Abstract
AML1/Runx1 is a frequent target of leukemia-associated gene aberration, and it encodes a transcription factor essential for definitive hematopoiesis. We previously reported that the AML1 molecules with trans-activation subdomains retained can rescue in vitro hematopoietic defects of AML1-deficient mouse embryonic stem (ES) cells when expressed by using a knock-in approach. Extending this notion to in vivo conditions, we found that the knock-in ES cell clones with AML1 mutants, which retain trans-activation subdomains but lack C-terminal repression subdomains including the conserved VWRPY motif, contribute to hematopoietic tissues in chimera mice. We also found that germline mice homozygous for the mutated AML1 allele, which lacks the VWRPY motif, exhibit a minimal effect on hematopoietic development, as was observed in control knock-in mice with full-length AML1. On the other hand, reduced cell numbers and deviant CD4 expression were observed during early T-lymphoid ontogeny in the VWRPY-deficient mice, whereas the contribution to the thymus by the corresponding ES cell clones was inadequate. These findings demonstrate that AML1 with its trans-activating subdomains is essential and sufficient for hematopoietic development in the context of the entire mouse. In addition, its trans-repression activity, depending on the C-terminal VWRPY motif, plays a role in early thymocyte development.
Introduction
Vertebrate hematopoietic development is characterized by the sequential appearance of two cell populations, further defined as primitive and definitive hematopoiesis.1 In mice, for example, primitive hematopoiesis is first seen in the form of blood islands in the yolk sac of the 7.5-day-old (E7.5) mouse embryo. It is thought that this cell population is directly differentiated from hemangioblasts, a bipotent precursor. Primitive hematopoiesis consists predominantly of a large and nucleated erythroid population containing embryonic-type hemoglobin. In contrast to the restricted and temporal development of this first wave, which diminishes at midgestation, definitive hematopoiesis originates from the aorta-gonad-mesonephros (AGM) region, where stem cells with long-term repopulating ability of multilineage hematopoiesis emerge at approximately E9.5, as a result of budding, from the ventral endothelial cells of the great vessels.2,3 The stem cells then migrate into the fetal liver and proliferate to rapidly establish the definitive hematopoiesis of all lineages, including progenitors for T- and B-lymphoid populations. Active sites for definitive hematopoiesis are transferred to bone marrow and spleen before birth and function throughout life within these organs.
These stem cells are equipped with a number of critical transcription factors that play pivotal roles in determining the fate of the cells at discrete developmental stages. Most of these molecules have been identified by isolating DNA-binding proteins to known cis-regulatory elements of lineage-specific genes or by cloning the DNA targets of leukemia-associated chromosomal translocations.4,5 Acute myeloid leukemia 1 (AML1, also known as runt-related transcription factor 1 [Runx1]) is a prototype of the latter group. This gene was originally isolated from the breakpoint of the t(8:21)(q22:q22) reciprocal translocation associated with 40% of patients with acute myelogenous leukemia of the French-British-American classification M2 subtype6-8 and later recognized as one of the most frequent targets of gene aberration associated with human leukemia.9,10 AML1 encodes the DNA-binding subunit of the core-binding factor transcription complex (CBF; also known as polyomavirus enhancer binding protein 2: PEBP2).11,12 Gene-targeting experiments have demonstrated that AML1 is essential for the early development of definitive hematopoiesis.13,14 AML1 is first detectable on E7.5 in the endoderm and in some parts of the extraembryonic mesoderm, and it is expressed in primitive erythrocytes in the yolk sac on E8.0.15 At approximately E10.5, AML1 is expressed in the ventral epithelium of the dorsal aorta and in the vitelline and umbilical arteries within the AGM region,15 where hematopoietic cell clusters emerge. AML1-deficient mice lack these hematopoietic clusters and die in utero after complete block of fetal liver hematopoiesis in midgestation,15-18 underscoring the pivotal role of AML1 in this initial stage of emerging definitive hematopoiesis. In addition, leukemia-associated fusion genes, such as AML1-MTG8 (myeloid translocation gene on chromosome 8, also called eight twenty-one [ETO]), formed by the t(8;21) translocation, have been shown to trans-dominantly repress this normal AML1 function within the entire animal19,20 and thus contribute to leukemic transformation.21
AML1 functions as a transcription activator and a transcription repressor in a context-dependent manner. AML1 protein has a modular structure, and its biochemical properties are mediated by the interaction with functional cofactors through its subdomains. For example, AML1 binds to the enhancer core DNA sequence, TGT/cGGT, which is found in the cis-regulatory sequences within a panel of hematopoiesis-related genes through the runt domain.22-24 This domain consists of 128 amino acid residues localized near the N-terminus with 69% identity to the Drosophila pair-rule gene known as runt.25 The runt domain also serves as the binding site for core-binding factor β (CBFβ, also known as PEBP2β),12,26,27 and this association is essential for the biologic activity of AML1.28-30 The trans-activation domain is localized to the region between residues 291 and 371 (451 residues represent one of the “full-length” isoforms, AML1b),31 and it serves as the binding site for transcription coactivators, such as p300/CBP.32 In contrast, the VWRPY motif, consisting of the 5 amino acids at the C-terminus, is thought to be involved in transcription repression through its association with a transcription corepressor, Groucho/transducin-like enhancer of split (TLE).33,34 The protein-protein interaction through the VWRPY motif appears to have been evolutionarily conserved,35,36 but little is known about its biologic roles in mammals. In addition, it has recently been demonstrated that residues 182 to 211 and 264 to 361 of AML1b also serve as repression subdomains in some cell types.37
We previously showed that the block of definitive hematopoiesis resulting from the loss of AML1 could be replicated in vitro by means of a murine embryonic stem (ES) cell differentiation system.13 The defect was rescued by re-expressing its cDNA from an artificial knock-in allele,38 thus providing evidence that this phenotype results solely from the absence of the gene. With this experimental system, we were also able to demonstrate that hematopoietic rescue required the trans-activation domain of AML1 but not the C-terminal trans-repression subdomain.38 To further define the biologic role(s) mediated through these functional subdomains of AML1, we performed 2-step in vitro culture experiments and chimera mouse analysis, for which we used a panel of ES cell clones with C-terminal deletion mutants of AML1. In addition, we used the knock-in approach to generate genome-manipulated mouse lines that express the AML1 molecule lacking the VWRPY motif. Our results indicate that transcriptionally active AML1 is essential for the early development of definitive hematopoiesis in the entire animal and that trans-repression activity through the VWRPY motif of the molecule plays a role in the early development of T-cell lineage.
Materials and methods
ES cell clones
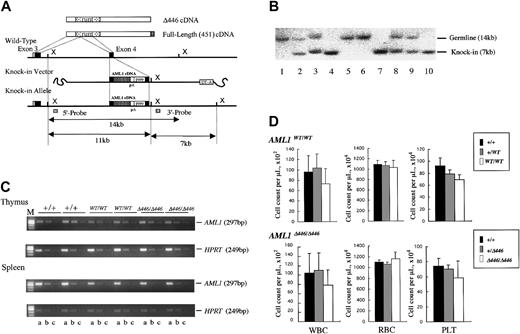
All replacement-type vectors were constructed based on a 12-kb genomic DNA fragment encompassing exon 4 of AML1, which corresponds to the middle of the runt domain as described elsewhere.38 Wild-type or one of the C-terminal–deleted cDNAs (Figure 1A) was inserted into exon 4 so as to keep the reading frame open, and polyadenylation (poly-A) signal sequences of the rabbit globin gene and a puromycin-resistance cassette for positive selection were inserted at the 3′ end of the artificial exon 4. A poly-A–less diphtheria toxin A suicide cassette was added at the utmost 3′ end of the construct for negative selection (Figures 1B, 3A). C-terminal–deletion mutants used for the vectors were constructed by inserting artificial stop codons into the mouse AML1b cDNA at residue 293, 320, 390, or 446, and the resultant mutant molecules were named Δ293, Δ320, Δ390, and Δ446 (Figure 1A). Each targeting vector was linearized and transfected into 9 million of the ES cells of either the wild-type (E14 or its derivatives) (Figure 3A) or the AML1-deficient (Figure 1B) genotype, and puromycin-resistant clones were selected. Homologous-recombinant clones were then serially assessed by means of Southern blot analysis with 5′ and 3′ outside probes, as previously described.38
Two-step culture experiments for in vitro hematopoietic rescue of knock-in ES cell clones. (A) Structure of the subdomain of the AML1 molecule and the C-terminal deletion mutants used for the knock-in experiments. Numbers represent positions of the residues. Runt indicates the runt domain; AD, trans-activation domain; ID, inhibitory domain; and VWRPY, VWRPY motif. (B) Targeted insertion of the cDNA into AML1-deficient ES cells, whose disrupted alleles are indicated by insertions of hygromycin-resistant (hygr) and neomycin-resistant (neo) cassettes at exon 4. Replacement-type vector for the targeted insertion and the schema of the resultant knock-in allele are shown at the bottom. pA indicates polyadenylation signal sequences; puro, puromycin-resistance cassette. (C) Capability for in vitro differentiation of the knock-in ES cell clones for each of the AML1 mutant cDNAs in a representative 2-step replating culture experiment (see “Materials and methods”). ES cells were cultured to form embryoid bodies. Hematopoietic progenitors of individual lineages that developed within embryoid bodies were then analyzed by the second-step culture on day 6 for primitive erythroid (Ery-P) and on day 10 for those of definitive origin, among them definitive erythroid (Ery-D), granulocyte-macrophage and macrophage (Myeloid), and mixed lineages including erythroid (E-Mix). Columns indicate numbers of progenitors per 105 disaggregated cells in culture.
Two-step culture experiments for in vitro hematopoietic rescue of knock-in ES cell clones. (A) Structure of the subdomain of the AML1 molecule and the C-terminal deletion mutants used for the knock-in experiments. Numbers represent positions of the residues. Runt indicates the runt domain; AD, trans-activation domain; ID, inhibitory domain; and VWRPY, VWRPY motif. (B) Targeted insertion of the cDNA into AML1-deficient ES cells, whose disrupted alleles are indicated by insertions of hygromycin-resistant (hygr) and neomycin-resistant (neo) cassettes at exon 4. Replacement-type vector for the targeted insertion and the schema of the resultant knock-in allele are shown at the bottom. pA indicates polyadenylation signal sequences; puro, puromycin-resistance cassette. (C) Capability for in vitro differentiation of the knock-in ES cell clones for each of the AML1 mutant cDNAs in a representative 2-step replating culture experiment (see “Materials and methods”). ES cells were cultured to form embryoid bodies. Hematopoietic progenitors of individual lineages that developed within embryoid bodies were then analyzed by the second-step culture on day 6 for primitive erythroid (Ery-P) and on day 10 for those of definitive origin, among them definitive erythroid (Ery-D), granulocyte-macrophage and macrophage (Myeloid), and mixed lineages including erythroid (E-Mix). Columns indicate numbers of progenitors per 105 disaggregated cells in culture.
Procedure for generating germline mice carrying the knock-in AML1 allele. (A) Procedure for the targeted insertion (knock-in) of full-length AML1 cDNA or of the Δ446 mutant into wild-type ES cells by homologous recombination. (B) Southern blot analysis of the knock-in allele using XbaI-digested genomic DNA obtained from a representative litter of crossed Δ446-heterozygotes. Lanes 1, 5, and 6: wild-type (+/+). Lanes 4, 7, and 10: homozygote (Δ446/Δ446). Remaining lanes: heterozygote (+/Δ446). (C) Semiquantitative RT-PCR for comparison of the expression of AML1 with that of the HPRT gene in wild-type germline mice (+/+), those homozygous for full-length cDNA (WT/WT), and those homozygous for Δ446 (Δ446/Δ446) genotypes (see “Materials and methods”). Results for 2 independent mice of each genotype are shown. Serially diluted cDNA pools were analyzed. Lanes a, b, and c: 5–2, 5–3, and 5–4 dilutions, respectively. M indicates marker. (D) Peripheral blood cell counts for the mutant mice compared with those for the matched-sibling control mice are indicated by columns. Bars signify standard deviations.
Procedure for generating germline mice carrying the knock-in AML1 allele. (A) Procedure for the targeted insertion (knock-in) of full-length AML1 cDNA or of the Δ446 mutant into wild-type ES cells by homologous recombination. (B) Southern blot analysis of the knock-in allele using XbaI-digested genomic DNA obtained from a representative litter of crossed Δ446-heterozygotes. Lanes 1, 5, and 6: wild-type (+/+). Lanes 4, 7, and 10: homozygote (Δ446/Δ446). Remaining lanes: heterozygote (+/Δ446). (C) Semiquantitative RT-PCR for comparison of the expression of AML1 with that of the HPRT gene in wild-type germline mice (+/+), those homozygous for full-length cDNA (WT/WT), and those homozygous for Δ446 (Δ446/Δ446) genotypes (see “Materials and methods”). Results for 2 independent mice of each genotype are shown. Serially diluted cDNA pools were analyzed. Lanes a, b, and c: 5–2, 5–3, and 5–4 dilutions, respectively. M indicates marker. (D) Peripheral blood cell counts for the mutant mice compared with those for the matched-sibling control mice are indicated by columns. Bars signify standard deviations.
Two-step culture analyses for hematopoietic differentiation of the ES cell clones
ES cells were first induced to form embryoid bodies in semisolid methylcellulose culture media, as described by Keller et al39 with modifications.38 Grown embryoid bodies were then disrupted by collagenase treatment, and 105 recovered single cells were replated in secondary methylcellulose cultures supplemented with human erythropoietin (EPO; Kirin Brewery, Tokyo, Japan) alone for primitive erythroid colonies or with a combination of human EPO, human granulocyte colony–stimulating factor (G-CSF; Kirin), murine granulocyte-macrophage colony-stimulating factor (GM-CSF), murine stem cell factor (SCF), and murine interleukin-3 (IL-3) (all from Genzyme, Cambridge, MA), which allowed for the growth of progenitors of definitive origin, as previously described.38
Generation of chimera and germline mice
ES cells were injected into the E3.5 blastocysts obtained from the C57BL6 strain, as described elsewhere,13 by using an inverted microscope (IX-70; Olympus, Tokyo, Japan) equipped with micromanipulators (Narishige, Tokyo, Japan). Manipulated blastocysts were then transferred into the uteri of pseudopregnant mother mice of the ICR strain. The resultant chimera mice were then crossed with C57BL6-strain mice to obtain ES-cell–derived offspring. Transmission of the mutated alleles and genotype of the progeny were determined by Southern blot analysis, as previously described.13,20,38 All procedures for mouse experiments performed in this study were approved by the Committee for Animal Research, Kyoto Prefectural University of Medicine.
Semiquantitative RT-PCR for AML1 expression
Semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) analysis to evaluate the expression of exogenous AML1 cDNA was performed as previously described.40 Five micrograms total RNA isolated from each of the tissue samples was used as the template for the random hexamer-primed RT reaction with Moloney murine leukemia virus (MMLV) reverse-transcriptase (Invitrogen, Carlsbad, CA), and serially diluted cDNA samples were assessed by PCR for a housekeeping gene, hypoxantin-guanine phosphoribosyl transferase (HPRT), to yield a standard representing the mRNA level within the samples (primers were 5′-GCTGGTGAAAAGGACCTCTCG-3′ for sense and 5′-CCACAGGACTAGAACACCTGC-3′ for antisense orientation). Equivalent dilutions of the cDNA samples were then analyzed for the AML1 message of a 292-bp fragment with a primer pair that encompassed intron 3: forward, 5′-CCAGCAAGCTGAGGAGCGGCG-3′; reverse, 5′-CCGACAAACCTGAGGTCGTTG-3′.40
GPI isoenzyme analysis
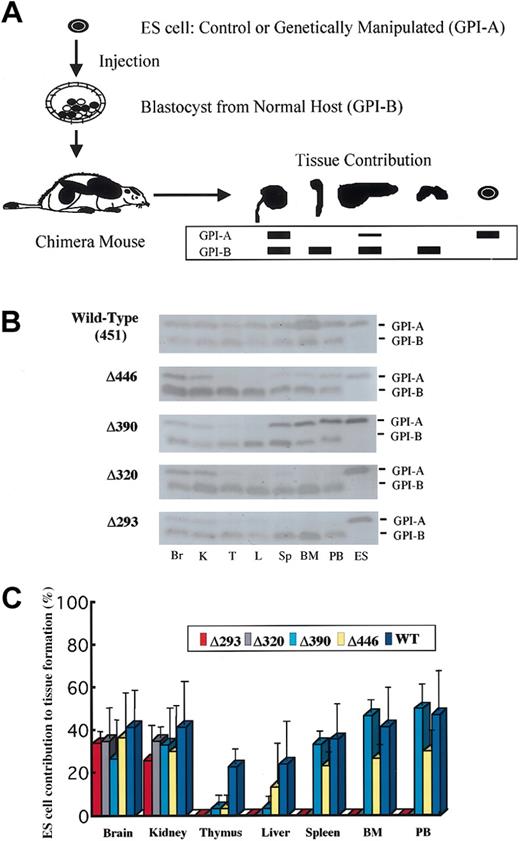
Organ or tissue samples obtained from chimera mice at the age of 5 weeks were homogenized, and the appropriately diluted supernatants were electrophoresed on cellulose acetate membranes (Helena Laboratories, Beaumont, TX) to separate glucose phosphate isomerase (GPI) isozymes (GPI-A from ES cells, 129 strain; GPI-B from the host, C57BL6 strain) according to the electrophoretic mobility of each (Figure 2A). The amount of each isozyme was then assayed by an on-the-membrane enzyme reaction, as described elsewhere.13,38 In parallel experiments, a panel of mixtures of 129-strain cells and C57BL6-strain cells in predetermined serial proportions (by increments of 10%) were analyzed to obtain a standard for determining the relative amount for the two isoforms in the samples of interest.
GPI isozyme analysis for the contribution by the knock-in ES clones to tissue in the chimera mice. (A) Schematic representation of the procedure for GPI analysis (see “Materials and methods”). (B) Representative results of the assay for chimera mice generated with knock-in clones for each of the AML1 mutant cDNAs. Lysates from tissues were separated by electrophoresis and stained for GPI activity. Contribution by ES cells is indicated by the GPI-A isoform, and that by host-derived cells is indicated by the GPI-B isoform. Results shown are for brain (Br), kidney (K), thymus (T), liver (L), spleen (Sp), bone marrow (BM), and peripheral blood (PB), with ES cells as GPI-A controls. (C) Columns with bars signifying standard deviation indicate quantification of tissue contributions by knock-in clones. Seven chimera mice were analyzed for full-length AML1 (WT: 451), 4 each for Δ446, Δ390, and Δ320, and 5 for Δ293 knock-in clones.
GPI isozyme analysis for the contribution by the knock-in ES clones to tissue in the chimera mice. (A) Schematic representation of the procedure for GPI analysis (see “Materials and methods”). (B) Representative results of the assay for chimera mice generated with knock-in clones for each of the AML1 mutant cDNAs. Lysates from tissues were separated by electrophoresis and stained for GPI activity. Contribution by ES cells is indicated by the GPI-A isoform, and that by host-derived cells is indicated by the GPI-B isoform. Results shown are for brain (Br), kidney (K), thymus (T), liver (L), spleen (Sp), bone marrow (BM), and peripheral blood (PB), with ES cells as GPI-A controls. (C) Columns with bars signifying standard deviation indicate quantification of tissue contributions by knock-in clones. Seven chimera mice were analyzed for full-length AML1 (WT: 451), 4 each for Δ446, Δ390, and Δ320, and 5 for Δ293 knock-in clones.
Flow cytometric analysis
Flow cytometric analysis was performed using conventional methods described elsewhere.20 The dissected thymus or spleen was mechanically disaggregated into single cells in RPMI 1640 medium (Invitrogen) containing 5% fetal calf serum (FCS) and were examined for live cells using the trypan blue dye exclusion test. Appropriate aliquots of the cells were then resuspended in phosphate-buffered saline (PBS) containing 3% FCS and 0.05% sodium azide. This was followed by incubation with each of the appropriately diluted monoclonal antibodies (mAbs) on ice for 60 minutes. The following fluorochrome-conjugated antibodies were used: anti-CD3 conjugated with fluorescein isothiocyanate (FITC) (Beckman Coulter, Fullerton, CA), anti-CD4 with phycoerythrin (PE) or with peridinin chlorophyll protein (Per-CP), anti-CD8a with FITC or with allophycocyanin (APC), anti-CD25 with FITC, anti-CD44 with PE, anti-B220 with PE, and anti-TCRβ with FITC (all from BD Biosciences, San Jose, CA). After 2 washings, samples were analyzed on a FACScalibur flow cytometer (BD Biosciences). Isotype-matched antibodies conjugated with the appropriate fluorochrome (BD Biosciences) at the same protein concentrations were used as negative controls for all experiments.
Hematopoietic progenitor assay for fetal liver cells
Fetal livers of E12.5 embryos were dissected under a stereoscopic microscope (SZX-12; Olympus) and then disaggregated by passing through a 26-gauge needle. In a triplicate experiment, 5000 cells were plated in a 1-mL mixture of Iscove modified Dulbecco medium (IMDM) with 1.2% methylcellulose supplemented with a combination of colony-stimulating factors, as described for the 2-step culture procedure.13,38 Colonies were counted under an inverted microscope (CK-2; Olympus), and the total number of the progenitors was estimated in proportion to the total number of cells obtained from each liver.
Results
AML1 transcription factor without its C-terminal repression subdomain rescues definitive myeloid lineages in vitro
AML1 is essential for the early development of definitive hematopoiesis but not for primitive erythropoiesis.13,14 The hematopoietic defect resulting from the loss of AML1 could be replicated in vitro with the murine ES cell differentiation system, and the phenotype was rescued by re-expressing its cDNA by using a knock-in approach, as previously reported.38 When definitive hematopoiesis is judged by the emergence of macrophages differentiated from the ES-cell–derived embryoid bodies, the AML1 molecule with its trans-activation subdomain appears to be sufficient for this rescue.38 In contrast, the C-terminal repression subdomain including the conserved VWRPY motif does not appear indispensable for rescue.38 To determine whether individual hematopoietic lineages showed any preference for rescue by specific deletion mutants, we subjected the constituent cells of the embryoid bodies to second-step culture in the presence of the appropriate colony-stimulating factors. AML1-deficient ES cell clones, which carried the knocked-in AML1 mutants that retained the trans-activating domain (wild-type AML1, Δ446, or Δ390) (Figure 1A-B), could rescue all lineages examined, including definitive erythroid (Ery-D), myeloid, and mixed lineage with erythroid (E-Mix) (Figure 1C). In contrast, mutants that lacked this domain (mutants Δ293 and Δ320) could not rescue any of the lineages (Figure 1C). In addition, examination of cytospin preparations by May-Grünwald-Giemsa staining showed that the appearance of the constituent cells from the colonies of definitive origin grown from knock-in clones was indistinguishable from that of the control heterozygous ES clones (not shown). Thus, in vitro hematopoietic rescue of individual definitive lineages requires the trans-activating domain of AML1, but the C-terminal repression subdomain does not appear to be essential.
In vivo rescue for myeloid lineages requires transcriptionally active AML1, whereas C-terminal repression subdomains may have a function in thymus, liver, or both
To define whether the trans-activating subdomain is required for in vivo hematopoietic development, as was observed in the in vitro experiments, we next evaluated the ability of ES cell clones to contribute to tissue formation in chimera mice. We generated chimera mice by injecting each of the knock-in ES cell clones into blastocysts obtained from a wild-type (C57BL6) host strain. ES cell contribution to tissue formation was assessed by means of GPI isoenzyme analysis (Figure 2A). For each mutation, 4 or more 5-week-old chimera mice were examined. As was seen in the full-length AML1b cDNA knock-in clone (Figure 2B-C), Δ446 and Δ390 mutants, which retain the activation subdomain but lack C-terminal repression subdomains, always contributed to myeloid tissues, including bone marrow, peripheral blood, and spleen. In contrast, Δ320 and Δ293 mutants did not contribute to hematopoietic tissues (Figure 2B-C), indicating that the activation subdomain is important for establishing in vivo hematopoiesis and that the C-terminal repression subdomain, including the VWRPY motif, is not required.
ES cell clones bearing the Δ320 or Δ293 mutant, which did not contribute to hematopoietic tissues, did not contribute to the liver either (Figure 2B-C). In addition, rescued ES clones with the Δ446 or Δ390 mutant, which did contribute to myeloid tissues in the chimera mice, contributed less to the formation of the liver (Figure 2B-C), suggesting that the cells with intact AML1 function were preferentially used to generate the liver. Similarly, rescued ES cell clones with the Δ446 or Δ390 molecule tended to contribute less to the thymus than to the myeloid tissues (Figure 2C), indicating that AML1 may play a role in thymus development through its C-terminal VWRPY motif.
Generation of germline mice bearing the knock-in allele of full-length or Δ446 cDNA for AML1
For the next step, we examined whether the findings from ES-cell–based experiments applied to the entire animal. For this purpose, we generated mouse lines that carried a knock-in allele of either the full-length or the Δ446 cDNA of the AML1 molecule in their germlines. As outlined in “Materials and methods” and illustrated in Figure 3, we introduced an exogenous cDNA into exon 4 of the AML1 gene locus by homologous recombination in wild-type mouse ES cell lines. These artificial alleles were designed to express 3′ sequences of AML1b cDNA downstream from its exon 4, under control of the endogenous cis-elements of transcription. Two independent mouse lines were established for each germline knock-in mutation (Table 1), and the phenotype of a mutation was recognized when it was observed in both lines. Assessment by means of semiquantitative RT-PCR analysis showed that the expression level of the knocked-in gene in thymus or spleen was indistinguishable from that of the endogenous gene (Figure 3C). The 2 mutant alleles were segregated according to Mendelian inheritance (Table 1), and homozygous mice carrying either of the knocked-in cDNAs (AML1WT/WT or AML1Δ446/Δ446) and kept under specific pathogen-free conditions were fertile and appeared healthy compared with the wild-type control mice. Thus, the knock-in procedure resulted in generating mice that expressed exogenous AML1 cDNA of full-length or of the Δ446 mutant, and each artificial allele could rescue these mice from the embryonic death that had been observed in the simple targeted disruption13,14 of this gene.
Genotypes of litters born by intercrossing heterozygous parents
AML1+/WT crossing . | . | . | . | AML1+/Δ446crossing . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone . | +/+ . | +/WT . | WT/WT . | Clone . | +/+ . | +/Δ446 . | Δ446/Δ446 . | ||||||
| 164 | 46 | 68 | 53 | 804 | 60 | 78 | 46 | ||||||
| 734 | 26 | 72 | 31 | 807 | 23 | 60 | 26 | ||||||
AML1+/WT crossing . | . | . | . | AML1+/Δ446crossing . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone . | +/+ . | +/WT . | WT/WT . | Clone . | +/+ . | +/Δ446 . | Δ446/Δ446 . | ||||||
| 164 | 46 | 68 | 53 | 804 | 60 | 78 | 46 | ||||||
| 734 | 26 | 72 | 31 | 807 | 23 | 60 | 26 | ||||||
+ indicates wild-type allele for the AML1 gene; WT, knock-in allele with full-length (451-residue) cDNA of AML1 gene; and Δ446, knock-in allele with Δ446-deletion mutant cDNA of AML1 gene.
Genetically modified mice of the AML1WT/WT or AML1Δ446/Δ446 genotype showed minimal abnormality in myeloid hematopoiesis
Homozygous mice had a tendency to have fewer white blood cells and lower platelet counts compared with those observed in the control wild-type or heterozygous littermates, regardless of which knock-in allele, AML1WT/WT or AML1Δ446/Δ446, they carried. However, these differences were not statistically significant (Figure 3D). In addition, red blood cell count, hemoglobin concentration, and hematocrit of the mutant mice were indistinguishable from those of control animals (Figure 3D). In agreement with these observations, histologic examinations revealed that the mutant mice had normal white blood cell differentials and indistinguishable architecture and cellularity of hematopoietic bone marrow compared with controls (negative data; not shown).
The biologic effect of AML1 on early hematopoietic development depends on the dosage. Mice heterozygous for the inactivating mutation of AML1 feature a precocious appearance of hematopoietic stem cells in the yolk sac41 and an approximately 50% reduction of hematopoietic progenitor cells in the fetal liver.14 To analyze fetal liver hematopoiesis in the knock-in animals, we cultured fetal liver cells obtained from the litter embryos of intercrossed heterozygotes at E12.5. In the presence of a combination of colony-stimulating factors that supported the growth of hematopoietic progenitors of definitive origin, approximately 2 to 3 × 104 colonies of various cell lineages per liver were detected in wild-type littermates. In contrast, the average number of progenitors for the mutant mice was approximately 20 000 for AML1WT/WT and 19 000 for AML1Δ446/Δ446, for an efficiency of 60% to 70% of that seen in wild-type littermates (Table 2). Mutant mice thus showed a tendency to have fewer progenitors, but the difference was not statistically significant.
Hematopoietic progenitors detected in fetal livers of knock-in embryos at E12.5
Genotype . | n . | BFU-Es . | CFU-GMs . | CFU-Mix's . | Total . |
|---|---|---|---|---|---|
| Wild-type | |||||
| +/+ | 11 | 5 ± 1 (1725) | 73 ± 20 (25 524) | 17 ± 3 (5635) | 103 ± 25 (36 450) |
| WT/WT | 11 | 7 ± 1 (1219) | 79 ± 19 (14 684) | 20 ± 4 (3706) | 110 ± 20 (20 347) |
| Δ446 mutant | |||||
| +/+ | 11 | 6 ± 2 (1693) | 75 ± 27 (23 464) | 17 ± 7 (5253) | 104 ± 35 (28 729) |
| Δ446/Δ446 | 11 | 9 ± 3 (1609) | 86 ± 38 (13 163) | 23 ± 10 (3503) | 123 ± 51 (19 112) |
Genotype . | n . | BFU-Es . | CFU-GMs . | CFU-Mix's . | Total . |
|---|---|---|---|---|---|
| Wild-type | |||||
| +/+ | 11 | 5 ± 1 (1725) | 73 ± 20 (25 524) | 17 ± 3 (5635) | 103 ± 25 (36 450) |
| WT/WT | 11 | 7 ± 1 (1219) | 79 ± 19 (14 684) | 20 ± 4 (3706) | 110 ± 20 (20 347) |
| Δ446 mutant | |||||
| +/+ | 11 | 6 ± 2 (1693) | 75 ± 27 (23 464) | 17 ± 7 (5253) | 104 ± 35 (28 729) |
| Δ446/Δ446 | 11 | 9 ± 3 (1609) | 86 ± 38 (13 163) | 23 ± 10 (3503) | 123 ± 51 (19 112) |
Numbers are mean ± SD per 5000 nucleated cells cultured. Mean numbers of cells per liver appear in parentheses. WT indicates wild-type AML1 cDNA knock-in; Δ446, Δ446 mutant AML1 CDNA knock-in; BFU-E, erythroid burst-forming unit; CFU-GM, granulocyte-macrophage colony-forming unit; and CFU-Mix, mixed-lineage colony-forming unit.
These results indicate that, as in the case of the knock-in expression of full-length AML1 cDNA, the expression of the exogenous AML1 cDNA of the Δ446 mutant, which retained its trans-activation subdomains but lacked the VWRPY motif, was sufficient to support myeloid hematopoiesis throughout the life of the entire mouse.
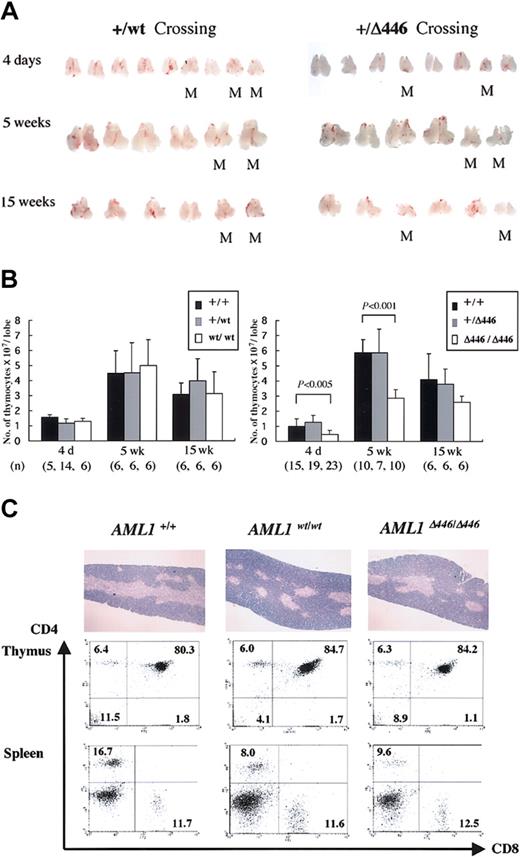
AML1Δ446/Δ446 mice have smaller thymus
Further examination showed that the thymi obtained from AML1Δ446/Δ446 mice were smaller than those from wild-type or heterozygous littermates or those from age-matched homozygotes with the full-length cDNA knock-in allele, AML1WT/WT. This difference was detectable at birth or when the mice were 4 days old, and it became more prominent at 5 weeks when this organ normally reaches its largest size. This size difference was less prominent in adult mice, however (Figure 4A). The average number of cells in the thymi from AML1Δ446/Δ446 mice was almost half that in the thymi from controls, a statistically significant difference (Figure 4B).
Reduced thymus size found in born AML1Δ446/Δ446 mice. (A) Macroscopic appearance of the dissected thymus from representative litters resulting from intercrossing of mutant mice heterozygous for either full-length cDNA knock-in (+/WT) or Δ446 mutant knock-in (+/Δ446) at the age of 4 days, 5 weeks, and 15 weeks. M indicates homozygous mutants. (B) Columns with bars for standard deviation indicate the number of cells per one lobe of thymus from littermate mice of each genotype. Numbers of mice examined are indicated in parentheses at the bottom. Statistically significant differences are shown as P values. (C) Microscopic appearance of hematoxylin-eosin–stained sections of the thymus are illustrated in the top panels (original magnification, × 13.2), and results of flow cytometric analysis of the CD4 and CD8 expression for thymocytes and splenic T cells are shown below.
Reduced thymus size found in born AML1Δ446/Δ446 mice. (A) Macroscopic appearance of the dissected thymus from representative litters resulting from intercrossing of mutant mice heterozygous for either full-length cDNA knock-in (+/WT) or Δ446 mutant knock-in (+/Δ446) at the age of 4 days, 5 weeks, and 15 weeks. M indicates homozygous mutants. (B) Columns with bars for standard deviation indicate the number of cells per one lobe of thymus from littermate mice of each genotype. Numbers of mice examined are indicated in parentheses at the bottom. Statistically significant differences are shown as P values. (C) Microscopic appearance of hematoxylin-eosin–stained sections of the thymus are illustrated in the top panels (original magnification, × 13.2), and results of flow cytometric analysis of the CD4 and CD8 expression for thymocytes and splenic T cells are shown below.
Despite this quantitative difference, histologic findings for the thymus of AML1Δ446/Δ446 mice, obtained from microscopic examination of hematoxylin-eosin–stained sections, were not significantly different from those for control animals (Figure 4C). Flow cytometric analysis showed a lower CD4/CD8 cell ratio of the splenic T lymphocytes for AML1Δ446/Δ446 and AML1WT/WT mice than for wild-type mice (Figure 4C), as was previously reported for mice heterozygous for an inactivated AML1 allele.42 However, the proportions of CD4- and CD8-expressing thymocytes in both knock-in strains were indistinguishable from those found in wild-type control mice (Figure 4C), in contrast to the findings for haploinsufficient mice, which showed reduced cell numbers for thymic single-positive (SP) cell populations.42 Thymocytes from AML1Δ446/Δ446 mice expressed the CD3 antigen, which suggests that mature T-cell receptors were expressed on the surfaces of the cells (not shown). Furthermore, there was no increase in terminal deoxynucleotidyl transferase–mediated nick-end-labeling (TUNEL)–positive apoptotic cells in the thymocytes of 4-day-old or 5-week-old AML1Δ446/Δ446 mice in comparison with those of control littermates (negative data; not shown).
Together with the findings obtained from chimera mouse analysis, these results indicate that AML1 plays an important in vivo role, somehow mediated by the C-terminal repression subdomain, in determining the number of cells in the developing thymus, most likely in a cell-autonomous fashion.
Δ446 mutation affects CD4 expression in early thymocyte development
Early T-cell development is a multistep process in the thymus, where hematopoietic stem cells migrate and are instructed and selected to eventually constitute mature peripheral T-cell populations of either CD4+ helper cells or CD8+ cytotoxic cells. The developmental stages are characterized by specific surface marker expression: in the very early stage, thymocytes are negative for CD4 or CD8 and are thus called a double-negative (DN) cell population. On successful rearrangement of one allele of the T-cell receptor β (TCRB) gene locus (β-selection), DN cells then express CD4 and CD8 antigens to become a double-positive (DP) cell population. Finally, DP cells lose the expression of either antigen to form mature SP cell populations. In addition, progression of thymocytes through the DN stage can be further categorized according to the expression of CD25 or CD44. The 4 stages of DN development, with increasing levels of maturity, are called DN1 (CD44+CD25–), DN2 (CD44+CD25+), DN3 (CD44–CD25+), and DN4 (CD44–CD25–), and β-selection occurs in DN3 cells.43 We analyzed the thymi of the mutant mice in the late gestational stage and found that differences in the size of the thymi of the Δ446 embryos compared with those of control littermates were detectable as early as E17.5. In addition, the CD4 SP fraction in the mutant embryos was higher than in the wild-type littermate embryos on E17.5 and E18.5 (Figure 5). It should be noted that these phenomena were not observed in AML1WT/WT embryos (Figure 5). Nevertheless, the AML1Δ446/Δ446 embryos regained the normal CD4-CD8 differential at birth or thereafter (Figure 4C), and there was no apparent block in the transition between the DN3 and DN4 stages in AML1Δ446/Δ446 embryos (Figure 5).
Effects of Δ446 mutation on CD4 and CD8 expression during T-cell ontogeny. Embryonic thymi from wild-type (AML1+/+), AML1WT/WT, and AML1Δ446/Δ446 genotypes at several developmental stages, including E16.5, E17.5, and E18.5, were analyzed for CD4 and CD8 expression by flow cytometric analysis. Δ446 mutation resulted in an increase in the CD4 SP cell population, as indicated. Total numbers of thymus cells are shown above the quadrant graphs. Double-negative populations at E18.5 were further analyzed by 4-color analysis for CD25 and CD44 expression, as shown in the panels on the right. Thymi from the Δ446 mutants showed no difference in DN-stage progression compared with those from controls.
Effects of Δ446 mutation on CD4 and CD8 expression during T-cell ontogeny. Embryonic thymi from wild-type (AML1+/+), AML1WT/WT, and AML1Δ446/Δ446 genotypes at several developmental stages, including E16.5, E17.5, and E18.5, were analyzed for CD4 and CD8 expression by flow cytometric analysis. Δ446 mutation resulted in an increase in the CD4 SP cell population, as indicated. Total numbers of thymus cells are shown above the quadrant graphs. Double-negative populations at E18.5 were further analyzed by 4-color analysis for CD25 and CD44 expression, as shown in the panels on the right. Thymi from the Δ446 mutants showed no difference in DN-stage progression compared with those from controls.
Discussion
It has been established that AML1 plays a pivotal role in definitive hematopoiesis because the loss of this gene in the mouse results in midgestational death caused by blockage of fetal liver hematopoiesis.13,14 We previously demonstrated that the hematopoietic defect can be replicated by using ES cells in an in vitro differentiation system and that this in vitro phenotype could be rescued by the re-expression of AML1 cDNA from a knock-in allele.38 In the study presented here, we focused on the biologic properties of AML1 mediated through its C-terminal subdomains as a result of hematopoietic rescue of AML1 deficiency at single-cell, tissue, and whole-animal levels. The findings for the C-terminal deletion mutants indicated that expression of the AML1 molecule, with its trans-activation subdomains retained, is necessary and sufficient for the in vitro differentiation of ES cells into definitive hematopoietic cells of all myeloid lineages and for the in vivo contribution of ES cells to myeloid tissues in chimera mice. These results further support our earlier observations in embryoid body experiments.38 In addition, our results provide genetic proof that the AML1 molecule, with its trans-activation subdomains retained, is sufficient for hematopoietic rescue within the entire animal. Germline mice that homozygously carried knock-in alleles that expressed full-length or C-terminally deleted cDNA and that lacked a trans-repression subdomain, including the VWRPY motif, were found to be viable and showed minimal abnormalities in myeloid hematopoiesis throughout life.
The VWRPY motif, localized at the C-terminus, is one of the functional subdomains for the trans-repression activity of the AML1 molecule, suggested as the binding site for the transcription corepressor Groucho/TLE.34-36 The orthologue of AML1 in Drosophila, Runt, functions as a transcriptional repressor for its target, the even-skipped gene, and in the maintenance of the repressed state for another target, the engrailed gene. Both these functions are realized in a VWRPY-motif/Groucho–binding-dependent fashion to mediate the segmentation process of the blastoderm.33,44 Although the trans-repression property of AML1 through the VWRPY motif is genetically conserved,35,36 the biologic importance of this activity in mammals has not yet been analyzed in detail. In this study, we observed that mice lacking this VWRPY motif of AML1 demonstrated less thymus cellularity and more CD4 SP cells than seen in controls, including heterozygous or wild-type littermates and age-matched mice homozygous for full-length AML1 cDNA. In addition, ES cell clones carrying AML1 mutants without the C-terminal trans-repression subdomains were found to make an inadequate contribution to the thymus in chimera mice. These findings indicate that mammalian AML1 does have biologic functions specific to this trans-repression subdomain in T-cell ontogeny. Data available thus far strongly suggest that the observed AML1 function is cell-autonomous. Nevertheless, further studies, such as complementation assay experiments involving T-cell–depleted Rag2-mutant45,46 mice that received transplanted bone marrow cells derived from VWRPY mutant mice, should result in more specific characterization of the thymic manifestation of the mutation and should contribute to defining the nature of the activity in greater detail.
A recent study describes T-cell–specific conditional disruption of AML1.47 A genetically modified mouse line was generated carrying floxed exon 4 of the AML1 gene locus and the transgene of lck-promoter–driven Cre. Thymocytes from the progeny mice are thus lacking in whole AML1 activity in the early DN stages of the T-cell ontogeny. Subsequently, reduced numbers of thymocytes and premature CD4 expression caused by the loss of appropriate repression of this gene48 were noted in the thymus during late gestation.47 In our study, AML1Δ446/Δ446 embryos showed reduced cell numbers and increased CD4 SP cell populations during early thymic development, both of which were observed when AML1 was conditionally disrupted in the thymus.47 Available data did not allow us to draw a conclusion regarding the cellular mechanism of AML1 (ie, whether premature CD4 expression occurred because of the loss of appropriate repression of this gene or CD8 expression was reversed in the DP cell population). Nevertheless, the results presented here at least indicate that these phenomena are mediated by AML1's trans-repression activity, which depends on its VWRPY motif. In contrast to T-cell–specific AML1 disruption, which also showed an impaired β-selection process leading to a block in the DN3 stage during late gestation and abnormal CD8 expression in young adults,47 AML1Δ446/Δ446 had no effect on these stages, suggesting that these biologic functions are mediated by the activity of AML1 and that this activity is independent of the VWRPY motif.
The contribution to the liver by ES cells with AML1, which lacked the C-terminal repression subdomain was found to be inadequate in the chimera mouse analysis, whereas AML1Δ446/Δ446 mice developed functioning livers. In addition, even though the contribution to the liver by ES cells lacking entire AML1 activity was even less,13,29,38 the AML1-knockout embryo does develop a liver at least up to midgestation.13,14 It is obvious that ES cells with intact AML1 function are preferentially used to form the liver, and conditional liver-specific disruption of this gene should make it clear whether this gene is also involved in liver development.
Consistent with our observation that AML1 performs biologic functions in the above-mentioned sites through its C-terminal VWRPY motif, the expression of its associated corepressor, TLE, has also been reported to occur in those tissues. The Drosophila orthologue, Groucho protein, which plays critical roles in multiple developmental processes by acting as a dedicated transcriptional corepressor for a number of transcription regulators—including Hairy, Dorsal, Engrailed, Tcf, and Runt—is encoded by a single gene and is ubiquitously expressed during early development of the fly.49,50 In contrast, at least 6 homologues have so far been identified in mammals: TLE1 through TLE6 in humans and Groucho-related genes (Grg)1 through Grg6 in mice.51-56 TLE/Grg1, 2, 3, 4, and 6 are thought to be full-length molecules with all 5 functional subdomains, whereas TLE/Grg5 is a truncated form that retains only the 2 N-terminal domains.50,52,55 Expression profiles of the TLE/Grg molecules have been extensively analyzed and reportedly occur in unique but overlapping patterns during embryonic development. Sites at which they are highly expressed include the central nervous system, somites, and proliferating epithelial tissues, which undergo mesenchymal induction.51-58 Expression of the genes was also detected in adult livers and thymi.51,56-58 Of note, a histologic study using in situ hybridization analyses showed Grg3 to be highly expressed in thymocytes at E16.5,55 which coincides with the developing stages of the thymus when the AML1Δ446/Δ446 mice manifested the phenotype. This observation may explain the stage-specific effect of the VWRPY mutation. However, so far only TLE/Grg1 and TLE/Grg2 have been experimentally proven to function as corepressors for AML1,35,36,59 whereas no evidence has been provided yet that other TLE/Grg family members could function in a similar fashion. Further analysis of the temporal and spatial expression patterns of the TLE/Grg family members in mouse development and their functional relationship with AML1 should make a significant contribution to a better understanding of the molecular basis for the AML1Δ446/Δ446 phenotype.
The AML1 gene locus is one of the most frequent targets of human leukemia-associated gene aberrations, and the leukemic oncoprotein is believed to be formed by 2 mechanisms. One is a chromosomal translocation that results in the formation of chimera proteins, such as AML1-MTG8 (ETO), AML1-Evi1, or Tel-AML1.9,10 These molecules acquire a strong ability to trans-dominantly repress AML1's transcriptional activities.60-65 The other mechanism is a genetic mutation often found within the runt domain sequences.66-69 Mechanisms of leukemogenesis caused by these point mutations of AML1 remain for the most part unknown. The experimental system with a knock-in approach used in this study to express exogenous mutant AML1 in germline mice by a knock-in approach is expected to become a valuable tool for probing the molecular basis for leukemogenesis mediated by subtle mutations in the AML1 gene locus found in leukemia and related disorders in humans.
To summarize, genetic dissection of the biologic function and the C-terminal function subdomain structure of AML1 allowed us to identify their respective roles as dependent on or independent of the C-terminal repression subdomain. In addition, we successfully developed an experimental system that enabled us to directly analyze the genetic consequences of AML1 mutations in the context of the entire animal. These findings and genetic tools can be expected to provide new insights into the molecular basis of hematopoietic regulation and leukemogenesis caused by alterations in the AML1 gene.
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-06-2109.
Supported by grants from the Ministry of Education, Sports, Culture, Science, and Technology, Japan, and by Grants-in-Aid for Cancer Research from the Ministry of Health, Welfare, and Labor, Japan. T.O. also was supported by a grant from the Mitsubishi Pharma Research Foundation for Advanced Medicine.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Shuji Takao, Nobuhide Kitai, Toshiko Nakano, Ayumi Takahashi, and Emi Saito for their outstanding technical assistance. We also thank Drs Kiyoshi Takeda (Osaka University), Ron DePinho (Harvard University), and the faculty members of the 4th Annual Mouse Developmental Genetics Course (held at the Albert Einstein College of Medicine) for their valuable comments on the set-up and maintenance of a transgenic mouse facility. We are grateful to the Core Animal Resource Center of Kyoto Prefectural University of Medicine, organized by Dr Masakazu Kita, for providing us with an excellent environment for the mouse resources.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal