright | 2 ± 0.5 | N/O | N/O | N/O | N/O | N/O |
| Sum of positive clones (%) | NA | 54/60 (90) | 15/60 (25) | 21/60 (35) | 60/60 (100) | 9/60 (15) |
NK clones were prepared from the indicated individuals. Sixty NK clones from each donor were analyzed for CEACAM1 expression and were classified into 3 groups according to CEACAM1 intensity (negative, low, and high). All NK clones in each CEACAM1 subgroup were further stained for various NK receptors and were subdivided according to intensity of the expression of the particular receptor (negative, dim, and bright). The mean MFI of the various groups ± SD is presented, with the number of the relevant NK clones of the total number of NK clones in each CEACAM1 subgroup indicated in parentheses. The sum of all positive (dim and bright) clones in each subgroup is indicated for each patient, with the percentage indicated in parentheses (total number of clones for each patient is 60). All staining was performed with antibodies in the form of F(ab′)2 and at the same time. Data are presented in all groups as means of MFI ± SD.
N/O indicates not observed; and NA, not available.
MFI = 3 (1/60 = 2%).
MFI = N/O.
MFI = 32 ± 13 (59/60 = 98%).
MFI = 2 ± 0.6 (17/60 = 29%).
MFI = 8 ± 2 (26/60 = 43%).
MFI = 25 ± 7 (17/60 = 28%).
MFI = 2 ± 0.6 (16/60 = 27%).
MFI = 8 ± 2 (14/60 = 23%).
MFI = 31 ± 13 (30/60 = 50%).
MFI = 3 ± 1 (53/60 = 88%).
MFI = 8 ± 1 (7/60 = 12%).
In addition, all of the NK clones were stained for the presence of CD16, KIR2DL1, KIR2DL2, CD94, or LIR1. In each individual, the total NK clones were further subclassified in each CEACAM1 subgroup according to the staining intensity of each receptor (negative, dim, and bright), and the mean MFI ± SD was calculated accordingly. We compared the overall percentages of NK clones from each individual expressing KIR2DL1, KIR2DL2, and LIR1. KIR2DL1 was expressed on 62%, 77%, and 72% of the NK clones obtained from patients A, B, and C, respectively, compared with only 25% of the NK clones from the healthy sister (Table 4). KIR2DL2 was expressed on 65%, 65%, and 85% of the NK clones obtained from patients A, B, and C, respectively, compared with only 35% of the NK clones from the healthy sister (Table 4). Finally, the LIR1 was expressed on 67%, 68%, and 60% of the NK clones obtained from patients A, B, and C, respectively, compared with only 15% of the NK clones from the healthy sister (Table 4). Thus, expression of all inhibitory NK receptors tested was up-regulated on the NK cells derived from the patients. Importantly, the increase in the percentage of NK clones derived from the patients that express class I MHC–recognizing inhibitory receptors is statistically significant when compared with the healthy sister: KIR2DL1 (P = .01), KIR2DL2 (P = .04), and LIR1 (P = .02). Further analysis reveals that the receptor expression level is also increased among NK clones obtained from the patients as compared with those obtained from the healthy sister. Bright expression of KIR2DL1 was observed on 24 of 60, 32 of 60, and 33 of 60 NK clones obtained from patients A, B, and C, respectively, but not on any of the 60 NK clones obtained from the healthy sister (P = .003) (Table 4). Similarly, bright expression of KIR2DL2 was observed on 21 of 60, 34 of 60, and 37 of 60 NK clones obtained from patients A, B, and C, respectively, as opposed to only 16 of 60 obtained from the healthy sister (P = .06) (Table 4). It is still largely unknown which factors determine the specific expression of the various NK receptors in a given individual. Our patients express the Cw7 protein, which is recognized by KIR2DL2.30 It is interesting to learn that a statistically significant bright expression of KIR2DL1 but not KIR2DL2 was observed in the patients' NK clones, suggesting that the expression level of the various NK receptors is somehow shaped by the appropriate MHC proteins (Table 4). Thus, the low levels of class I MHC proteins in the patients have resulted in an impaired repertoire of inhibitory receptors, manifested not only in the increased percentages of positive clones but also in the higher expression levels.
In contrast to these results, expression of the CD94 receptor was observed on 95%, 97%, and 92% of the NK clones obtained from patients A, B, and C, respectively, and on 100% of the NK clones from the healthy sister (P = .66) (Table 4). We have previously demonstrated that expression of CEACAM1 is confined mainly to the CD16– subset of NK cells.15,18 Expression of CD16 was observed on 83%, 98%, and 87% of the NK clones obtained from patients A, B, and C, respectively, and on 90% of the NK clones from the healthy sister (P = .29) (Table 4). Notably, however, CEACAM1 expression on NK clones derived from the healthy sister was restricted mainly to the CD16– cells (6 of 7 CEACAM1dim NK clones; Table 4). In contrast, 50 of 59, 42 of 43, and 37 of 44 NK clones derived from patients A, B, and C, respectively, expressed both CD16 and CEACAM1 (P = .007) (Table 4). Thus, an as-yet-unknown mechanism permits the expression of CEACAM1 on both CD16– and CD16+ NK clones derived from the patients (Table 4).
CEACAM1 interactions protect autologous PHA-induced T-cell blasts from NK cell–mediated killing
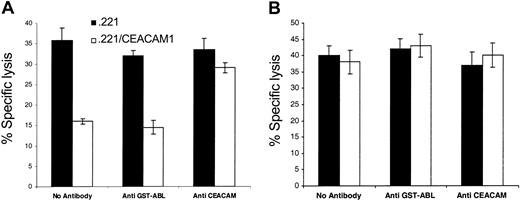
The functional significance of CEACAM1 expression was assayed with clones capable of killing .221 cells (mainly from patient B; Table 3). As the CEACAM1 protein binds via homophilic interactions to other CEACAM1 proteins,15,18,31,32 the various NK clones were tested for killing against .221 cells expressing the CEACAM1 protein.15,18 Inhibition of killing was observed when CEACAM1+ NK clones were used (representative clone in Figure 3A). This inhibition was the result of CEACAM1 interactions, as lysis was restored when anti-CEACAM F(ab′)2 antibodies were included in the assay (Figure 3). No inhibition was observed when CEACAM1– NK clones were used (Figure 3B).
Inhibition of NK-mediated killing by homophilic CEACAM1 interactions. Killing of .221 and .221/CEACAM1 cells, incubated with or without polyclonal anti-CEACAM antibodies, by a representative CEACAM1+ NK clone (panel A) or by a CEACAM1– NK clone (panel B). As control, anti–glutathion S-transferase (GST)–ABL polyclonal antibodies were used. The effector-to-target (E/T) ratio was 2:1. All antibodies used were in the form of F(ab′)2. Figures show the average of 3 independent experiments. The data represent means of the percentage of killing ± SDs.
Inhibition of NK-mediated killing by homophilic CEACAM1 interactions. Killing of .221 and .221/CEACAM1 cells, incubated with or without polyclonal anti-CEACAM antibodies, by a representative CEACAM1+ NK clone (panel A) or by a CEACAM1– NK clone (panel B). As control, anti–glutathion S-transferase (GST)–ABL polyclonal antibodies were used. The effector-to-target (E/T) ratio was 2:1. All antibodies used were in the form of F(ab′)2. Figures show the average of 3 independent experiments. The data represent means of the percentage of killing ± SDs.
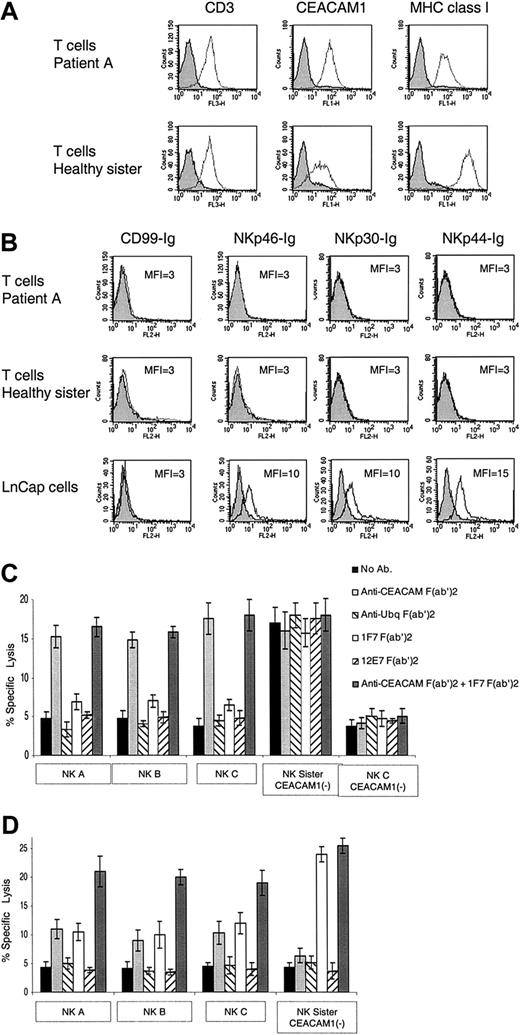
We next tested whether normal, nonvirally infected, PHA-induced T-cell blasts will be killed by the patients' NK cells. In agreement with previous reports demonstrating that activated T cells express the CEACAM1 protein,23,29 expression of CEACAM1 was observed on all PHA-induced T-cell blasts derived from all patients (Figure 4A). The expression level of the CEACAM1 protein on the PHA-induced T-cell blasts derived from the TAP2-deficient patients was approximately 5-fold higher as compared with the PHA-induced T-cell blasts obtained from the healthy sister (Figure 4A). In agreement with the results presented in Tables 1 and 2, low levels of class I MHC protein expression were observed on PHA-induced T-cell blasts derived from patients compared with the healthy sister (Figure 4A). Staining of the various PHA-induced T-cell blasts for the presence of ligands for the NKp46, NKp44, and NKp30 receptors with the use of immunoglobulin-fusion proteins was negative (Figure 4B). NKp46-Ig, NKp30-Ig, and NKp44-Ig did, however, recognize tumor targets such as LnCap (Figure 4B) or other cell lines.7,8 These results suggest that the cellular ligands for these receptors either are not expressed or are expressed in low levels on the surface of the PHA-induced T-cell blasts. Indeed, the expression of other lysis ligands such as MICA was previously reported to be present on the PHA-induced T-cell blasts.33
Killing of PHA-induced T-cell blasts. (A) Staining of PHA-induced T-cell blasts with various mAbs. Staining of PHA-induced T-cell blasts derived from patient A and from the healthy sister was performed with the F(ab′)2 fragments of anti-CD3, anti-CEACAM1, and anti–MHC class I mAb HP-1F7. (B) Staining of PHA-induced T-cell blasts and of the LnCap cell line with various fusion proteins. Staining was performed with the NKp46-Ig, NKp30-Ig, NKp44-Ig, and the control CD99-Ig fusion proteins. (C) NK clones derived from patients A, B, and C were assayed for cytotoxic activity against autologous PHA-induced T-cell blasts. The NK clones obtained from the healthy sister were assayed against PHA-induced T-cell blasts derived from patient A. NK clones were preincubated with or without F(ab′)2 fragments of polyclonal anti-CEACAM or the control polyclonal antiubiquitin antibodies. The targets, autologous PHA-induced T-cell blasts, were incubated with or without the F(ab′)2 fragments of HP-1F7 or the control 12E7 mAb. Assays were performed at an E/T ratio of 2:1. Shown are the mean results of several NK clones that were obtained from 3 independent experiments. The data represent the mean percentage of killing ± SD. (D) NK clones derived either from the healthy sister or from patients A, B, and C were assayed for killing of PHA-induced T-cell blasts derived from the healthy sister. NK clones and target PHA-induced T-cell blasts were pretreated as described for panel C. Assays were performed at an E/T ratio of 2:1. Shown are the mean results of several NK clones that were obtained from 3 independent experiments. All mAbs used were in the form of F(ab′)2. The data represent the mean percentage of killing ± SD.
Killing of PHA-induced T-cell blasts. (A) Staining of PHA-induced T-cell blasts with various mAbs. Staining of PHA-induced T-cell blasts derived from patient A and from the healthy sister was performed with the F(ab′)2 fragments of anti-CD3, anti-CEACAM1, and anti–MHC class I mAb HP-1F7. (B) Staining of PHA-induced T-cell blasts and of the LnCap cell line with various fusion proteins. Staining was performed with the NKp46-Ig, NKp30-Ig, NKp44-Ig, and the control CD99-Ig fusion proteins. (C) NK clones derived from patients A, B, and C were assayed for cytotoxic activity against autologous PHA-induced T-cell blasts. The NK clones obtained from the healthy sister were assayed against PHA-induced T-cell blasts derived from patient A. NK clones were preincubated with or without F(ab′)2 fragments of polyclonal anti-CEACAM or the control polyclonal antiubiquitin antibodies. The targets, autologous PHA-induced T-cell blasts, were incubated with or without the F(ab′)2 fragments of HP-1F7 or the control 12E7 mAb. Assays were performed at an E/T ratio of 2:1. Shown are the mean results of several NK clones that were obtained from 3 independent experiments. The data represent the mean percentage of killing ± SD. (D) NK clones derived either from the healthy sister or from patients A, B, and C were assayed for killing of PHA-induced T-cell blasts derived from the healthy sister. NK clones and target PHA-induced T-cell blasts were pretreated as described for panel C. Assays were performed at an E/T ratio of 2:1. Shown are the mean results of several NK clones that were obtained from 3 independent experiments. All mAbs used were in the form of F(ab′)2. The data represent the mean percentage of killing ± SD.
CEACAM1+ NK clones from each patient were assayed for lysis against the various PHA-induced T-cell blasts. All CEACAM1– NK clones and bulk cultures derived from the healthy sister killed the TAP2-deficient PHA-induced T-cell blasts (see, eg, “NK Sister CEACAM1–” in Figure 4C). Similar results were obtained with bulk NK cells and clones obtained from other healthy donors (data not shown). Remarkably, none of the tested NK clones or bulk cultures derived from the patients killed their autologous PHA-induced T-cell blasts (see, eg, NK A, NK B, and NK C in Figure 4C). To test whether the lack of self-killing is because of CEACAM1- or class I MHC–mediated inhibition, blocking antibodies were included in the assay. The PHA-induced T-cell blasts were preincubated with or without the anti-CEACAM antibodies, the control antiubiquitin antibodies, HP-1F7 mAb, or the control 12E7 mAb. A significant enhancement of the killing activity of the patients' NK clones (A, B, and C) was observed when the F(ab′)2 fragments of anti-CEACAM antibodies were included in the assay, either alone or in combination with the anti–class I MHC mAb HP-1F7 (Figure 4C). Similar results were obtained regardless of whether the NK clones tested expressed the NKp46 receptor (data not shown). Importantly, no effect was observed when the F(ab′)2 fragments of the anti–class I MHC mAb HP-1F7 were included (Figure 4C). Thus, the low expression level of class I MHC proteins is not enough to confer protection. Killing was not restored when the control F(ab′)2 fragments of either the polyclonal antiubiquitin antibodies or the 12E7 mAb were used (Figure 4C). These results indicate that self-attack of the autologous PHA-induced T-cell blasts by NK clones derived from the patients is prevented by the homophilic CEACAM1 inhibitory interactions.
Strikingly, autologous NK clones negative for CEACAM1 expression were unable to kill the autologous PHA-induced T-cell blasts (see, eg, patient C's CEACAM1– NK cells, which express NKp46; Figure 4C). This property is unique to NK cells derived from the patients, as CEACAM1– NK cells from the healthy sibling efficiently attacked self-cells following MHC class I blocking (Figure 4C-D).
We next tested the NK clones and bulk cultures obtained from patients A, B, and C and the healthy sister in killing assays against PHA-induced T-cell blasts derived from the healthy sister. The various cells were treated as described in the text discussion of Figure 4C. CEACAM1– NK clones derived from the healthy sister were unable to kill the autologous PHA-induced T-cell blasts. The inhibition was the result of class I MHC interactions, as the F(ab′)2 fragments of HP-1F7 mAb included in the assay, either alone or in combination with the F(ab′)2 fragments of anti-CEACAM antibodies, abolished this inhibition (Figure 4D). In contrast, when CEACAM1+ NK clones derived from the TAP2-deficient patients were used, lysis of the sister's PHA-induced T-cell blasts could be completely restored only when the F(ab′)2 fragments of both the HP-1F7 and anti-CEACAM antibodies were used (Figure 4D). Partial restoration of killing was observed when either the CEACAM1 or the class I MHC interactions were disrupted, indicating that both inhibitory mechanisms prevent the killing of normal PHA-induced T-cell blasts. Similar results were obtained when CEACAM1+ NK clones derived from the healthy sister were used (data not shown).
We have previously shown that CEACAM1 protein is up-regulated on NK cells derived from some melanoma patients and from decidua.15,18 Here, we show that the vast majority of activated NK cells derived from TAP2-deficient patients express the CEACAM1 protein in high levels, that the expression of the CEACAM1 protein is restricted to patient-derived NK cells with the ability to kill self-cells, and that the CEACAM1 protein is capable of inhibiting NK killing. Thus, NK cells derived from TAP2-deficient patients have developed or acquired a unique mechanism to control the killing of self-cells by using the CEACAM1 interactions.
Discussion
The question of why TAP2-deficient patients do not frequently suffer from autoimmune problems was particularly puzzling. A substantial amount of work was performed with MHC class I–deficient mice to address this problem.21 Studies with mice lacking the β2-microglobulin, TAP1, or both have established that NK cells develop in the absence of MHC class I proteins, yet acquire tolerance toward MHC class I– cells.34
Our experiments demonstrate that NK cell autoreactivity is controlled in the TAP2-deficient patients by the inhibition mediated via the CEACAM1 protein. In addition, a significant reduction in NKp46 expression was observed in most of the NK cells derived from these patients.
It is possible that the low levels of NKp46 expression observed here might be due to variability among different donors.5 However, NK clones in the same patient are either positive or negative for NKp46 expression. In addition, the NKp46 expression may gradually be lost in the TAP2-deficient patients in such a way that no expression was observed on NK cells derived from the older sister but some of the NK clones derived from the younger brothers still express NKp46.
It was shown in vitro that the NKp46 and NKp30 receptors are expressed early during the development of NK cells,35 so it is possible that the reduced NKp46 expression is the result of impaired NK cell development in our patients. In support of this hypothesis, we observed that unusually high percentages of the patients' NK cells express various KIRs. Furthermore, bright expression of the various NK receptors was observed in the patients. Similar results were observed in other TAP2-deficient patients.36 These observations, together with the fact that there was no correlation between the increased percentages of NK clones positive for the various inhibitory receptors and the appropriate class I MHC proteins on the patient's cells, suggest improper NK development.
A slight expression of class I MHC detected by W6/32 was observed on the patient EBV cells. In contrast, expression of HLA-A3 could not be detected (Tables 1, 2). It will be interesting to test which of the class I MHC proteins are expressed on the surface of the EBV-transformed cells.
It is well established that NK cells are able to kill class I MHC–deficient targets. Therefore, killing of self-cells in the TAP2-deficient patients should occur because some NK clones expressing NKp46 were observed in patients B and C (Table 3) and lysis ligands for NKG2D may be expressed on the PHA-induced T-cell blasts.33 Here we demonstrate that the CEACAM1-mediated inhibition protects self-cells from killing by the patients NK cells (Figure 4). It is still unknown which factors control the expression of CEACAM1 either on normal NK cells, which are mostly CD16–, or in the TAP2-deficient patients, which are mostly CD16+. The CEACAM1 protein might be involved in additional biologic activities in the TAP2-deficient patients, such as hepatic insulin clearance,37 adhesion,38 and epithelial cell polarization.39
Strikingly, CEACAM1– NK clones derived from the TAP2-deficient patients were unable to kill their autologous T cells (Figure 4). It is possible that another, yet-unidentified, inhibitory mechanism of NK cell killing is used by the CEACAM1– NK clones or that other lysis receptors are not expressed on such clones. We have also analyzed the expression of other killing receptors such as 2B4 (detected with the C1.7 mAb) and NKG2D (detected with anti–human NKG2D [anti-hNKG2D] mAb). All bulk NK cultures tested displayed similar expression level of the 2B4 receptor (approximately MFI = 12). Remarkably, a significant decrease in the NKG2D expression was observed in all of the NK cells derived from the patients (MFI = 7, 6, and 3 in patients A, B, and C, respectively, as compared with MFI = 31 for the healthy mother). Similar patterns of decreased expression were also observed by using anti-NKp44 and anti-NKp30 sera obtained from mice immunized with NKp30-Ig and NKp44-Ig (data not shown). Thus, it seems as if the expression of other NK-activating receptors is reduced in the TAP2-deficient patients. It will be interesting to test whether the lack of lysis receptors other than NKp46 is the reason for inability of the CEACAM1– NK cells to kill autologous cells.
The TAP2-deficient patients present a major enigma to immunologists, as NK-mediated autoimmune manifestations are observed in some of these patients only during the third and fourth decades of life.40 It seems that the NK cells in these patients developed different mechanisms to overcome the NK-mediated autoimmunity. The acquisition of these mechanisms might be influenced by various factors such as exposure to different environmental pathogens. This could be the reason why, for example, the impaired expression of the NKp46 receptor was not observed in 2 other TAP2-deficient patients.20 Understanding of the molecular mechanisms responsible for the reduced NKp46 expression and the up-regulation of CEACAM1 might facilitate the development of novel therapy for these patients. Such treatments might also be useful in controlling NK cell killing activity in other medical situations such as bone marrow transplantations.
Prepublished online as Blood First Edition Paper, November 6, 2003; DOI 10.1182/blood-2003-06-2114.
O.M. is supported by research grants from the Israel Cancer Research Foundation (ICRF), The Cooperation Program in Cancer Research of the Deutsches Krebsforchungzentrum (DKFZ), the Israel Ministry of Science (MOS), the Israel Science Foundation (ISF), the Cancer Research Institute (CRI), and the European Commission (QLK2-CT-2002-011112); R.S.B. is supported by National Institutes of Health (NIH) grant NIH DK51362 and by the Harvard Digestive Diseases Center; V.C. is supported by a research grant from the Cancer Research Campaign.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr David Cosman (Immunex) for providing the LIR-1 Ig plasmid and fusion protein, Dr Miguel López-Botet for providing the HP-1F7 and the HP-F1 mAbs, and Dr Ygal Haupt for providing the control rabbit polyclonal antibodies.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal