Abstract
The AML1 gene (recently renamed Runx1), which encodes the DNA-binding subunit of a transcription factor of the core binding factor (CBF) family, is required for the establishment of definitive hematopoiesis. We have previously demonstrated that Runx1 is expressed in yolk sac mesodermal cells prior to the establishment of the blood islands and in the embryoid body (EB)–derived blast-colony–forming cells (BL-CFCs), the in vitro equivalent of the hemangioblast. Analysis of Runx1-deficient embryonic stem (ES) cells demonstrated that this gene is essential for the generation of normal numbers of blast colonies, the progeny of the BL-CFCs. In the present study, we analyzed the potential of Runx1+/– ES cells to determine if heterozygosity at the Runx1 locus impacts early developmental events leading to the commitment of the BL-CFCs. Our results indicate that Runx1 heterozygosity leads to an acceleration of mesodermal commitment and specification to the BL-CFCs and to the hematopoietic lineages in EBs.
Introduction
The AML/Runx genes belong to a family of heterodimeric transcription factors named core binding factors (CBFs). These genes encode an α protein that contains a highly conserved region of 128 amino acids designated the runt domain (RD), due to its homology to a region in the drosophila RUNT protein.1 The RD mediates both heterodimerization with the CBFβ protein and binding to DNA. In human and mice, 3 Runx genes have been identified and all have been shown to play important roles in normal development processes as well as in carcinogenesis.2,3 Runx1, originally named AML1, was initially identified through its frequent translocations associated with human leukemia. In addition to a role in leukemic transformation, targeting studies in mice have demonstrated that Runx1 is essential for the establishment of the definitive hematopoietic system in embryonic life.4,5 In the absence of definitive hematopoiesis, Runx–/– embryos progress through the yolk sac stage of development and die around day 12 to 13 of gestation, at the time hematopoiesis shifts to the fetal liver. Recent studies have demonstrated that Runx1 is also haploinsufficient and that heterozygosity at this locus results in a dramatic change in the temporal and spatial generation of hematopoietic stem cells (HSCs).6 In Runx1+/– embryos, HSCs are found 1 day earlier than normal in the extraembryonic yolk sac and the intraembryonic aorta-gonadmesonephros (AGM) region.
We have recently defined a role for Runx1 at the hemangioblast stage of hematopoietic and endothelial development in embryonic stem (ES) cell–derived embryoid bodies.7 In that study, we demonstrated that Runx1 is expressed in embryoid bodies (EBs) during the appearance of blast-colony–forming cells (BL-CFCs) and that it is essential for the development of a normal number of blast colonies. In the present study we analyzed the impact of heterozygosity at the Runx1 locus on BL-CFC development in EBs. Our results indicate that Runx1 heterozygosity results in the acceleration of mesodermal commitment and specification to BL-CFCs and to the hematopoietic lineages in Runx1+/– EBs.
Study design
Embryonic stem cell growth and differentiation, colony assays, and gene expression were performed as previously described.7 The sequences of the oligonucleotides used for the polymerase chain reaction (PCR) are Rex1 sense, CGTGTAACATACACCATCCG; Rex1 antisense, GAAATCCTCTTCCAGAATGG; NeuroD sense, GGAGTAGGGATGCACCGGGAA; NeuroD antisense, CTTGGCCAAGAACTAGATCTGG; Fgf5 sense, AAAGTCAATGGCTCCCACGAA; Fgf5 antisense, CTTCAGTCTGTACTTCACTGG; brachyury sense, CATGTACTCT TCTTGCTGG; brachyury antisense, GGTCTCGGGAAAGCAGTGGC; goosecoid sense, CGGAGAAGAGGGAAGAGGAAGGTAAAAG; goosecoid antisense, ATAAATACTACGGTGGGGGACGAGGCTCAC; Flk1 sense, CACCTGGCACTCTCCACCTTC; Flk1 antisense, GATTTCATCCCACTACCGAAAG; Gata1 sense, CATTGGCCCCTTGTGAGGCCAGAGA; Gata1 antisense, ACCTGATGGAGCTTGAAATAGAGGC; β-H1 sense AGTCCCCATGGAGTCAAAGA; β-H1 antisense, CTCAAGGAGACCTTTGCTCA; Eklf sense, TCGCCGGAGACGCAGGCT; Eklf antisense, CCCAGTCCTTGTGCAGGA; β-major sense, CTGACAGATGCTCTCTTGGG; β-major antisense CACAACCCCAGAAACAGACA; c-Fms sense, GCGATGTGTCAGCAATGGCAGT; c-Fms antisense, AGACCGTTTTGCGTAAGACCTG; β-actin sense, ATGAAGATCCTGACCGAGCG; and β-actin antisense, TACTTGCGCTCAGGAGGAGC.
Results and discussion
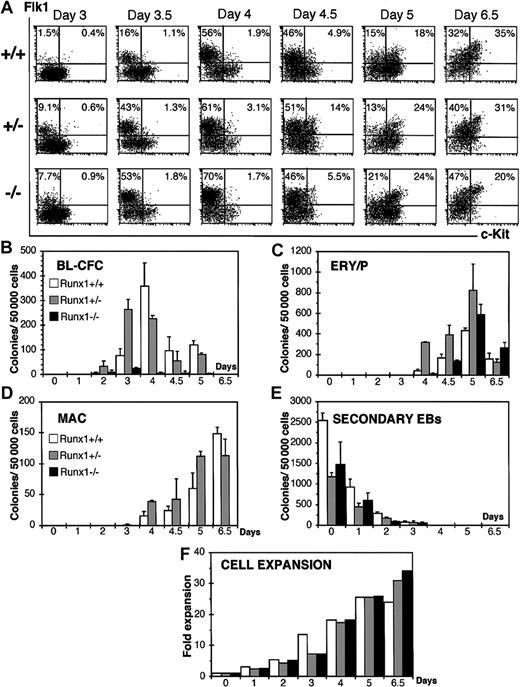
To determine if Runx1 heterozygosity or deficiency affects the in vitro differentiation potential of ES cells we analyzed the temporal expression patterns of the cell surface markers c-Kit and Flk1 following the in vitro differentiation of wild-type (Runx1+/+), heterozygous (Runx1+/–), and deficient (Runx1–/–) ES cells. We evaluated Flk1 expression, the receptor 2 for vascular endothelial growth factor (VEGF-R2), as previous results have demonstrated that it is expressed on BL-CFCs8,9 and that the onset of Flk1 corresponds to BL-CFC development. C-Kit is expressed on ES cells and on cell populations found at the early stages of EB development.9,10 Its expression is down-regulated following differentiation. Beyond the hemangioblast stage, c-Kit is coexpressed with Flk1 marking the establishment of hematopoiesis (Kennedy and G.K., unpublished observation, 2003). As shown in Figure 1A, a distinct Flk1+, c-Kit– cell population (Flk1+/c-Kit–) emerged in wild-type EBs by day 3.5 of differentiation. The frequency of these Flk1+/c-Kit– cells increased during the next 24 hours of culture. This window defines the hemangioblast stage of development as demonstrated by the presence of BL-CFCs at these time points (Figure 1B). By day 4.5, a double-positive Flk1+/c-Kit+ cell population was detected that increased in size over the next 48 hours of differentiation. Primitive erythroid and macrophage progenitors were detected in day 4.5, 5, and 6.5 wild-type EBs (Figure 1C-D) defining the hematopoietic stage of EB development.
Kinetics analysis of EBs generated from wild-type (Runx1+/+), heterozygous (Runx1+/–), and deficient (Runx1–/–) ES cells. (A) FACS analysis of Flk1 and c-Kit expression on Runx1+/+, Runx1+/–, and Runx1–/– EB-derived cells. The days of differentiation are indicated. Numbers in each quadrant represent the percent of total population in each fraction. Number of blast colonies (B), primitive erythroid colonies (C), macrophage colonies (D), and secondary EB colonies (E) generated by Runx1+/+, Runx1+/–, and Runx1–/– EB cells. Days of differentiation are indicated. Bars represent standard error of the mean number of colonies from at least 3 cultures. (F) Expansion of cell numbers following differentiation of Runx1+/+, Runx1+/–, and Runx1–/– ES cells.
Kinetics analysis of EBs generated from wild-type (Runx1+/+), heterozygous (Runx1+/–), and deficient (Runx1–/–) ES cells. (A) FACS analysis of Flk1 and c-Kit expression on Runx1+/+, Runx1+/–, and Runx1–/– EB-derived cells. The days of differentiation are indicated. Numbers in each quadrant represent the percent of total population in each fraction. Number of blast colonies (B), primitive erythroid colonies (C), macrophage colonies (D), and secondary EB colonies (E) generated by Runx1+/+, Runx1+/–, and Runx1–/– EB cells. Days of differentiation are indicated. Bars represent standard error of the mean number of colonies from at least 3 cultures. (F) Expansion of cell numbers following differentiation of Runx1+/+, Runx1+/–, and Runx1–/– ES cells.
In Runx1+/– and Runx1–/– EBs, the emergence of a distinct Flk1+/c-Kit– population was reproducibly detected at least 12 hours earlier than in wild-type EBs. Similar patterns were detected in 3 independent experiments. The average percentage and standard error of the mean of Flk1+ cells in these studies was 3.5 ± 2.1 in Runx1+/+ EBs, 14 ± 2.2 in Runx1+/– EBs, and 9.1 ± 2 in Runx1–/– EBs at day 3 of differentiation and 13 ± 4.4 in Runx1+/+ EBs, 48.9 ± 4.1 in Runx1+/– EBs, and 48 ± 6.5 in Runx1–/– EBs at day 3.5 of differentiation. The pattern of BL-CFC development in Runx1+/– EBs was also accelerated (Figure 1B), consistent with the earlier emergence of the Flk1+/c-Kit– cell population (Figure 1A).
As described previously,7 Runx1–/– EBs generated significantly lower numbers of blast colonies than wild-type or heterozygous EBs. In addition to acceleration of hemangioblast development, Runx1+/– EBs displayed accelerated hematopoietic commitment as demonstrated by the earlier generation of Flk1+/c-Kit+ cells and the presence of macrophage progenitors by day 4 of differentiation. The primitive erythroid progenitor numbers were higher in Runx1+/– EBs at all time points tested, reflecting expansion and possible accelerated development of this population as well. Runx1–/– ES cells generated primitive erythroid but no definitive hematopoietic precursors. Together, these findings indicate that commitment to the hematopoietic and vascular lineages, as defined by the development of the BL-CFCs and the generation of Flk1+ cells, is accelerated in Runx1+/– EBs.
This observed acceleration does not appear to result from a more rapid differentiation of the Runx1+/– ES cells as the kinetics of decline in secondary EB potential, an indication of undifferentiated ES cells, is similar to that of wild-type ES cells (Figure 1E). It is also not due to dramatic changes in the proliferative potential of the ES cells or early progeny as the expansion of the EB cell number is the same in wild-type, heterozygous, and Runx1–/– cultures (Figure 1F). Finally, the acceleration does not appear to reflect differences in cell cycle as flow cytometric analysis did not detect differences at the ES cell level or in early developing EBs between wild-type, heterozygous, and Runx1–/– ES cells (data not shown).
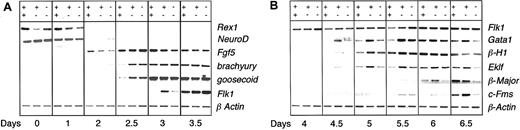
To further define the earliest developmental stage impacted by heterozygosity at the Runx1 locus, we evaluated the expression of genes indicative of the loss of ES cell potential, the development of epiblast-like cells, the onset of mesoderm development, and the commitment to the hematopoietic lineages during the differentiation of wild-type, heterozygous, and deficient (Runx1–/–) ES cells. The expression of Rex1, a zinc finger transcription factor found in ES cells but not in their differentiated progeny,11 was rapidly shut down upon differentiation of the 3 ES cell lines, consistent with the rapid loss of potential to generate secondary EBs (Figure 1E). NeuroD,12 a gene expressed in ES cells but down-regulated upon differentiation in conditions that do not support neuronal development, was also rapidly down-regulated in the EBs generated from the 3 different ES cell lines. Fgf5, a gene expressed in the epiblast of the early embryo,13 displayed a similar pattern in EBs from all 3 cell lines. In contrast to genes marking the early stages of development, brachyury14 and goosecoid15 expression, indicative of mesoderm commitment and development, was accelerated by at least 12 hours (Figure 2A) in Runx1+/– and Runx1–/– EBs compared with wild-type EBs. The onset of Flk1 expression was also accelerated in the Runx1+/– EBs (Figure 2A) confirming our flow cytometry analysis. As expected from the progenitor studies, the expression of genes associated with hematopoietic development, including the zinc finger transcription factor Gata1,16 the embryonic β-H1 globin, the adult β-major globin, the erythroid Kruppel-like transcription factor Eklf,17 and c-Fms was consistently detected at earlier time points in Runx1+/– EBs compared with wild-type EBs (Figure 2B). Low levels of β-major and c-Fms expression were detected at day 5.5 in Runx1–/– EBs. These levels did not, however, increase at later time points (Figure 2B; day 6 and 6.5), an observation consistent with the early defect in definitive hematopoiesis.7
Gene expression analysis of Runx1+/+, Runx1+/–Runx1–/– EBs. Gene expression of Runx1+/+ (++), Runx1+/–, (+–), and Runx1–/– (––) EBs from day 0 to day 3.5 (A) and from day 4 to day 6.5 (B) of differentiation. Day 0 represents undifferentiated ES cells.
Gene expression analysis of Runx1+/+, Runx1+/–Runx1–/– EBs. Gene expression of Runx1+/+ (++), Runx1+/–, (+–), and Runx1–/– (––) EBs from day 0 to day 3.5 (A) and from day 4 to day 6.5 (B) of differentiation. Day 0 represents undifferentiated ES cells.
The findings reported here indicate that Runx1 heterozygosity leads to an acceleration of mesodermal commitment and specification to the hemangioblast and to the hematopoietic lineages in Runx1+/– EBs. These observations suggest that Runx1 may play a negative role in mesoderm commitment at a stage of development earlier than predicted by the knock-out studies. We have previously demonstrated the presence of Runx1 transcripts in ES cells and in early EBs at stages corresponding to mesoderm development.7 An early role for Runx1 in mesoderm induction and specification in EBs is consistent with the role of the drosophila ortholog Runt in segmentation.18 In contrast to its potential negative role in mesoderm formation, Runx1 is essential for the establishment of definitive hematopoiesis at later stages. These apparent opposite functions of Runx1 in the context of mesoderm induction and hematopoietic commitment would be consistent with previous reported findings showing that Runx1 can act as an activator or a repressor of transcription in a cell-dependent context.19,20 Alternatively, repression and activation could be mediated by the expression of different isoforms of Runx1 with opposite transactivation potentials at specific stages of development.21-23 Further studies will be required to evaluate these possibilities. Our results are consistent with the acceleration of HSC development in Runx1 heterozygous embryos recently described.6 Our study and that of Cai et al6 have been conducted on Runx1+/– ES cells5 and Runx1+/– ES embryos,4 respectively generated by independent targeting events demonstrating that this acceleration of hematopoietic development is not limited to a single Runx1+/– ES cell line. In our previous study we did not detect dramatic differences in the kinetics of development of primitive erythroid, definitive erythroid, and macrophage precursors between the yolk sac of wild-type, Runx1+/–, and Runx1–/– embryos. It may be difficult to observe these changes in mice as the kinetics of hematopoietic development are rapid and as such, the differences between wild-type and heterozygous mice may be limited to a narrow window at the onset of this developmental program. Subtle changes in hematopoietic development that may be difficult to detect could, however, significantly impact later stages as suggested by the acceleration of HSC appearance and their more rapid exhaustion observed in Runx1+/– embryos.6
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-06-2149.
Supported by National Institutes of Health grants R01 HL18834, R01 HL65169, and R01 DK/HL60627.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal