Abstract
Depsipeptide (FK228) is a novel histone deacetylase inhibitor currently in clinical trials and the first to demonstrate clinical activity in patients. Responses have been observed in patients with T-cell lymphomas, despite prior treatment with multiple chemotherapeutic agents. To better understand the effects of histone deacetylase inhibitors on T-cell lymphoma, the human T-cell lymphoma cell line HUT78 was tested for sensitivity and molecular response to depsipeptide. Treatment with depsipeptide, as well as other histone deacetylase inhibitors, caused induction of histone acetylation, induction of p21 expression, and substantial apoptosis without significant cell cycle arrest. Treatment with the caspase inhibitor z-VAD-fmk significantly inhibited depsipeptide-induced apoptosis, enabling detection of cell cycle arrest. Treatment with depsipeptide increased expression of the interleukin-2 (IL-2) receptor, and combination with the IL-2 toxin conjugate denileukin diftitox resulted in more than additive toxicity. Cells selected for resistance to depsipeptide overexpressed the multidrug resistance pump, P-glycoprotein (Pgp). However, cells selected for resistance to depsipeptide in the presence of a Pgp inhibitor had a Pgp-independent mechanism of resistance. These studies confirm the activity of depsipeptide in a T-cell lymphoma model and suggest a general sensitivity of T-cell lymphoma to histone deacetylase inhibitors, an emerging new class of anticancer agents. (Blood. 2004;103:4636-4643)
Introduction
The histone deacetylase inhibitors (HDIs) are a new class of antineoplastic agents currently being evaluated in clinical trials. HDIs induce growth arrest usually associated with cellular differentiation or apoptosis. Alterations in the enzymes controlling histone acetylation and deacetylation have been shown to be a direct mechanism of transformation in some malignancies.1 The consequence of a decrease in histone acetylation is a decreased expression of cell cycle inhibitors and other genes involved in regulating a differentiated phenotype.2
Depsipeptide (FK228) is an HDI that has in vitro and in vivo cytotoxic activity.3 Several families of HDIs have been characterized. These include the short-chain fatty acids, such as sodium butyrate and valproic acid; the organic hydroxamic acids, such as trichostatin A (TSA) and suberanilohydroxamic acid (SAHA); the benzamides, such as CI-994 and MS-27-275; the cyclic tetrapeptides, such as trapoxin A; and the bicyclic depsipeptides, such as depsipeptide. Similar to other HDIs, depsipeptide has been shown to induce cell cycle arrest, cellular differentiation, and apoptosis. Depsipeptide induces a p53-independent/p21-dependent G1 arrest and a p21-independent G2/M arrest. In addition, depsipeptide causes alterations in gene expression, including increased expression of p21 and cyclin E and decreased expression of cyclin D1 and c-myc.4-6
T-cell lymphomas are composed of a spectrum of clinical phenotypes ranging from low-grade, cutaneous T-cell lymphomas (CTCLs) to highly aggressive peripheral T-cell lymphomas. Except for early-stage CTCL, response to chemotherapy is typically limited. In a phase 1 trial of depsipeptide conducted at the National Cancer Institute (NCI), responses were observed in patients with T-cell lymphoma, and a phase 2 trial of depsipeptide in these patients is ongoing.7,8
The interleukin-2 (IL-2) receptor is a marker of T-cell differentiation and is expressed in some T-cell malignancies.9,10 The IL-2 receptor, IL-2R, is composed of the α, β, and γ subunits. All 3 subunits complexed together, IL-2Rαβγ, confer high-affinity binding to IL-2, while the combined βγ subunits, IL-2Rβγ, have intermediate affinity.11,12 The β subunit, CD122 or p75, is important for ligand internalization, and the γ subunit, CD132 or p64, is important for signal transduction.13 Therapies that target the IL-2 receptor have been developed for the treatment of T-cell lymphoma. Denileukin diftitox (DAB389IL-2 or Ontak) is a fusion molecule of IL-2 to diphtheria toxin and has been shown to have efficacy in patients with T-cell lymphoma.14,15 In addition, antibodies to the IL-2 receptor, specifically the α subunit (CD25 or Tac), are also in clinical trials. Differentiating agents, such as retinoids, have been shown to increase the expression of the IL-2 receptor subunits in malignant T cells.16 Recent studies have demonstrated that both retinoids and the histone deacetylase inhibitor arginine butyrate increase sensitivity of the HUT78 cutaneous T-cell lymphoma cell line to denileukin diftitox.17,18 Since depsipeptide, like other HDIs, is able to induce markers of differentiation, we postulated that it could also increase expression of the IL-2 receptor subunits. Induction of the IL-2 receptor could allow for combination therapies with depsipeptide.
Since depsipeptide appears to be effective clinically, it is important to understand mechanisms that could limit its efficacy. Using the COMPARE analysis of the NCI drug screen, depsipeptide was found to segregate with other agents that are transported by P-glycoprotein (Pgp).19 Cell lines that overexpress Pgp are resistant to depsipeptide, and this resistance is reversed by Pgp inhibitors.19 Differentiating agents, including the HDIs, have been shown to induce the expression of Pgp.20-22 These studies suggest that depsipeptide could induce its own mechanism of resistance. To evaluate resistance mechanisms other than Pgp, cells were selected for resistance to depsipeptide in combination with an inhibitor of Pgp. This strategy has previously been successful at identification of non-Pgp mechanisms of resistance.23-26
We thus initiated laboratory studies to evaluate the molecular effects of depsipeptide in a human T-cell lymphoma cell line as a corollary to ongoing clinical studies. Our goal in these studies was to identify molecular changes following exposure to depsipeptide that could be used as biomarkers of response to depsipeptide, or other HDIs, in tumors of patients being treated with these agents. Histone acetylation, gene expression, cell cycle arrest, and apoptosis were evaluated to demonstrate the effects of depsipeptide. With the goal of testing HDIs in combination with other antineoplastic agents, we tested the effects of depsipeptide on the expression of the IL-2 receptor subunits. These were chosen since agents targeting the IL-2 receptor are already in clinical use or in clinical development for patients with T-cell lymphoma. A final goal was to evaluate mechanisms of resistance that may play a role in limiting response to depsipeptide. The HDIs induce expression of P-glycoprotein, which has been shown to confer resistance to depsipeptide. We asked whether depsipeptide induced or selected for multidrug resistance protein 1 (MDR-1) expression in the HUT78 cell line. To investigate non-Pgp mechanisms of resistance to HDIs, we also selected cells in the presence of a Pgp inhibitor.
Materials and methods
Cell lines and materials
The human HUT78 cells were purchased from the American Type Culture Collection (Manassas, VA). The cells were maintained in RPMI 1640 supplemented with 10 mM Hepes (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 1 mM pyruvate, penicillin 100 U/mL, streptomycin 100 μg/mL (all from Biofluids, Camarillo, CA), and 10% fetal calf serum (GIBCO, Grand Island, NY) at 37°Cin5%CO2. Depsipeptide, provided by Fujisawa Pharmaceutical (Osaka, Japan), was dissolved to 5 mg/mL in 4:1 propylene glycol and ethanol and then diluted to 100 μg/mL in dimethyl sulfoxide (DMSO) and stored at -20°C. For experiments, depsipeptide was further diluted in cell culture media. The z-VAD-fmk (Enzyme Systems Products, Livermore, CA) was dissolved in DMSO. Cells were treated with 100 μM, with control cells being treated with the same concentration of DMSO, for 1 hour prior to the addition of depsipeptide. Cells cycle arrest and annexin V analysis were performed at 48 hours.
Selection of depsipeptide-resistant cell lines
HUT78 cells were selected in a stepwise fashion with increasing concentrations of depsipeptide in the absence (Dp) or presence (DpVp) of 5 μM verapamil. For experiments with these cell lines, depsipeptide and verapamil were removed from the culture media 2 weeks prior to plating.
Flow cytometry
Apoptosis was measured using the annexin V-fluorescein isothiocyanate (annexin V-FITC) Apoptosis Detection Kit (BD Biosciences, San Diego, CA) according to the manufacturer's instructions. For surface protein expression of IL-2Rα and IL-2Rβ, cells were incubated for 30 minutes with phycoerythrin-labeled antihuman CD25 (IL-2Rα) or CD122 (IL-2Rβ) antibody (both from BD Biosciences) or a negative control antibody (BD Biosciences) in phosphate-buffered saline (PBS) containing 2% bovine serum albumin (BSA) in PBS for 30 minutes. Cell surface Pgp expression was determined by incubating cells in the anti-Pgp antibody, MRK-16 (Kamiya Biomedical, Seattle, WA), or a negative control antibody in 2% BSA in PBS for 30 minutes, washing the cells, then incubating them with a phycoerythrin secondary antibody (Vector Laboratories, Burlingame, CA) for 30 minutes. Dead cells were excluded based on propidium iodide staining. For cell cycle studies, HUT78 cells were washed with PBS, centrifuged, and resuspended in 500 μL staining solution (0.1 mg/mL propidium iodide and 0.6% Triton-X in PBS) to which 500 μL RNAse A solution (200 U/mL in PBS) was added. The cells were allowed to incubate for 30 minutes at room temperature and were subsequently analyzed using a FACSort flow cytometer (Becton Dickinson, San José, CA). The program ModFit LT version 2.0 (Verity Software, Topsham, ME) was used to determine the percentage of cells in each phase of the cell cycle.
Rhodamine 123 efflux analysis was performed as previously described.19,27 Briefly, cells were resuspended in complete media (phenol red-free Iscove modified Eagle medium [IMEM] with 10% fetal calf serum) containing 0.5 μg/mL rhodamine 123 (Sigma Chemical, St Louis, MO), with or without 2.5 μg/mL valspodar, a Pgp inhibitor, and incubated for 30 minutes at 37°C in 5% CO2. Cells were then washed once in cold complete medium and then incubated for 1 hour at 37°C in substrate-free media continuing with or without valspodar. Subsequently, cells were washed twice with cold Dulbecco PBS and placed on ice in the dark until analyzed. Cells were analyzed on a FACSort flow cytometer. For all samples, at least 10 000 events were collected. Debris was eliminated by gating on forward versus side scatter and dead cells were excluded based on propidium iodide staining.
Cytotoxicity assay
Cytotoxicity assays were performed in 96-well dishes with 5 × 103 cells per well. For experiments testing sensitivity to depsipeptide, sodium butyrate, TSA, and MS-275, cells were placed in varying concentrations of depsipeptide for 72 hours. Combination studies with denileukin diftitox were performed by incubating cells in depsipeptide for 48 hours, adding denileukin diftitox, and continuing with depsipeptide for an additional 72 hours. Cell viability was assayed using Owen reagent (MTS [3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium]) purchased as the Cell Titer 96 Aqueous One Solution Cell Proliferation Assay reagent (Promega, Madison, WI). Assays were performed according to the manufacturer's directions. Experiments were performed in triplicate.
Protein isolation and immunoblot analysis
Protein was prepared by washing treated HUT78 cells in PBS and resuspending in lysis buffer (0.02 M Tris [pH 7.4], 1% Triton X-100, and 0.02% beta-mercaptoethanol) with 2 ng/mL aprotinin and sonicating at 50% for 2 minutes in a water bath (Misonix, Farmingdale, NY). Protein concentration was measured using the BCA Protein Assay (Pierce, Rockford, IL). Extracts were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA). Blots were incubated in primary antibody overnight at 4°C. Antibodies used were antiacetylated histone H3 and anti-PARP (anti-poly(adenosine diphosphate-ribose) polymerase; Upstate Biotechnology, Waltham, MA), anti-CD25 and anti-CD122 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-p21 (Transduction Laboratories, Bedford, MA), and anti-glyceraldehyde phosphate dehydrogenase (anti-GAPDH; American Research Products, Belmont, MA). Blots were incubated in secondary antibody for 1 hour at room temperature. Secondary antibodies used were horseradish peroxidase-conjugated antirabbit or antimouse antibody (Amersham, Piscataway, NJ). Chemiluminescence was carried out using an enhanced chemiluminescence (ECL) detection kit (SuperSignal West Pico Chemiluminescent Substrate; Pierce or Amersham) according to manufacturer's instructions.
RNA isolation and RT-PCR
RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Reverse transcription of 1 μg of total RNA using 0.1 μg/μL random primer was performed with cDNA equivalent to 250 ng total RNA used for polymerase chain reaction (PCR). Amplification was performed for 30 cycles as previously described.28 Water was amplified a total of 40 cycles to detect possible contamination. Forty microliters of each PCR product was separated by gel electrophoresis and stained with 2 μg/mL ethidium bromide for analysis on a Fotoeclipse densitometer (Fotodyne, Hartland, WI). The primers used for PCR are as follows: MDR-128 forward: GCCTGGCAGCTGGAAGACAAATACACAAAATT, reverse: CAGACAGCAGCTGACAGTCCAAGAACAGGACT; IL-2Rα29 forward: GAATTTATCATTTCGTGGTGGGGCA, reverse: TCTTCTACTCTTCCTCTGTCTCCG; IL-2Rβ29 forward: ATCTCCCTCCAAGTTGTCC, reverse: TGCTTCTGCTTGAGAGTCAGC; and rRNA30 forward: AAACTCTGGTGGAGGTCCGT, reverse: CTTACCAAAAGTGGCCCACTA.
Results
Depsipeptide cell cycle effects and cytotoxicity
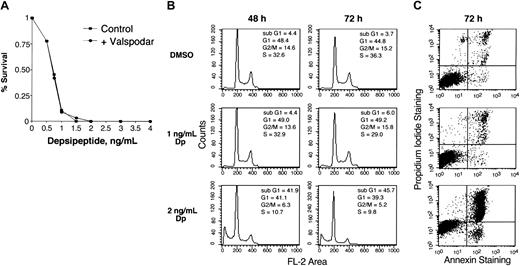
The HUT78 T-cell lymphoma cell line was established from cells isolated from a patient with cutaneous T-cell lymphoma31 and, unlike other available T-cell lymphoma cell lines, has no evidence of human T-cell lymphotropic virus 1 (HTLV-1) infection. Figure 1A shows the results of cytotoxicity assays with depsipeptide performed on these cells. An observed 50% inhibitory concentration (IC50) of 0.75 ng/mL (1.4 nM) following a 72-hour exposure indicates a marked sensitivity to depsipeptide. Valspodar did not increase sensitivity to depsipeptide, indicating that this cell line does not express Pgp, a known mechanism of resistance to depsipeptide.19
Cytotoxicity assay, cell cycle, and annexin V analysis of HUT78 cell line treated with depsipeptide. (A) HUT78 cells were treated for 3 days with varying concentrations of depsipeptide. Cytotoxicity assay was performed using Owen reagent as described in “Materials and methods.” Data shown represent more than 3 experiments and are graphed on a linear scale. The cytotoxicity assay was performed in the absence or presence of 1 μg/mL valspodar, an inhibitor of Pgp. (B) Cell cycle analysis was performed using cells treated with or without depsipeptide (DP) at 0, 1, or 2 ng/mL for 48 and 72 hours. (C) Plot of annexin V staining of cells from the same experiment treated for 72 hours.
Cytotoxicity assay, cell cycle, and annexin V analysis of HUT78 cell line treated with depsipeptide. (A) HUT78 cells were treated for 3 days with varying concentrations of depsipeptide. Cytotoxicity assay was performed using Owen reagent as described in “Materials and methods.” Data shown represent more than 3 experiments and are graphed on a linear scale. The cytotoxicity assay was performed in the absence or presence of 1 μg/mL valspodar, an inhibitor of Pgp. (B) Cell cycle analysis was performed using cells treated with or without depsipeptide (DP) at 0, 1, or 2 ng/mL for 48 and 72 hours. (C) Plot of annexin V staining of cells from the same experiment treated for 72 hours.
While depsipeptide, and other HDIs, cause cell cycle arrest in many cell lines, only minimal G1 and G2/M cell cycle arrest was observed in the HUT78 cell line. Figure 1B shows a cell cycle analysis with increasing time and concentration of depsipeptide (DP), where any suggestion of cell cycle arrest occurred concurrently with significant apoptosis. After 48 hours, for example, the G1 population decreased from 48% to 41%, while the G2/M population decreased from 14.6% to 6.3% following treatment with 2 ng/mL depsipeptide. More than 40% of cells were undergoing apoptosis, as detected by an increasing sub-G1 fraction. This was confirmed by increasing annexin V staining by flow cytometry (Figure 1C). At higher concentrations of depsipeptide, apoptosis was observed at earlier time points. Thus, instead of the cell cycle arrest frequently observed in cell lines derived from malignant epithelial cells, apoptosis was favored in the HUT78 cells.
Molecular response to depsipeptide
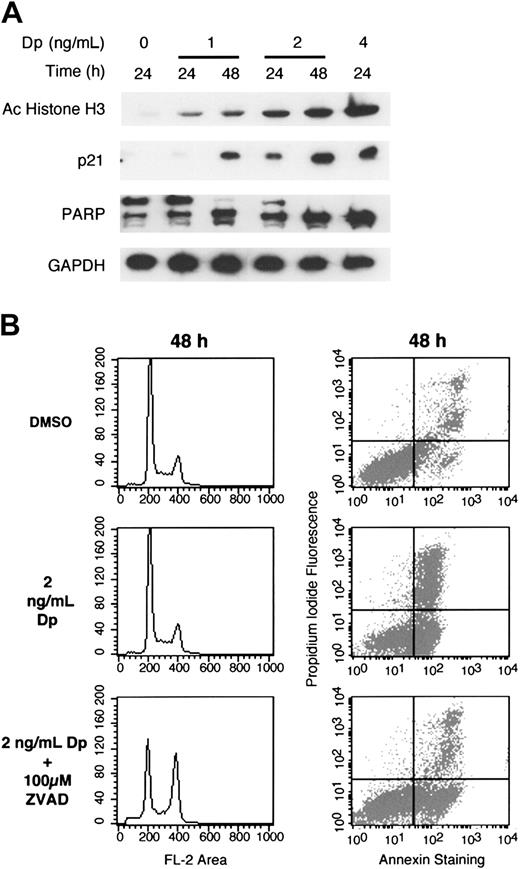
Protein was isolated at several time points from cells treated with varying concentrations of depsipeptide for evaluation of molecular response. As shown in Figure 2A, depsipeptide induced histone acetylation. Induction of p21 was observed, with increasing levels noted with increasing time and concentration. Furthermore, expression of p21 paralleled the level of histone acetylation, with higher levels of histone acetylation and p21 expression detected with increasing concentrations of depsipeptide. GAPDH, previously demonstrated to be unaffected by HDIs, was used as a control for gel loading.32 As observed with annexin V staining, increasing PARP degradation was observed with higher concentrations of depsipeptide. Treatment of cells with 4 ng/mL depsipeptide for 24 hours led to significant cell death. To understand whether cell death in this cell line was caspase mediated, the multi-caspase inhibitor z-VAD-fmk was used. Cells were treated with z-VAD-fmk for 1 hour prior to the addition of depsipeptide and were exposed to both agents together for 48 hours. Treatment with z-VAD-fmk resulted in a 2-fold increase in survival (from 25% to 50%) in cells treated with 2 ng/mL (Figure 2B) and 3-fold (from 16% to 50%) in cells treated with 4 ng/mL (data not shown). Interestingly, with the inhibition of apoptosis, a G2/M arrest, previously undetected following exposure to depsipeptide alone, became apparent.
Immunoblot analysis of depsipeptide-treated HUT78 cells. (A) HUT78 cells were treated for 24 to 48 hours with 0, 1, 2, or 4 ng/mL depsipeptide, as indicated. Whole cell lysates were prepared and 7 μg/mL proteins were separated by PAGE. Immunoblots were performed as described in “Materials and methods” using antibodies against acetylated histone H3, demonstrating the direct effect of depsipeptide on the cells; p21, indicating that depsipeptide is able to induce gene expression, as previously described; and PARP, to demonstrate the apoptotic effect of depsipeptide. GAPDH confirms equivalent loading of proteins. (B) HUT78 cells were pretreated with caspase inhibitor z-VAD-fmk (100 μM) or equal amount of DMSO as control for 1 hour prior to the addition of 2 ng/mL depsipeptide. Cells were exposed to both agents together for an additional 48 hours. Cells cycle arrest and annexin V analysis was performed as described in “Materials and methods.”
Immunoblot analysis of depsipeptide-treated HUT78 cells. (A) HUT78 cells were treated for 24 to 48 hours with 0, 1, 2, or 4 ng/mL depsipeptide, as indicated. Whole cell lysates were prepared and 7 μg/mL proteins were separated by PAGE. Immunoblots were performed as described in “Materials and methods” using antibodies against acetylated histone H3, demonstrating the direct effect of depsipeptide on the cells; p21, indicating that depsipeptide is able to induce gene expression, as previously described; and PARP, to demonstrate the apoptotic effect of depsipeptide. GAPDH confirms equivalent loading of proteins. (B) HUT78 cells were pretreated with caspase inhibitor z-VAD-fmk (100 μM) or equal amount of DMSO as control for 1 hour prior to the addition of 2 ng/mL depsipeptide. Cells were exposed to both agents together for an additional 48 hours. Cells cycle arrest and annexin V analysis was performed as described in “Materials and methods.”
Sensitivity of HUT78 cell line to other HDIs
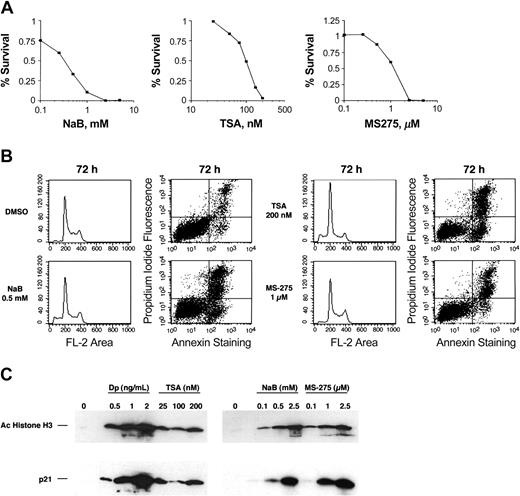
To evaluate whether the sensitivity of the HUT78 cells to depsipeptide represents a general susceptibility to HDIs, we treated HUT78 cells with sodium butyrate, TSA, or MS-275. The observed IC50s were 0.2 mM for sodium butyrate, 100 nM for TSA, and 1 μM for MS-275 (Figure 3A). Figure 3B shows cell cycle effects and annexin V staining of HUT78 cells treated for 48 hours at IC50 doses. As observed for depsipeptide, these agents induced apoptosis with little to no effect on cell cycle. Figure 3C shows histone acetylation and p21 induction in the cells following treatment with each of the 3 HDIs.
Cytotoxicity assay, cell cycle, and annexin V analysis of HUT78 cell line treated with other HDIs. (A) Cytotoxicity assay was performed on HUT78 cells treated for 3 days with varying concentrations of sodium butyrate (NaBu), trichostatin A (TSA), or MS-275 in the presence or absence of valspodar. Data are shown on a logarithmic scale. (B) Cell cycle analysis and annexin V staining shown for cells treated for 48 hours. (C) HUT78 cells were treated for 48 hours with the amount of depsipeptide, TSA, NaBu, or MS-275 as indicated. Whole cell lysates were prepared, and 10 μg/mL of proteins were separated by PAGE. Immunoblots were performed using antibodies against acetylated histone H3, demonstrating histone deacetylase activity of the individual HDIs, and p21, demonstrating the induction of gene expression by each individual HDI.
Cytotoxicity assay, cell cycle, and annexin V analysis of HUT78 cell line treated with other HDIs. (A) Cytotoxicity assay was performed on HUT78 cells treated for 3 days with varying concentrations of sodium butyrate (NaBu), trichostatin A (TSA), or MS-275 in the presence or absence of valspodar. Data are shown on a logarithmic scale. (B) Cell cycle analysis and annexin V staining shown for cells treated for 48 hours. (C) HUT78 cells were treated for 48 hours with the amount of depsipeptide, TSA, NaBu, or MS-275 as indicated. Whole cell lysates were prepared, and 10 μg/mL of proteins were separated by PAGE. Immunoblots were performed using antibodies against acetylated histone H3, demonstrating histone deacetylase activity of the individual HDIs, and p21, demonstrating the induction of gene expression by each individual HDI.
Alterations in gene expression
Next, reverse transcriptase-PCR (RT-PCR) analysis was used to evaluate alterations in gene expression in HUT78 cells treated with depsipeptide. One gene of interest is the MDR-1 gene, previously shown to be induced by HDIs and other differentiating agents. Table 1 shows that, at concentrations and time points associated with induction of p21 expression, there was a greater than 30-fold increase in MDR-1 expression, based on a quantitative RT-PCR assay and normalization of MDR-1 expression to that of rRNA expression.
Induction of MDR-1, CD25, and CD122 mRNA by depsipeptide
Depsipeptide, ng/mL . | MDR-1 . | . | IL-2Rα . | . | IL-2Rβ . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | 24 h . | 48 h . | 24 h . | 48 h . | 24 h . | 48 h . | |||
| 0 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| 1 | 4.62 | 7.66 | 19.33 | 1.45 | 1.8 | 2.4 | |||
| 2 | 18.47 | 31.03 | 10.19 | 0.65 | 2 | 2.81 | |||
Depsipeptide, ng/mL . | MDR-1 . | . | IL-2Rα . | . | IL-2Rβ . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | 24 h . | 48 h . | 24 h . | 48 h . | 24 h . | 48 h . | |||
| 0 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| 1 | 4.62 | 7.66 | 19.33 | 1.45 | 1.8 | 2.4 | |||
| 2 | 18.47 | 31.03 | 10.19 | 0.65 | 2 | 2.81 | |||
HUT78 cells were treated for 24 to 48 hours with 0, 1, 2, or 4 ng/mL of depsipeptide, as indicated, and RNA was isolated using RNeasy. RT-PCR was performed as described in “Materials and methods.” Values represent the fold induction of mRNA detected by RT-PCR after normalization by the RT-PCR product of rRNA.
Since the IL-2 receptor is a target for the treatment of CTCL and differentiating agents have been demonstrated to increase both the expression of the IL-2 receptor and sensitivity to agents that target the IL-2 receptor, we examined the effect of depsipeptide on the expression of the IL-2 receptor subunits. By RT-PCR there was a 19-fold induction of the IL-2Rα subunit mRNA and a greater than 2-fold increase of IL-2Rβ subunit mRNA (Table 1). For reasons not yet understood, the kinetics of gene induction for the IL-2 receptor subunits vary from the kinetics of the MDR-1 gene.
Induction of sensitivity to other antineoplastic agents
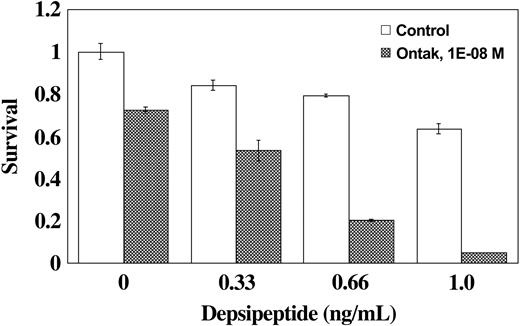
Considering the observed induction of IL-2Rα and IL-2Rβ mRNA, we evaluated the effect of IL-2-targeted therapy in combination with depsipeptide. DAB389IL-2 (denileukin diftitox) is a fusion protein that combines the active domains of diphtheria toxin with IL-2 and is currently used in the treatment of T-cell lymphomas. As shown in Figure 4, we observed a more than additive effect when the HUT78 cells were pretreated with depsipeptide for 48 hours followed by treatment with combined depsipeptide with DAB389IL-2 for an additional 72 hours. When HUT78 cells were treated with DAB389IL-2 alone, a 72% cell survival was observed. When cells were treated with 0.66 ng/mL of depsipeptide without the addition of DAB389IL-2, there was a 79% cell survival. In contrast, when the cells were treated with 0.66 ng/mL of depsipeptide for 2 days prior to the addition of DAB389IL-2, only 20% of the cells survived following the combined treatment.
Sensitization of HUT78 cells to DAB389IL-2 (denileukin diftitox) by depsipeptide. HUT78 cells were first exposed to depsipeptide or control treatment for 48 hours; denileukin diftitox (DAB389IL-2) was then added for 72 hours. Cytotoxicity assay was performed as described in “Materials and methods.” Error bars indicate standard error of triplicates.
Sensitization of HUT78 cells to DAB389IL-2 (denileukin diftitox) by depsipeptide. HUT78 cells were first exposed to depsipeptide or control treatment for 48 hours; denileukin diftitox (DAB389IL-2) was then added for 72 hours. Cytotoxicity assay was performed as described in “Materials and methods.” Error bars indicate standard error of triplicates.
Drug resistance
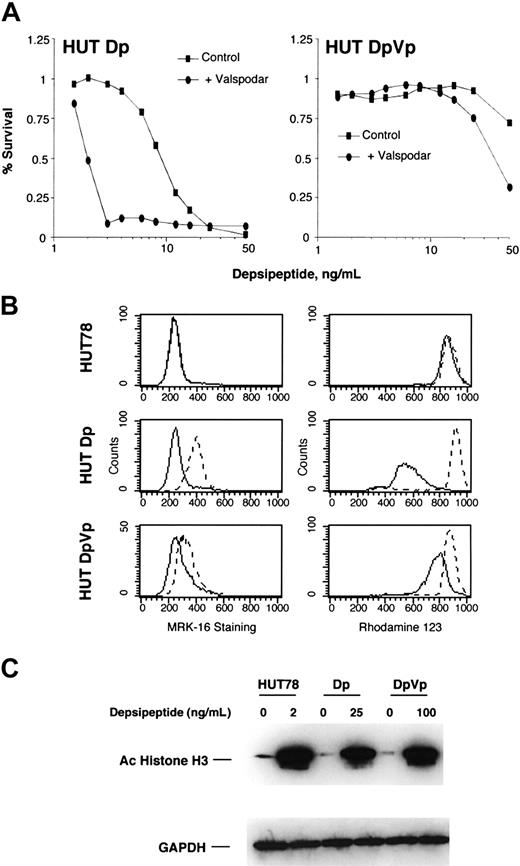
In order to characterize potential mechanisms of resistance to depsipeptide in CTCL, we selected HUT78 cells for resistance to depsipeptide. Since depsipeptide induced the expression of MDR-1 in HUT78 cells and is transported by the MDR-1 gene product Pgp, we selected cells in the presence and absence of a Pgp inhibitor, verapamil. Cell lines were selected in a stepwise process for approximately 1 year and were maintained in 12 ng/mL depsipeptide. For these analyses, cells were cultured in the absence of drug for 2 weeks. In cell lines selected in depsipeptide alone (Dp cells), the observed IC50 was approximately 8 ng/mL for cells, representing a greater than 10-fold resistance to depsipeptide. These cells were sensitized to depsipeptide when treated with the Pgp inhibitor, valspodar (Figure 5A). The observed IC50 for the cells selected in the presence of verapamil (DpVp cells) exceeded 50 ng/mL. Combination with valspodar did not significantly increase sensitivity to depsipeptide. Expression of Pgp was evaluated by using an antibody, MRK-16, that detects Pgp on the cell surface by flow cytometry, and function of Pgp was evaluated by the efflux of rhodamine 123. No Pgp was detected in the parent cell line, consistent with the observation that valspodar did not increase sensitivity to depsipeptide by cytotoxicity assay. As expected, Dp cells selected for resistance to depsipeptide alone had significant levels of detectable Pgp and rhodamine 123 efflux. Cells selected for resistance to depsipeptide in the presence of verapamil had slight expression of Pgp and minimal rhodamine efflux, indicating that resistance is not mediated by Pgp. Increased histone acetylation was observed in both the Dp and DpVp cells following treatment with 25 and 100 ng/mL depsipeptide, respectively, suggesting that in both cell lines, any reduction in histone acetylation that might result from drug resistance mechanisms can be overcome by increased doses of depsipeptide.
Cytotoxicity assay, MRK-16 staining, rhodamine 123 efflux, and molecular response to depsipeptide of resistant HUT78 cells. HUT78 cells were selected for resistance to depsipeptide in the absence (HUT78Dp) or presence of 5 μg/mL of verapamil (HUT78DpVp). For these experiments, cells were first grown out of depsipeptide and verapamil for 2 weeks. (A) Cells were treated with depsipeptide for 3 days with or without 1 μg/mL of valspodar, an inhibitor of Pgp, and cytotoxicity assays were performed. Data are shown on a logarithmic scale. (B) The panels on the left show the results of cells stained with the Pgp-specific antibody, MRK-16 (dashed line), or with an isotype-specific antibody as a control (solid line). In the right panel, Pgp function was assayed by the ability of the cells to efflux rhodamine. Cells were first incubated with rhodamine 123 and then incubated for 1 hour in the absence of rhodamine, both steps in the absence (solid line) or presence (dashed line) of valspodar. (C) HUT78, HUT78Dp, and HUT78DpVp cells were treated overnight with the indicated amount of depsipeptide. Whole cell lysates were prepared, and 10 μg/mL of proteins were separated by PAGE. Immunoblots were performed using antibodies against acetylated histone H3, demonstrating histone deacetylase activity of the individual HDIs, and GAPDH, to confirm equivalent loading of proteins.
Cytotoxicity assay, MRK-16 staining, rhodamine 123 efflux, and molecular response to depsipeptide of resistant HUT78 cells. HUT78 cells were selected for resistance to depsipeptide in the absence (HUT78Dp) or presence of 5 μg/mL of verapamil (HUT78DpVp). For these experiments, cells were first grown out of depsipeptide and verapamil for 2 weeks. (A) Cells were treated with depsipeptide for 3 days with or without 1 μg/mL of valspodar, an inhibitor of Pgp, and cytotoxicity assays were performed. Data are shown on a logarithmic scale. (B) The panels on the left show the results of cells stained with the Pgp-specific antibody, MRK-16 (dashed line), or with an isotype-specific antibody as a control (solid line). In the right panel, Pgp function was assayed by the ability of the cells to efflux rhodamine. Cells were first incubated with rhodamine 123 and then incubated for 1 hour in the absence of rhodamine, both steps in the absence (solid line) or presence (dashed line) of valspodar. (C) HUT78, HUT78Dp, and HUT78DpVp cells were treated overnight with the indicated amount of depsipeptide. Whole cell lysates were prepared, and 10 μg/mL of proteins were separated by PAGE. Immunoblots were performed using antibodies against acetylated histone H3, demonstrating histone deacetylase activity of the individual HDIs, and GAPDH, to confirm equivalent loading of proteins.
Discussion
To extend the observed responses to depsipeptide in patients with T-cell lymphoma, we sought an in vitro model to examine the molecular effects of depsipeptide. HUT78, a T-cell lymphoma cell line derived from a patient with cutaneous T-cell lymphoma, responds to depsipeptide treatment with increased histone acetylation, induced p21 expression, and marked apoptosis without significant cell cycle arrest. These changes were found to be a general response of HUT78 cells to structurally distinct histone deacetylase inhibitors. Gene expression studies revealed increased expression of IL-2 receptor subunits and MDR-1, and we were able to exploit the induction of IL-2 receptor subunits to sensitize cells to denileukin diftitox.
While the precise regulators of growth arrest or apoptosis observed in cancer lines exposed to HDIs remain unclear, it has been demonstrated that growth arrest is the predominant effect in cell lines where HDIs induce the expression of p21.33 The HDIs have a direct effect, increasing p21 expression through its promoter region.34 Cell lines in which p21 expression is inhibited due to knock out or antisense expression do not undergo growth arrest; instead these p21-deficient cells preferentially undergo apoptosis.5,33,35-37 While the precise apoptotic pathway remains to be elucidated, the HUT78 T-cell lymphoma line appears to be different, since depsipeptide causes early apoptosis without significant cell cycle arrest despite p21 induction, indicating that the apoptotic effects of depsipeptide predominate. Of note, when apoptosis is inhibited with z-VAD-fmk, the cell cycle arrest effects of depsipeptide are observed. This has been described previously in other experimental models.38,39 The increase in the surviving population by 2- to 3-fold implicates one or more of the caspases in the apoptosis induced by depsipeptide. The residual 50% mortality in the depsipeptide-treated cells pretreated with z-VAD-fmk indicates that apoptosis was not completely blocked. We hypothesize that this occurs as a result of either the activation of caspases not inhibited by z-VAD-fmk or through the activation of non-caspase-mediated apoptotic pathways. Further studies will need to be performed to dissect the precise steps in the apoptotic pathway that are activated by depsipeptide. Induction of p21 by HDIs without concomitant cell cycle arrest was also previously observed in the A549 lung cancer cell line.40 Although induction of p21 was noted after treatment with trapoxin, no cell cycle arrest was observed, and marked apoptosis was noted.40 The results presented here are consistent with a previous study reporting that depsipeptide induced apoptosis in IL-2-dependent malignant T-cell lines.41
Two aspects of the development of depsipeptide for clinical use were evaluated in the laboratory studies presented here. First, seeking strategies for combination therapy, we evaluated the effect of depsipeptide on the expression of the IL-2 receptor. Denileukin diftitox has recently been shown to be effective in both cutaneous and peripheral T-cell lymphomas.15,42 Even though the HUT78 cells readily underwent apoptosis, at low concentrations of depsipeptide we found that there was some increase in expression of the IL-2 receptor subunits and increased sensitivity to denileukin diftitox. This may occur through increased activation of the same or of an alternate apoptotic pathway. Previously, a 6-fold-increased expression of IL-2Rα and a 50%-increased expression of IL-2Rβ were reported in HUT78 cells treated with retinoids.16
Beyond agents that target the IL-2 receptor, a rationale also exists for other combinations. Retinoids have demonstrated efficacy in T-cell lymphoma and the selective retinoid bexarotene is in clinical use for this indication.43-45 Furthermore, retinoids have demonstrated synergy with HDIs in neuroblastoma and colorectal carcinoma cell lines, suggesting that the combination of an HDI with a retinoid could be further evaluated for patients with T-cell lymphomas.46,47
The second aspect was to evaluate potential mechanisms of resistance. Limited evidence suggests that expression of Pgp plays a role in mediating resistance to chemotherapy in lymphomas.48 In addition, Pgp was detected by immunohistochemistry in 18 of 25 patients with advanced CTCL and some form of prior therapy.49 We have previously observed that depsipeptide is transported by Pgp and that cells that overexpress Pgp are resistant to depsipeptide.19 Furthermore, several HDIs have been shown to induce the expression of MDR-1.20,21 This appears to occur through HDI-sensitive elements in the promoter region.50 The studies presented here demonstrate that depsipeptide induces the expression of MDR-1, that cells selected for resistance to depsipeptide overexpress Pgp, and that this resistance is reversed with a Pgp inhibitor. Thus, depsipeptide has the ability to induce its own mechanism of resistance, a finding that could explain the eventual failure of depsipeptide following initial clinical activity. Of note, since the HDIs as a class increase the expression of Pgp, it may be difficult to combine the use of these agents with drugs that are transported by Pgp, such as doxorubicin or etoposide.
In contrast, cells selected for resistance to depsipeptide in the presence of a Pgp inhibitor did not significantly increase Pgp expression and were not sensitized by inhibitors of Pgp. Therefore, these cells appear to have an alternate mechanism of resistance. Molecular mechanisms of resistance to HDIs have not, as yet, been elucidated. Previously, 2 models of resistance to sodium butyrate were described. In one model, K562 erythroleukemic cells were first exposed to mutagens and then selected for resistance to sodium butyrate. Upon treatment with sodium butyrate, the resistant cells had lower levels of histone acetylation, less growth inhibitory effects, and less induction of cellular hemoglobin.51 More recently, HeLa cells selected for resistance to sodium butyrate were found to have an increased basal level of p21 expression. In the resistant cells, sodium butyrate did not down-regulate cyclin D1 levels, as observed in the parent cell line.52 Cells resistant to the hydroxamic acid, TSA, were also selected by first treating mouse mammary tumor cells with mutagens.53 Exposure to higher levels of TSA did not increase histone acetylation. Evaluation of histone deacetylase enzymatic activity in total nuclear extracts found that while the measured Km was the same as the parent cells, the Ki of TSA was 9-fold higher in the resistant cells. Our DpVp-selected cell line had increased histone acetylation following exposure to increased concentrations of depsipeptide, suggesting that the resistance mechanism is downstream of this immediate molecular target.
Depsipeptide has shown single-agent activity in early clinical trials in malignancies of T-lymphocyte origin. In nonmalignant T lymphocytes, sodium butyrate, TSA, and depsipeptide have been shown to inhibit antigen-induced T-cell stimulation.54-56 Apart from its activity in nonmalignant T cells, why would depsipeptide have such potent activity in patients with T-cell lymphoma? One answer may lie in our current understanding that CTCL develops from a clone of T cells in a hyperproliferative state, failing to trigger an impaired apoptotic pathway.57 Recent studies have demonstrated that fas-mediated apoptosis in T cells requires an induction of p21.58 Furthermore, HDIs have been shown to increase the expression of both fas receptor and fas ligand.59,60 Thus, depsipeptide and other HDIs may exert profound effects on T-cell malignancies by directly activating the T-cell apoptotic pathway by increasing expression of the fas receptor or its ligand or by activating a downstream activator of apoptosis, such as the increased expression of p21. Of note, a novel splice variant of the fas receptor, associated with decreased surface membrane expression, has recently been identified in a number of patients with CTCL.61 If depsipeptide exerts its effect by acting directly on the expression of fas receptor and ligand, this mutation may explain why some patients do not respond to therapy. Alternatively, observed responses may be understood if depsipeptide exerts its effect downstream of the fas receptor. This will require further analysis.
In conclusion, these studies demonstrate that response to depsipeptide in the HUT78 human T-cell lymphoma cell line is associated with an induction of histone acetylation, p21 expression, and detection of markers of apoptosis. This cell line is also sensitive to other HDIs, suggesting that several members of this new class of anticancer agents are likely to have clinical efficacy in T-cell lymphoma. In addition, treatment with depsipeptide increased the sensitivity of this cell line to denileukin diftitox. As denileukin diftitox is currently in clinical use for cutaneous T-cell lymphoma, these results suggest a strategy for combination therapy in this disease. Furthermore, depsipeptide increased the expression of the MDR-1 gene, and cells selected for resistance overexpressed Pgp. Of note, this resistance was reversed with a Pgp inhibitor. A non-Pgp mechanism of resistance has developed in the cells selected in the presence of a Pgp inhibitor, although the precise nature of this resistance remains to be determined. Taken together, these results support the therapeutic activity of the histone deacetylase inhibitors, such as depsipeptide in cutaneous T-cell lymphoma, and suggest that the induction of molecular targets by depsipeptide may allow the design of rational combination therapies.
Prepublished online as Blood First Edition Paper, March 2, 2004; DOI 10.1182/blood-2003-09-3068.
Supported in part by research funding from Fujisawa Pharmaceutical Co, Osaka, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank John Wright and Naoki Sogo for their support of our studies with depsipeptide in T-cell lymphoma.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal