Abstract
Thymosin β4(Tβ4), a 4.9-kDa polypeptide primarily known as a main G-actin–sequestering peptide, is present in high concentrations in various cells and in the circulation. We have found that Tβ4 upregulates the expression of plasminogen activator inhibitor 1 (PAI-1) in endothelial cells measured both at the level of mRNA and protein synthesis. This effect seems to be cell specific and was not observed when other cells such as human fibroblasts, PC3, and U937 were tested. Tβ4 significantly activated the PAI-1 promoter in EA.hy 926 cells transiently transfected either with plasmid p800LUC containing PAI-1 promoter fragment (–800 to +71) or the PAI-1 promoter linked with green fluorescent protein. Tβ4 mediated up-regulation of PAI-1 involved activation of the mitogen-activated protein kinase cascade. Furthermore, Tβ4 enhanced c-Fos/c-Jun DNA-binding activity to the activator protein 1 (AP-1)–like element (–59 to –52). The specificity of this binding activity was demonstrated by competition electrophoretic mobility shift assay and after transfection of EA.hy 926 cells with the mutated PAI-1 promoter. Taken together, these data indicate that, in response to Tβ4 stimulation, AP-1 activity increases to enhance PAI-1 transcription through its unique AP-1–like element at –59 to –52 in the PAI-1 promoter.
Introduction
Thymosin β4(Tβ4) is the most abundant member of the β-thymosins, a family of highly conserved polar 5-kDa peptides. Biologic functions and the mechanism by which Tβ4 regulates mobility of the cells was recently reviewed by Huff et al.1 This 43–amino acid oligopeptide does not possess a signal sequence for secretion and is very soluble in water, but nothing is known about the molecular mechanisms mediating the effects attributed to its extracellular activities. Tβ4 is detected outside of cells in blood plasma or in wound fluid. Extracellular concentrations of Tβ4 can be significantly increased on activation of platelets by thrombin. Tβ4 is also released by other cells, including bone marrow endothelial cells.2 The extracellular Tβ4 may serve as a glutaminyl substrate of tissue transglutaminase and can be specifically cross-linked to some proteins including fibrin, collagen, and actin. This provides a mechanism to increase the local concentration of Tβ4 near sites of clots and tissue damage, where it may contribute to wound healing, angiogenesis, and inflammatory responses.3 Tβ4 was described to induce expression of metalloproteinases4 and migration of human umbilical vein endothelial cells.5 It was also found to inhibit proliferation of bone marrow stem cells.1 Interestingly, Tβ4 topically applied increases collagen deposition and stimulates keratinocyte migration. When used in the chick chorioallantoic membrane in vivo angiogenesis model, Tβ4 enhanced angiogenesis, whereas Tβ10 exhibited an inhibitory effect on the angiogenesis process.6
The β-thymosins rapidly concentrate around sites of infection, as demonstrated in the animal model, and thus were proposed to be useful for the detection of both infections and inflammatory lesions.7 Usually, in the same area there is a significant increase in plasminogen activator inhibitor type 1 (PAI-1) expression. Therefore, in this study we attempted to evaluate whether extracellular Tβ4 shows any effect on synthesis and release of PAI-1 from endothelial cells.
Materials and methods
Reagents
Tβ4, synthetic whole length of 4.9-kDa peptide, was a generous gift from Dr Metcalf (Laboratory of Allergic Diseases, National Institutes of Health, Bethesda, MD). Tβ10 was a gift from Dr Pawlikowski (Institute of Endocrinology, Medical University in Lodz). The Elisa-Imulyse PAI-1 immunoenzymatic kit was from Biopool (Umeå, Sweden). Rabbit polyclonal antibodies against active extracellular signal-regulated kinases such as extracellular signal–regulated kinases 1 and 2 (ERK1/2), c-Jun N-terminal kinases 1, 2, and 3 (JNK1/2/3), and phosphor-c-Jun(Ser63) were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-(rabbit IgG) Ig–horseradish peroxidase conjugates were from Promega (Madison, WI). All standard tissue culture reagents including Dulbecco modified Eagle Medium (DMEM), fetal bovine serum (FBS), and LipofectAMINE Plus reagent were from Gibco (Karlsruhe, Germany). The Luciferase Assay System was from Promega. Plasmid p800LUC with PAI-1 promoter was a gift from Dr David J. Loskutoff (Department of Cell Biology, Scripps Research Institute, La Jolla, CA). Protein assay reagents and polyacrylamide gel chemicals were from BioRad (Hercules, CA). Antibodies to protein kinases were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Culture media and supplements were purchased from Sigma-Aldrich (St Louis, MO). H-D-Val-Leu-Lys-AMC was from Bachem (Weil am Rhein, Germany).
Cell culture
Human umbilical vein endothelial cells (HUVECs) were isolated from freshly collected umbilical cords by collagenase treatment8 and cell cultures were maintained as described previously.9 For the experiments, confluent cultures were used at the second passage. Human endothelial cell line EA.hy 926 was derived by fusion of HUVECs with continuous human lung carcinoma cell line A549.10 This endothelial hybrid cell line is presently the best characterized macrovascular endothelial cell (EC) line (for a review, see Bouis et al11 ). EA.hy 926 cells produce large amounts of tissue-type plasminogen activator (t-PA), PAI-1, and a small amount of urokinase. Their fibrinolytic characteristics are stable for at least 30 passages.10 Similarly, adhesive properties of these cells are almost the same as those of primary ECs.9,12 These hybrid cells (ECs) are currently used as in vitro model systems for various physiologic and pathologic processes, especially in angiogenesis research.13 The cells were cultured in DMEM with high glucose, supplemented with 10% FBS, HAT (100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine), and antibiotics in a 90% to 95% humidified atmosphere of 5% CO2 at 37°C.
Human skin fibroblasts, isolated by the explant technique from normal skin biopsy specimens, were provided by Dr B. Rozga (Lodz University). PC3, a hormone-resistant line derived from a bone metastasis of prostatic cancer was a gift from Dr J. Niewiarowska (Medical University in Lodz). U937, a human promonocyte cell line was kindly donated by Dr A. Sobota (Nencki's Institute, Warsaw, Poland).
For microscopic examination, cells were plated at a density of 5 × 104 cells/well on Thermanox coverslips in 8-well tissue culture chamber slides (Nunc, Roskilde, Denmark) with detachable chambered upper structures.
To measure cytotoxic effects, ECs were treated for 20 hours at 37°C in medium supplemented with FBS with Tβ4 at concentrations up to 100 nM. Cell viability was determined microscopically by trypan blue exclusion. In addition, the apoptotic effect of Tβ4 within the concentrations used was evaluated by flow cytometry using annexin V–fluorescein isothiocyanate (FITC) apoptosis detection kit (Sigma, St Louis, MO). Only cell cultures having less than 1% dead cells were included in the study.
Measurements of PAI-1 antigen
HUVECs or EA.hy 926 cells were planted in 48-well microplates at a density of 2 × 105 cells/mL. On the next day, they were starved overnight in medium containing 0.1% FBS, and then stimulated for 20 hours with different concentrations of Tβ4 or Tβ10 (0-160 nM). Afterward, the supernatants were collected and assayed for PAI-1 antigen by enzyme-linked immunosorbent assay (ELISA) using the Elisa-Imulyse PAI-1 kit.
Measurements of PAI-1, t-PA, and VWF mRNA
For this purpose, EA.hy 926 cells were stimulated for 4 and 24 hours with different concentrations of Tβ4 (0-160 nM), then total cellular RNA was extracted by the Trizol reagent method (Invitrogen, Carlsbad, CA) using a single-step purification protocol.14 RNA pellets were dissolved in water, and their concentrations and purity were determined by spectrophotometer readings at 260 and 280 nm. The relative amounts of specific mRNAs were quantified by reverse transcription-polymerase chain reaction (RT-PCR) as described previously.15 The following primers were used: 5′GTCGAATTCCTGGAGCTCAG3′ and 5′CTGCGCCACCTGCTGAAACA3′, 5′GCTGCTGGACAATGGTGCC3′ and 5′GGATGTCACAGCTTGGGC3′, 5′GCAATCTGCGTGTCTCAAG3′ and 5′TCATTGTAAATATTCTTKTGG3′ specific for mRNA of PAI-1, von Willebrand factor (VWF), and t-PA, respectively. In the same samples, β-actin mRNA was amplified with primers 5′GTGGGGCGCCCCAGGCACCA3′ and 5′CTCCTTAATGTCACGCACGATTTC3′ and used as an intrinsic control of mRNA quantity PCR amplification. The final products were separated by electrophoresis in 7% polyacrylamide gels in Tris-acetate-EDTA buffer (TAE; tris(hydroxymethyl)aminomethane; ethylenediaminetetraacetic acid) using the genetic size marker 100–base pair (bp) DNA ladder (Promega). Bands were visualized by UV light; the results were recorded photographically and analyzed densitometrically using an LKB Ultrascan XL Enhanced Laser Densitometer (Bromma, Sweden).
Kinase assays
EA.hy 926 cells were grown in 6-well plates at a density of 2 × 105 cells/mL. The day after they were starved in DMEM supplemented with 0.5% FBS and were then stimulated with different concentrations of thymosin β4 (0-80 nM) for 20 minutes or overnight in the case of ERKs and JNKs or c-Jun, respectively. After treatment, cell lysates were produced and analyzed for protein kinases as described previously.16 Immunodetection was accomplished using the enhanced chemiluminescence kit (ECL kit; Amersham Pharmacia, Buckinghamshire, United Kingdom), then films were scanned and protein bands quantitated by Gel Doc 2000 Gel Documentation System (BioRad).
Transfection of cells
Semiconfluent cell cultures (EA.hy 926) in 6-well tissue culture plates were transfected with DNA constructs (plasmid p800LUC with PAI-1 promoter and pGFP-PAI-1) using the LipofectAMINE method. To generate the expression vector pGFP-PAI-1 with GFP reporter gene, the 871-bp fragment of PAI-1 promoter was excised from p800LUC plasmid. The following sites were used: the EcoRI site at the position +71 and HindIII site at the position –800. After digestion and purification with Wizard PCR Preps DNA Purification System (Promega), the PAI-1 promoter region was then ligated into the multiple cloning sites of the expression vector pGFP-N1 (Clontech). Escherichia coli JM110 strain was used as a host for transformation and maintenance of the expression construct pGFP-PAI-1. The activator protein 1 (AP-1) consensus sequence was modified by point mutations introduced by using mutagenic primers (5′ CTGGAACATGTGTTTGTCTATTT 3′ and 5′ GACCTTGTACACAAACAGATAAA 3′). Thus, the wild-type binding site from PAI-1 promoter (TGAGTTCA; –60 to –52) was substituted by mutated version TGTGTTTG as confirmed by DNA sequencing. Cells were transfected with 4 μg p800LUC with PAI-1 promoter or its mutated form (p800LUCmut), and 5 μg pSV vector containing the β-galactosidase gene to evaluate transfection efficiency. For Tβ4 studies, increasing concentrations of this oligopeptide (0-80 nM) were added to wells, and the cells were incubated for 24 hours, washed, and harvested in the lysis buffer. After centrifugation for 5 minutes at 4°C, the supernatants were transferred into fresh vials and used for enzymatic assays. Luciferase assay kits (Gibco) were used according to the manufacturer's instructions, using a 1420 multilabel counter-Victor (Wallac, Perkin Elmer, Shelton, CT). β-Galactosidase activity from a constitutively expressed internal control was assayed with a β-galactosidase enzyme assay system (Gibco) according to the manufacturer's instruction and with an EL340 plate reader (Bio-Tek Instruments, Winooski, VT). In parallel experiments, EA.hy 926 cells were transfected with luciferase reporter vector pGL3 (Promega) and used as control cells to test whether the effect of inhibitors was specific for the PAI-1 promoter.
EA.hy 926 cells grown in slide chambers at the density of 2 × 105 cells/mL were transfected with pGFP-PAI1 using 1 μg of the plasmid DNA and LipofectAMINE (Gibco) according to the manufacturer's instructions. Both reporter vectors pGFPN1 and pGL3 were used to detect the effect of Tβ4 on the activity of GFP and luciferase promoters, respectively.
Electrophoretic mobility shift assay
Confluent EA.hy 926 cell cultures were stimulated for 2 hours either with Tβ4 (40 nM) or tumor necrosis factor α (TNF-α; 10 ng/mL), and then nuclear extracts were prepared as described earlier.17 DNA-protein complexes were formed by incubating 2 or 8 μg nuclear proteins with 80 000 cpm 32P-labeled oligodeoxynucleotide probe in 20 μL binding buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]–KOH, pH 7.9, 1 mM EDTA, 5 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol, 10% glycerol) in the presence of a nonspecific competitor poly(dI:dC) (Pharmacia, Uppsala, Sweden) for 20 minutes at room temperature. For competition experiments, unlabeled double-stranded competitor oligodeoxynucleotides in 200-fold molar excess were added to the reaction mixtures. Sequences of these oligodeoxynucleotides, except non-specific competitor sequence (Sp1, 5′ AATTCCCCGCCCCCGCCCCC 3′) are shown below. Where indicated, 1 μg mouse monoclonal antibodies, p-c-Jun (KM-1) or c-Fos (6-2H-2F), both obtained from Santa Cruz Biotechnology, were added to the reaction mixture and incubation was continued for an additional 20 minutes. DNA-protein complexes were resolved from unbound oligodeoxynucleotides on a 5% polyacrylamide gel using 0.5 × TBE buffer (25 mM Tris borate, pH 8.3, 0.5 mM EDTA) at a constant voltage (150 V for 2 hours) and detected by autoradiography.
The double-stranded oligodeoxynucleotides (AP-1/PAI-1, 5′ AATTGGAACATGAGTTCATCTATTT 3′, representing the PAI-1 promoter region between –65 and –45 and AP-1con, 5′-AATTGGAACATGAGTCATCTATTT-3′ consisting of the AP-1 consensus sequence [TGAGTCA] flanked by 13 bp from the PAI-1 promoter) were radioactively labeled by filling in the overhangs with Sequenase 2.0 (Amersham) in the presence of α-[32P] deoxyadenosine triphosphate (Amersham) and purified on 7% polyacrylamide gels.
Results
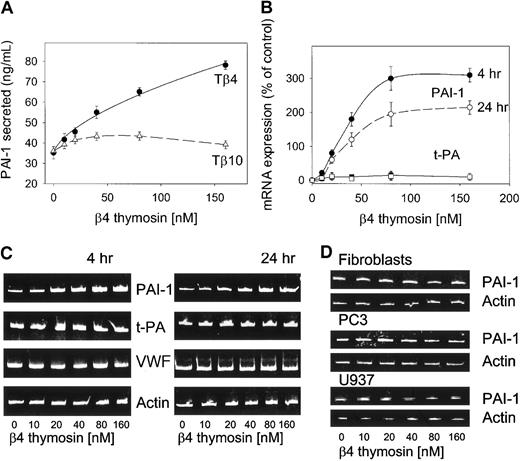
Because the concentration of Tβ4 significantly increases outside of cells in blood plasma under inflammation conditions, as well as in prethrombotic states, in the present studies we attempted to explain whether this low-molecular-weight protein can regulate PAI-1 expression in ECs. In preliminary experiments, the immortalized ECs, EA.hy 926 or HUVECs, were incubated with increasing concentrations of Tβ4 and Tβ10, and aliquots of the conditioned media were collected at different time points then subjected to ELISA. Figure 1A shows that Tβ4 significantly increased the levels of PAI-1 secreted from HUVECs into the culture medium in a concentration-dependent manner. The maximum of PAI-1 release was approached after treatment of the cells with extracellular Tβ4 concentrations as low as 160 nM. The same extent of the PAI-1 secretion was observed when EA.hy 926 cells were tested (not shown). This effect was specific for Tβ4 because Tβ10 did not stimulate HUVECs to secrete PAI-1. At the same dose there was a maximal induction of PAI-1 mRNA in ECs measured by RT-PCR observed after 4 and 24 hours of treatment of cells with Tβ4 (Figure 1B). Specificity of this effect was further evidenced by the fact that there was no increase in mRNA of t-PA, VWF, or actin when analyzed in the same samples (Figure 1C). The effect of Tβ4 on the EC PAI-1 appears to be cell specific because there was no change in PAI-1 mRNA expression on treatment of other cells such as human fibroblasts, PC3, and U937 cells (Figure 1D).
Tβ4 induces expression of PAI-1 in ECs. (A) Release of PAI-1 from HUVECs. These cells planted in 48-well microplates were starved overnight in medium containing 0.1% FBS and then stimulated for 20 hours with different concentrations of Tβ4 or Tβ10 (0-160 nM). Afterward, the supernatants were collected and assayed for PAI-1 antigen by ELISA using an Elisa-Imulyse PAI-1 (Biopool). The graph shows the average ± SD of duplicate measurements performed during 3 experiments. (B) Relative quantitative RT-PCR analysis of PAI-1 mRNA ± SD. In these experiments, EA.hy 926 cells were stimulated for 4 and 24 hours with different concentrations of Tβ4 (0-160 nM), then total cellular RNA was extracted. RT-PCR followed by staining with ethidium bromide was used to determine PAI-1 and t-PA mRNA levels. This experiment has been repeated 3 times with similar results. (C) Increase in PAI-1 mRNA but not in t-PA, VWF, and actin mRNAs on incubation of EA.hy 926 cells with different concentrations of Tβ4 for 4 and 24 hours. (D) When Tβ4 is used in the same concentration range as in the case of EA.hy 926 cells, it has no effect on the PAI-1 expression in human fibroblasts, PC3, and U937 cells.
Tβ4 induces expression of PAI-1 in ECs. (A) Release of PAI-1 from HUVECs. These cells planted in 48-well microplates were starved overnight in medium containing 0.1% FBS and then stimulated for 20 hours with different concentrations of Tβ4 or Tβ10 (0-160 nM). Afterward, the supernatants were collected and assayed for PAI-1 antigen by ELISA using an Elisa-Imulyse PAI-1 (Biopool). The graph shows the average ± SD of duplicate measurements performed during 3 experiments. (B) Relative quantitative RT-PCR analysis of PAI-1 mRNA ± SD. In these experiments, EA.hy 926 cells were stimulated for 4 and 24 hours with different concentrations of Tβ4 (0-160 nM), then total cellular RNA was extracted. RT-PCR followed by staining with ethidium bromide was used to determine PAI-1 and t-PA mRNA levels. This experiment has been repeated 3 times with similar results. (C) Increase in PAI-1 mRNA but not in t-PA, VWF, and actin mRNAs on incubation of EA.hy 926 cells with different concentrations of Tβ4 for 4 and 24 hours. (D) When Tβ4 is used in the same concentration range as in the case of EA.hy 926 cells, it has no effect on the PAI-1 expression in human fibroblasts, PC3, and U937 cells.
To determine the molecular level at which PAI-1 expression can be regulated, we have used a transient transfection approach to ask if Tβ4 can influence transcription of the PAI-1 promoter. For this purpose, a PAI-1–green fluorescent protein (GFP) promoter/reporter construct was expressed in EA.hy 926. This construct contained the PAI-1 promoter fragment corresponding to positions from –800 to +71 to driving the expression of GFP. The cells were transfected with the high efficiency (> 80%) using conventional techniques, such as a lipid-mediated transfection of EA.hy 926 cells. For this purpose, EA.hy 926 cells grown in slide chambers with a density of 2 × 105 cells/mL were treated with 1 μg of the plasmid DNA (pGFP-PAI-1) and LipofectAMINE (Gibco) according to manufacturer's instructions. Within hours after infection, most of the cells in each culture were GFP+. On treating cultures with Tβ4 there was a concentration-dependent increase in GFP expression (Figure 2) indicating activation of the PAI-1 promoter in the transfected EA.hy 926 cells. These studies demonstrate that the regulation of PAI-1 transcription, as well as PAI-1 mRNA and protein by Tβ4, is similar in terms of the kinetics and magnitude of induction, indicating that Tβ4 primarily regulates PAI-1 gene expression at the point of transcription.
Tβ4 induces transcription of PAI-1 promoter in EA.hy 926 cells. GFP expression in cultured EA.hy 926 cells was analyzed after transfection with pGFP-PAI-1 reporter constructs. EA.hy 926 cells grown in slide chambers at a density of 2 × 105cells/mL were transfected with 1 μg pGFP-PAI-1 using LipofectAMINE and incubated with 0, 40, and 80 nM Tβ4 for 24 hours. The top panels are fluorescent images, and the bottom panels are the corresponding phase-contrast micrographs. This experiment has been repeated 3 times with similar results.
Tβ4 induces transcription of PAI-1 promoter in EA.hy 926 cells. GFP expression in cultured EA.hy 926 cells was analyzed after transfection with pGFP-PAI-1 reporter constructs. EA.hy 926 cells grown in slide chambers at a density of 2 × 105cells/mL were transfected with 1 μg pGFP-PAI-1 using LipofectAMINE and incubated with 0, 40, and 80 nM Tβ4 for 24 hours. The top panels are fluorescent images, and the bottom panels are the corresponding phase-contrast micrographs. This experiment has been repeated 3 times with similar results.
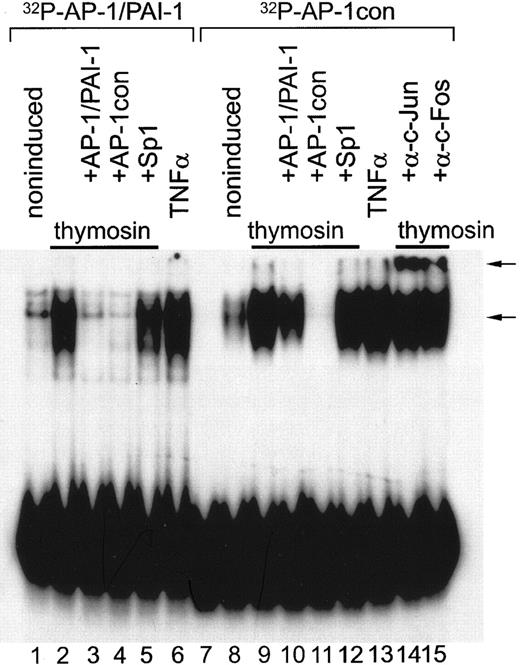
Many of the stimulators of PAI-1 synthesis in HepG2 or ECs, including phorbol myristate acetate (PMA), serum, and interleukin 1 (IL-1), may be classified as agents that increase the abundance or activity of the transcription factor, AP-1. AP-1 is a collection of homodimeric or heterodimeric complexes composed of jun and fos gene products. To see whether induction of PAI-1 transcription occurs via the same mechanism, lysates were prepared from EA.hy 926 cells stimulated with different concentrations of Tβ4 for varying times and then analyzed by Western blot analysis using a phospho-specific antibody. As shown in Figure 3, Tβ4 stimulates JNK1 kinase activity rapidly and potently, resulting in phosphorylation of Ser63 on c-Jun. Tβ4 stimulated phosphorylation of JNK, the only known kinase that phosphorylates c-Jun. There was also activation of ERK1/2, however, to a much lower extent than that of JNK1. It should be noted that these assays are performed on cells in serum-containing media, indicating that Tβ4 is capable of stimulating JNK1 activity in exponentially growing cells. These data suggest that Tβ4 regulates PAI-1 gene expression via up-regulating c-Jun levels and subsequent binding of c-Fos/c-Jun heterodimers to the proximal element of the PAI-1 gene. To confirm the functionality of the c-Jun complexes induced by Tβ4, we performed electrophoretic mobility shift assays (EMSAs) using nuclear extracts of EA.hy 926 cells. Two 32P-labeled oligonucleotide probes containing AP-1–binding motifs were used, the first one consisting of the wild-type PAI-1 gene sequence (PAI-1/AP-1; –65 to –45) with the TGAGTTCA motif located between –59 and –52, and the second one corresponding to the AP-1 consensus sequence (AP-1con) containing the TGAGTCA motif. As shown in Figure 4, specific DNA/protein complexes can be observed when nuclear extracts of EA.hy 926 cells treated with Tβ4 were incubated with either the PAI-1/AP-1 or the AP-1con probe. Because the DNA element within the PAI-1 gene (–59 to –52) varies from the AP-1 consensus by a 1-bp insert (TGAGTTCA versus TGAGTCA), the binding affinity of the induced nuclear proteins toward the consensus AP-1 sequence and the AP-1–like sequence in the PAI-1 gene is different. Our data confirm the previous observations showing that the 32P-labeled PAI-1/AP-1 probe has much lower binding affinity than the AP-1con. Competition experiments with 100-fold excess unlabeled, double-stranded AP-1/PAI-1 and AP-1con probes inhibited the formation of the complexes. However, they differed in the inhibitory efficiency when 32P-AP-1con was used as a probe (Figure 5, lanes 10 and 11). Nuclear extracts obtained from the cells activated by TNF-α showed significantly higher binding of both probes when compared to those produced from the Tβ4-stimulated cells (Figure 5, lanes 6 and 13). There was no inhibition when other oligodeoxynucleotides, for example, with the Sp1-binding sequence, were added. To identify the transcription factors that were induced to bind to these probes on Tβ4 exposure, EMSAs were performed on the same nuclear extracts in the presence of factor-specific antibodies. Supershifting of the complex was noted with antibodies to both c-Fos and c-Jun family proteins, but not those raised against irrelevant antibody (not shown). The autoradiogram shown in Figure 4 is slightly overexposed to clearly present the supershift produced by antibodies to p-c-Jun and c-Fos. Further decreasing the time of exposure or decreasing the amount of complex formed prior to incubation with antibodies caused the reduction of the band, representing DNA-protein complexes produced on incubation with the antibodies. This strongly indicated that the complex consisted of DNA-bound c-Fos/c-Jun heterodimers and suggested that Tβ4 significantly induced its formation.
Tβ4 induces phosphorylation of JNK1, ERK, and phospho-c-Jun protein. Confluent EA.hy 926 cells were treated with increasing concentrations of Tβ4 (0-80 nM) for 20 minutes or overnight in the case of ERKs and JNK1 or c-Jun, respectively. Total protein was harvested in 2 × sodium dodecyl sulfate (SDS) buffer. After polyacrylamide gel electrophoresis (PAGE) separation and transfer to polyvinylidene difluoride membranes, Western blot analysis was performed with an antibody to ERK, JNK, and phospho-(p)-c-Jun (Ser63), and enhanced chemiluminescence was performed for detection of bound secondary antibody. All data are representative of 3 replicate experiments.
Tβ4 induces phosphorylation of JNK1, ERK, and phospho-c-Jun protein. Confluent EA.hy 926 cells were treated with increasing concentrations of Tβ4 (0-80 nM) for 20 minutes or overnight in the case of ERKs and JNK1 or c-Jun, respectively. Total protein was harvested in 2 × sodium dodecyl sulfate (SDS) buffer. After polyacrylamide gel electrophoresis (PAGE) separation and transfer to polyvinylidene difluoride membranes, Western blot analysis was performed with an antibody to ERK, JNK, and phospho-(p)-c-Jun (Ser63), and enhanced chemiluminescence was performed for detection of bound secondary antibody. All data are representative of 3 replicate experiments.
Binding of Tβ4-induced nuclear proteins to the PAI-1/AP-1–like sequence and the AP-1 consensus sequence. EMSA was performed using labeled oligodeoxynucleotides containing the AP-1–binding sequence from the PAI-1 promoter (AP-1/PAI-1, lanes 1-6) or consensus AP-1–binding sequence (TGAGTCA) flanked by 13 bp from PAI-1 promoter (AP-1con, lanes 7-15). Nuclear extracts used in these studies were prepared from confluent EA.hy 926 cell cultures unstimulated (lanes 1 and 8) or stimulated for 2 hours either with Tβ4 (40 nM; lanes 2-5, 9-12, and 14-15) or TNF-α (10 ng/mL; lanes 6 and 13). For competition experiments, unlabeled double-stranded competitor oligodeoxynucleotides in 200-fold molar excess were added to the reaction mixtures as indicated (lanes 3-5 and lanes 10-12). Immunologic identification of Tβ4-induced DNA-binding protein was performed with mouse monoclonal antibodies specific to fos/jun family members: c-Jun phosphorylated on Ser63 (lane 14) and c-Fos (lane 15). Protein-DNA complexes were resolved on a native 5% polyacrylamide gel and detected by autoradiography. Protein-DNA complexes and supershifts with antibodies are marked by arrows. Lane 7 contains only DNA probe.
Binding of Tβ4-induced nuclear proteins to the PAI-1/AP-1–like sequence and the AP-1 consensus sequence. EMSA was performed using labeled oligodeoxynucleotides containing the AP-1–binding sequence from the PAI-1 promoter (AP-1/PAI-1, lanes 1-6) or consensus AP-1–binding sequence (TGAGTCA) flanked by 13 bp from PAI-1 promoter (AP-1con, lanes 7-15). Nuclear extracts used in these studies were prepared from confluent EA.hy 926 cell cultures unstimulated (lanes 1 and 8) or stimulated for 2 hours either with Tβ4 (40 nM; lanes 2-5, 9-12, and 14-15) or TNF-α (10 ng/mL; lanes 6 and 13). For competition experiments, unlabeled double-stranded competitor oligodeoxynucleotides in 200-fold molar excess were added to the reaction mixtures as indicated (lanes 3-5 and lanes 10-12). Immunologic identification of Tβ4-induced DNA-binding protein was performed with mouse monoclonal antibodies specific to fos/jun family members: c-Jun phosphorylated on Ser63 (lane 14) and c-Fos (lane 15). Protein-DNA complexes were resolved on a native 5% polyacrylamide gel and detected by autoradiography. Protein-DNA complexes and supershifts with antibodies are marked by arrows. Lane 7 contains only DNA probe.
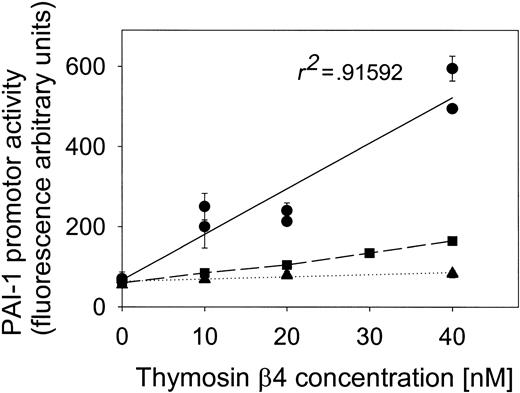
Point mutations of the AP-1–binding site within the PAI-1 promoter abolish its sensitivity to Tβ4-induced transcriptional activity. EA.hy 926 cells were transiently transfected either with p800LUC (•) or its mutated version (▪), in which the wild-type AP-1 consensus motif (TGAGTTCA) was substituted by its mutated version, TGTGTTTG. Eighteen hours following transfection, cells were washed and transferred to DMEM media containing 0.5% bovine serum albumin. After 24 hours, Tβ4 was added at the indicated concentration and the cells where harvested after an additional 7 hours. Luciferase and β-gal activities were assayed as described in “Materials and methods.” Values are expressed as relative light units (RLU) of luciferase normalized to β-gal activity with data of 2 experiments shown as the mean of 3 separate samples plus the SE. In addition, data obtained using EA.hy 926 cells transfected with the empty vector, pGL2, are shown (▴).
Point mutations of the AP-1–binding site within the PAI-1 promoter abolish its sensitivity to Tβ4-induced transcriptional activity. EA.hy 926 cells were transiently transfected either with p800LUC (•) or its mutated version (▪), in which the wild-type AP-1 consensus motif (TGAGTTCA) was substituted by its mutated version, TGTGTTTG. Eighteen hours following transfection, cells were washed and transferred to DMEM media containing 0.5% bovine serum albumin. After 24 hours, Tβ4 was added at the indicated concentration and the cells where harvested after an additional 7 hours. Luciferase and β-gal activities were assayed as described in “Materials and methods.” Values are expressed as relative light units (RLU) of luciferase normalized to β-gal activity with data of 2 experiments shown as the mean of 3 separate samples plus the SE. In addition, data obtained using EA.hy 926 cells transfected with the empty vector, pGL2, are shown (▴).
Finally, to prove that the AP-1 binds to the PAI-1 promoter on induction of cells with Tβ4, the wild-type AP-1 consensus motif (TGAGTTCA; –59 to –52) present in the PAI-1 promoter fragment (–800 to +71) was modified by point mutations, and thus substituted by its mutated version TGTGTTTG. Both, the wild-type and mutated PAI-1 promoter were placed in p800LUC containing the firefly luciferase gene as a reporter. Then, the responsiveness of the PAI-1 promoter to Tβ4 was compared in subconfluent EA.hy 926 cells transfected with both p800LUC and p800LUCmut. Incubation of cells with the increasing doses of Tβ4 for 24 hours resulted in a significant activation of the PAI-1 promoter as reported by luciferase activity (Figure 5). The addition of 40 nM Tβ4to cells transiently transfected with p800LUC caused a 2.5-fold increase in PAI-1 promoter transcription. This effect was almost abolished when the cells were transfected with p800LUCmut, thus supporting the role of AP-1 in activation of the PAI-1 promotor by Tβ4.
Discussion
The function of β-thymosins in the organization of the cytoskeleton is well documented. However, there are increasing numbers of reports suggesting participation of β-thymosins in biologic processes such as angiogenesis, inflammation, wound healing apoptosis, and carcinogenesis. It raises the question whether all these events result from the G-actin sequestering by β-thymosins or from the cytokine-like activity of these low-molecular-weight proteins. The present studies provide evidence that extracellular Tβ4, via unknown receptor mechanisms, can induce the transcription of the PAI-1 gene, thus stimulating PAI-1 release from ECs. This conclusion is supported by the following observations: (1) Tβ4 stimulates PAI-1 promoter activity in EA.hy 926 cells, measured both by analysis of expression of the reporting genes such as GFP or luciferase, and at the level of PAI-1 mRNA. Interestingly, this effect appears to be cell specific because it was not produced when other cells such as human fibroblasts, PC3, and U937 were tested. (2) Tβ4 induces the secretion of PAI-1 from ECs. (3) This effect is preceded by activation of JNK1 and phosphorylation of c-Jun, a component of AP-1 transcription factor. (4) Tβ4 induced binding of c-Fos/p-c-Jun to the highly conserved and unique AP-1–like element in the PAI-1 gene. The PAI-1 gene contains AP-1–like element sequences in the region from –60 to the TATA box (–31), and this proximal region has been implicated in the PAI-1 transcriptional response to transforming growth factor β (TGF-β) and to PMA in several cell types.18-20 These data suggest that Tβ4 regulates the PAI-1 gene expression via up-regulating c-Jun levels and subsequent binding of c-Fos/c-Jun heterodimers to the proximal element of the PAI-1 gene. Therefore, Tβ4 appears to stimulate the PAI-1 promoter in a manner similar to most PAI-1–activating cytokines interacting with a common DNA-binding site, the PMA-responsive element (tPA responsive element [TRE]), and activating gene transcription in response to activators of protein kinase C (PKC), growth factors, and cytokines. 21-23
It is noteworthy that this effect is produced by low concentrations of Tβ4 and is detectable at 100 ng/mL, that is, at a much lower concentration than that which can be achieved extracellularly. For example, Tβ4 was found in amounts approaching 13 μg/mL (2.6 μM) of wound fluid,24 and blood platelets, which contain large amounts of Tβ4 (200-500 μM), can be its major source.25,26 Tβ4 is present in supernatants of stimulated platelet supernatants and together with other low-molecular-weight proteins released from platelets, for example, in response to trauma or mediators of inflammation, was found to play a direct antimicrobial role.27 Whether this new biologic activity of the extracellular Tβ4 is connected to its intracellular G-actin–sequestering function or to other previously described roles such as being a substrate for tissue transglutaminase, stimulation of matrix metalloproteinase (MMP) production, modulation of bone marrow cell growth, deposition of collagen, enhanced keratinocyte migration, and angiogenesis is unknown and requires further investigations.
To summarize, our data indicate that Tβ4, when released from cells by either secretion or cell damage, may significantly influence expression of AP-1–controlled genes by the mechanism similar to that characteristic for known cytokines. The detailed mechanism by which extracellular Tβ4 can stimulate transcription of PAI-1 gene is not known because receptors for this protein subfamily are not yet identified. However, it may be speculated that similar to α-thymosins, to which high- and low-affinity binding sites were recently localized in lymphoid cells,28 there are also membrane receptors for β-thymosins. Their identification would open the way toward the understanding of the molecular mechanism of action of Tβ4.
Prepublished online as Blood First Edition Paper, October 30, 2003; DOI 10.1182/blood-2003-04-1015.
Supported by projects KBN 3/PO4A 068 23 (C.S.C.) and 3 P05A 039 23 (J.W.) from the Polish Committee for Scientific Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal