Abstract
Sequence analysis of the immunoglobulin heavy chain genes (IgH) has demonstrated preferential usage of specific variable (V), diversity (D), and joining (J) genes at different stages of B-cell development and in B-cell malignancies, and this has provided insight into B-cell maturation and selection. Knowledge of the association between rearrangement patterns based on updated databases and clinical characteristics of pediatric acute lymphoblastic leukemia (ALL) is limited. We analyzed 381 IgH sequences identified at presentation in 317 children with B-lineage ALL and assessed the VHDHJH gene utilization profiles. The DHJH-proximal VH segments and the DH2 gene family were significantly overrepresented. Only 21% of VH-JH joinings were potentially productive, a finding associated with a trend toward an increased risk of relapse. These results suggest that physical location at the VH locus is involved in preferential usage of DHJH-proximal VH segments whereas DH and JH segment usage is governed by position-independent molecular mechanisms. Molecular pathophysiology appears relevant to clinical outcome in patients who have only productive rearrangements, and specific rearrangement patterns are associated with differences in the tumor biology of childhood ALL. (Blood. 2004;103:4602-4609)
Introduction
Immunoglobulin (Ig) genes are assembled from germ line variable (V), diversity (D), and joining (J) gene segments during early B-cell differentiation by a site-directed DNA rearrangement mechanism known as VDJ recombination.1 Further recombination at the heavy chain (H) locus is prevented by a productive VHDHJH rearrangement that also triggers rearrangements at the light chain (L) loci, most often κ followed by λ.2,3 During these joining steps, antibody diversity is generated by the following mechanisms: (1) somatic recombination of multiple VH, DH, and JH segments; (2) nucleotide deletions of the 3′-end VH segment; (3) non-germ line-encoded (N) nucleotide insertions by terminal deoxynucleotidyl transferase (TdT); (4) germ line-encoded palindromic (P) nucleotide additions; (5) transcription of D regions in any of 3 potential open reading frames; (6) fusion and inversion of D regions; and (7) somatic mutation.4-8
A complete map of the human Ig VHDHJH loci has now been constructed on chromosome 14q32.2 in a telomeric-to-centromeric direction.9,10 The VH region contains 123 VH segments, of which 79 are pseudogenes and 44 have an open reading frame.10 The VH genes are grouped into 7 VH families based on their high sequence homology and not by their location on chromosome 14q32. VH3 is the largest family followed by VH4 and VH1.9,10 VH6-1 is the most proximal to the DHJH loci.9,10 The DH region contains 27 DH segments, of which 25 have been shown to be involved in creation of human antibody.11 Seven DH families are classified based on sequence homology. DH7-27 is closest to the JH locus.11 Six JH segments are functional.12,13 The rearranged heavy chain consists of the 3 high variable regions, complementarity determining regions (CDR1, CDR2, and CDR3), flanked by less variable framework regions (FR1, FR2, FR3, and FR4).4 The CDRs, especially CDR3, are considered as the core portion responsible for antigen recognition.4 The CDR3 sequence is unique to each rearrangement and therefore identifies individual B cells or clonal B-cell expansion.4,14,15
The VHDHJH gene repertoires are restricted and developmentally regulated at early stages of differentiation.3,16 In the most immature B-cell precursors (pro-B cells), the IgH genes remain germ line or there is only DHJH joining.2,17 At the next stage, in early pre-B cells, VH genes join to the joined DHJH to complete the IgH rearrangement.2,18 Numerous studies in both mice and humans have shown stage-specific trends in usage of V, D, and J genes, the degree of N nucleotide addition, and the rate of somatic mutation.8,16-20 In the murine system, a biased use of JH-proximal VH segments and a high frequency of absence of N sequences at DHJH joining were demonstrated at fetal stages of development.19-21 In humans, a similar trend was found by a marked overrepresentation of some VH (VH3, VH5, and VH6), DH (DH7-27), and JH (JH3 and JH4) segments and by a short CDR3 length in fetal liver B cells and in immature B cells.16-18
Previous studies have demonstrated a preferential usage of specific VH genes in B-cell malignancies.15,22,23 In mantle cell lymphoma, VH3-21, VH3-23, VH4-34, VH4-59, and VH5-51 segments have shown to be most widely used.24,25 In B-cell chronic lymphocytic leukemia (CLL), patients with unmutated compared with somatically mutated VH genes have a worse prognosis.26,27 A biased utilization of VH1-69 combined with selected DH gene segments and JH6 has been found in unmutated cases,28 and patients with mutated Ig VH3-21 genes had significantly shorter survival than other mutated patients.29 In B-lineage acute lymphoblastic leukemia (B-ALL), a privileged usage of VH6 gene has been shown in both adult and childhood patients.30-32 Sequence analysis of the CDR region demonstrated that DH6 (DN1), JH4, and JH6 appeared to be overrepresented compared with the expected frequency of use according to the size of each DH or JH gene family.14,33 Several studies have suggested that childhood B-ALL is derived from early fetal life or immature B cells.31-33 In one study, in-frame and out-of-frame CDR3 joinings were observed in one third and two thirds of the rearrangements in pre-B-ALL, similar to the frequency of occurrence in nonmalignant early pre-B cells,33 whereas another study found that in-frame CDR3 rearrangement occurred in 78% in children and 64% in adults, similar to that observed in healthy B cells (75%).34
Most previous studies of V gene usage in B-ALL have been performed using databases with limitations for the newly identified germ line genes and the completed gene map and in particular have focused on gene family usage rather than location on the chromosome. In particular, awareness of connections among the IgH rearrangement patterns and clinical characteristics has been relatively limited in ALL compared with the more recent studies in CLL. To derive clone-specific oligonucleotides from IgH rearrangements for minimal residual disease (MRD) detection, we prospectively sequenced VHDHJH regions at presentation in 317 children with B-cell lineage ALL treated sequentially in a single protocol. The aims of this study were to describe VHDHJH rearrangement profiles in children with B-lineage ALL, to evaluate biologic and structural features in clonal expanded B cells by comparison with previous findings of the human Ig VH repertoire,9,10 and to investigate the association between rearrangement patterns and clinical characteristics.
Materials and methods
Patient samples
Bone marrow (BM) and/or peripheral blood (PB) samples were obtained at presentation and IgH rearrangements sequenced in 317 children with B-lineage ALL enrolled consecutively in Dana-Farber Cancer Institute (DFCI) ALL Consortium protocol 95-01. Institutional Review Board approval and informed consent were obtained for treatment and for procurement of the samples in all cases.
DNA preparation
Mononuclear cells were isolated by Ficoll gradient centrifugation (Pharmacia, Uppsala, Sweden), lysed, and DNA extracted and purified according to the manufacturer's instructions using the NucleoSpin kit (BD Biosciences, Palo Alto, CA).
PCR analysis of IgH gene rearrangements
To identify patient leukemia-related IgH gene rearrangements, diagnostic BM and/or PB samples were polymerase chain reaction (PCR) amplified using a series of 7 VH family FR1 consensus primers and a JH consensus primer in a modification of a method previously described.35 Primers were purchased from a commercial supplier (Invitrogen, Carlsbad, CA). The PCR conditions and methods used for detection of PCR products have been previously described.36
Direct sequencing of PCR product
Clonal PCR products were excised and purified using QIAquick gel extraction kits (QIAGEN, Valencia, CA). Purified PCR fragments were sequenced directly by the Dana-Farber/Harvard Cancer Center Core Sequencing Facility (Boston, MA). Sequence reactions were analyzed on an Applied Biosystems 3700 capillary sequencer using Big Dye Terminator Chemistry version 2 (Applied Biosystems, Foster City, CA). The relevant consensus forward and reverse primers were used as sequence primers to obtain the sequence of both strands. Nucleotide sequences were aligned using the DNAstar software (DNASTAR, Madison, WI).
Interpretation of sequence data
VH,DH, and JH segments were identified with a closest matching known human germ line genes using the ImMunoGeneTics (IMGT) Database (http://imgt.cines.fr, IMGT, European Bioinformatics Institute, Montepellier, France), the IGBlast search (http://www.ncbi.nlm.nih.gov/igblast/, National Center for Biotechnology Information, Bethesda, MD), or V BASE directory using DNAPLOT (http://www.mrc-cpe.cam.ac.uk/DNAPLOT.php?menu=901, Center for Protein Engineering, Cambridge, United Kingdom). The following criteria were used for DH gene determination: a minimal homology of 6 matches in a row or 7 matches interrupted by 1 mismatch. The CDR3 length was calculated according to previously described criteria.6
Statistical analysis
Descriptive statistics (percentages, medians, and ranges) were used to describe VDJ gene utilization profiles. The χ2 test was used to compare 2 categoric variables, and for 2 × 2 tables we used the Fisher exact test.37 The Wilcoxon rank-sum and the Kruskal-Wallis tests were used to compare a continuous variable with a categoric variable with 2 or more categories, respectively.38 The exact binomial distribution was used to assess differences between a specific observed percentage and an expected percentage. The Kaplan-Meier method and the log-rank test were used to estimate and compare time to relapse according to productivity of the rearrangements.39,40 All tests were 2-sided except the exact binomial, which was 1-sided.
Results
A total of 381 IgH sequences were identified at presentation from 317 children with B-lineage ALL enrolled in DFCI ALL Consortium protocol 95-01. A high identity (more than 98%) to the human germ line gene segments was found in 375 (98.4%) of the 381 sequences. We identified only 6 sequences (1.5%) with identity in a range of 86% to 96%. Of note, 4 of these cases used V3-11, raising the possibility of polymorphisms at this locus.
VH gene usage
The frequency of the usage of the specific VH segments is shown by their position on chromosome 14 in a telomeric-to-centromeric direction in Figure 1.
VH gene segment usage profile by physical location on chromosome 14 in 381 rearrangements from 317 children with B-lineage ALL showing a privileged use of the DHJH-proximal VH segments. p indicates pseudogenes.
VH gene segment usage profile by physical location on chromosome 14 in 381 rearrangements from 317 children with B-lineage ALL showing a privileged use of the DHJH-proximal VH segments. p indicates pseudogenes.
When this region was divided into 4 clusters each of approximately 200 kilobase (kb), we observed a privileged use of the DHJH-proximal VH segments in cluster D, with 47% versus expected 25% use (P < .001 using the binomial test). Fifty-two germ line VH segments were used by the 381 IgH sequences. VH6-1 (35, 9.19%), VH3-13 (32, 8.4%), and VH4-34 (22, 5.77%) were the 3 most overused VH segments in pediatric B-ALL. Of the 52 VH segments used, 9 were pseudogenes used by 24 sequences. In the DHJH-proximal VH segments, VH6-1 segment was used in 35 of the 177 sequences, followed by VH3-13 (n = 32), VH3-11 (n = 19), VH1-2 (n = 18), VH2-5 (n = 16), VH1-3 (n = 12), VH3-9 (n = 12), and VH3-15 (n = 10) segment. The rest of these VH segments (VH1-14, VH5-a, VH2-10, VH1-8, VH2-5, VH3-7, and VH4-4) in this region were used by fewer than 10 sequences.
Of the 381 IgH sequences, VH3 family gene segments were identified in 192 clones (50%), followed by VH1 (62, 16%), VH4 (60, 16%), VH6 (35, 9%), VH2 (22, 6%), and VH5 (10, 3%). VH3 is the largest family in children with B-lineage ALL, with its usage occurring at frequency commensurate with its germ line family size.10 We observed a privileged usage of VH6 (9%) in children with B-lineage ALL compared with peripheral B lymphocytes (PBLs) (0.8%, P = .006 using the χ2 test). Although murine data suggest that this gene segment is overused also in normal B cells in early life,30 there are no published human studies to assess whether VH6 use in ALL is different from what would be seen in fetal and childhood B cells.
DH gene usage
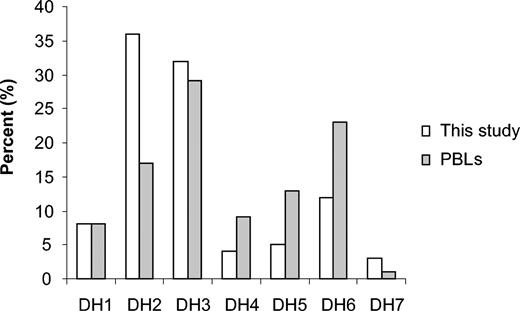
Of the 381 IgH sequences, DH segments could be identified in 304 sequences. For 77 (20%) of the 381 sequences insufficient D segment sequence was available for definitive identification of the DH gene segment used, either because of aberrant VDJ recombinations or from exonuclease activity. A single DH gene was identified in 301 sequences, and the unusual DH-DH joining sequences were found in only 3 sequences. For these 3 sequences using more than one DH gene, the JH-proximal DH segment was considered for gene usage analysis to assess DJ recombination. The DH2 gene family was used most frequently at 35% (105), followed by DH3 (32%, 97), DH6 (12%, 37), DH1 (8%, 23), DH5 (5%, 15), DH7 (4%, 12), and DH4 (4%, 12), respectively. The DH2 and DH7 genes were overrepresented whereas DH4 and DH5 gene segments were underrepresented in B-lineage ALL CDR3 regions compared with usage in PBLs12 (P < .001 using χ2 test) as shown in Figure 2.
DH gene family usage in 381 rearrangements in ALL compared with published results from PBLs.
DH gene family usage in 381 rearrangements in ALL compared with published results from PBLs.
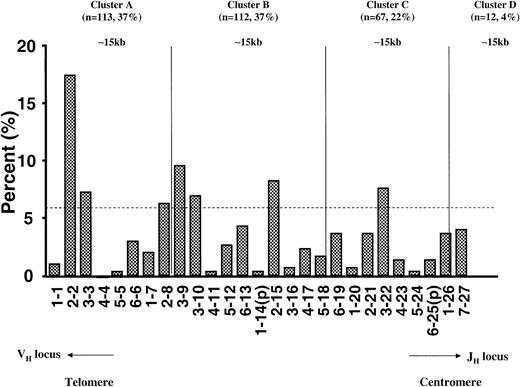
Similar to the VH gene families, assignment to DH gene families is based upon sequence homology and not their chromosomal loci. The telomeric-to-centromeric position of these genes is shown in Figure 3 with the percentage of utilization of each gene. Four clusters were assigned, each of approximately 15 kb. A privileged usage of JH-distal DH segments was observed in cluster A (37%) and in cluster B (37%) versus expected 25% (P < .001 using the exact binomial test). Of 27 human germ line DH gene segments, 26 were used by the B-ALL IgH sequences including 2 pseudogenes (DH1-14 and DH6-25). None of the sequences used DH4-4 segment. Seven DH segments (DH2-2, DH3-3, DH2-8, DH3-9, DH3-10, DH2-15, and DH3-22) were overrepresented in comparison to individual family size as shown in Figure 3 (P < .001 using the exact binomial distribution).
DH gene segment usage profile by physical location on chromosome 14 in 304 rearrangements from children with B-lineage ALL showing a privilege utilization of the JH-distal DH segments. The dotted line indicates statistical significance of D gene segment utilization compared with expected use by the exact binomial distribution (P < .001); p indicates pseudogenes.
DH gene segment usage profile by physical location on chromosome 14 in 304 rearrangements from children with B-lineage ALL showing a privilege utilization of the JH-distal DH segments. The dotted line indicates statistical significance of D gene segment utilization compared with expected use by the exact binomial distribution (P < .001); p indicates pseudogenes.
Joining gene usage
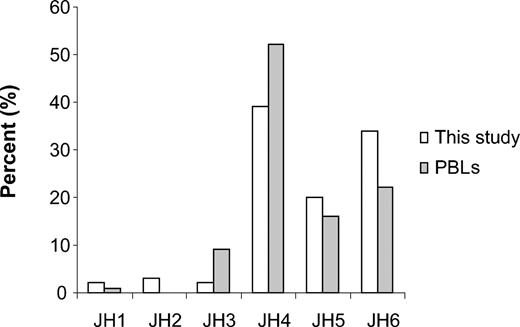
Usage of the JH4 gene family was 39% (148), followed by JH6 (34%, 131), JH5 (20%, 78), JH2 (3%, 11), JH3 (2%, 7), and JH1 (2%, 6), respectively. The JH gene usage in children with B-lineage ALL is significantly different from the JH usage found in normal PBLs12 where JH4 is the most prominent JH gene, used in 52.5%, versus JH6, in 22.2% (P < .001 using the χ2 test; Figure 4).
JH gene family usage in 381 rearrangements in this study compared with published results from PBLs.
JH gene family usage in 381 rearrangements in this study compared with published results from PBLs.
CDR3 length
The CDR3 length was calculated in size of base pairs (bp) for 381 sequences showing a median of 30.0 bp. Excessively long CDR3 length (more than 100 bp) was found in 4 sequences, and by basic local alignment sequence tool (BLAST) search aberrant VDJ recombination was identified, including incorporation of DH intron sequences in one case. For patients with multiple IgH sequences, the average of the CDR3 lengths was considered. We observed a significant association between CDR3 length and the VH or DH family utilization but not by their JH genes (Table 1). A short CDR3 was correlated with utilization of VH6 and DH7-27 segments (P < .01 using the Kruskal-Wallis test).
Correlation of CDR3 length and VHDHJH utilization in childhood B-lineage ALL
Gene family . | . | . | . | CDR3 length, bp . | P . |
|---|---|---|---|---|---|
| No. | Mean | SD | Median | ||
| VH | .006 | ||||
| VH1 | 62 | 29.7 | 15.9 | 26.5 | |
| VH2 | 22 | 31.9 | 12.0 | 31.5 | |
| VH3 | 192 | 36.8 | 37.7 | 30.0 | |
| VH4 | 60 | 32.1 | 16.3 | 31.5 | |
| VH5 | 10 | 33.4 | 15.2 | 33.5 | |
| VH6 | 35 | 22.3 | 11.9 | 21.0 | |
| DH | < .0001 | ||||
| DH1 | 23 | 25.3 | 10.1 | 24.0 | |
| DH2 | 108 | 41.4 | 29.4 | 36.5 | |
| DH3 | 97 | 37.1 | 24.5 | 34.0 | |
| DH4 | 12 | 31.3 | 12.8 | 28.5 | |
| DH5 | 15 | 27.9 | 9.8 | 27.0 | |
| DH6 | 37 | 35.0 | 36.8 | 26.0 | |
| DH7 | 12 | 20.9 | 8.7 | 20.5 | |
| JH | .16 | ||||
| JH1 | 6 | 48.0 | 21.4 | 44.0 | |
| JH2 | 11 | 33.5 | 19.4 | 34.0 | |
| JH3 | 7 | 31.1 | 16.6 | 33.0 | |
| JH4 | 148 | 29.9 | 15.3 | 27.0 | |
| JH5 | 78 | 32.0 | 28.4 | 30.0 | |
| JH6 | 131 | 37.1 | 40.3 | 31.0 |
Gene family . | . | . | . | CDR3 length, bp . | P . |
|---|---|---|---|---|---|
| No. | Mean | SD | Median | ||
| VH | .006 | ||||
| VH1 | 62 | 29.7 | 15.9 | 26.5 | |
| VH2 | 22 | 31.9 | 12.0 | 31.5 | |
| VH3 | 192 | 36.8 | 37.7 | 30.0 | |
| VH4 | 60 | 32.1 | 16.3 | 31.5 | |
| VH5 | 10 | 33.4 | 15.2 | 33.5 | |
| VH6 | 35 | 22.3 | 11.9 | 21.0 | |
| DH | < .0001 | ||||
| DH1 | 23 | 25.3 | 10.1 | 24.0 | |
| DH2 | 108 | 41.4 | 29.4 | 36.5 | |
| DH3 | 97 | 37.1 | 24.5 | 34.0 | |
| DH4 | 12 | 31.3 | 12.8 | 28.5 | |
| DH5 | 15 | 27.9 | 9.8 | 27.0 | |
| DH6 | 37 | 35.0 | 36.8 | 26.0 | |
| DH7 | 12 | 20.9 | 8.7 | 20.5 | |
| JH | .16 | ||||
| JH1 | 6 | 48.0 | 21.4 | 44.0 | |
| JH2 | 11 | 33.5 | 19.4 | 34.0 | |
| JH3 | 7 | 31.1 | 16.6 | 33.0 | |
| JH4 | 148 | 29.9 | 15.3 | 27.0 | |
| JH5 | 78 | 32.0 | 28.4 | 30.0 | |
| JH6 | 131 | 37.1 | 40.3 | 31.0 |
P values were obtained using the Kruskal-Wallis test, 2-sided.
CDR indicates complementarity-determining region; and bp, base pairs.
Biclonality and oligoclonality
A single IgH sequence was identified in 256 cases from diagnostic tumor samples, whereas 2 sequences were found in 58 cases and 3 sequences were found in 3 children. However, the technical approach we use identifies only major clonal rearrangements and does not detect small subclones. In this study we observed no correlation between biclonality or oligoclonality and clinical features except chromosome 9 deletions and in particular noted no association with relapse (Table 2).
Clonality and clinical characteristics of children with B-lineage ALL
. | Overall (%) . | Patients with 1 sequence (%) . | Patients with more than 1 sequence (%) . | P* . |
|---|---|---|---|---|
| No. | 317 | 256 (81) | 61 (19) | — |
| Median age, y (range) | 4.3 (0.01-17.9) | 4.3 (0.01-17.9) | 4.1 (0.9-16.5) | .56 |
| Sex | .89 | |||
| Male | 167 (53) | 134 (52) | 33 (54) | |
| Female | 150 (47) | 122 (48) | 28 (46) | |
| WBC, × 109/L | .38 | |||
| 0 to less than 20 | 211 (67) | 174 (68) | 37 (60) | |
| 20 to less than 50 | 54 (17) | 40 (16) | 14 (23) | |
| 50 to less than 100 | 28 (9) | 21 (8) | 7 (11) | |
| 100 or more | 23 (7) | 20 (8) | 3 (5) | |
| Relapse | .21 | |||
| No | 275 (87) | 225 (88) | 50 (82) | |
| Yes | 42 (13) | 31 (12) | 11 (18) | |
| Cytogenetic analysis | ||||
| Hyperdiploid more than 50 chromosomes | 53 (17) | 40 (16) | 13 (21) | .34 |
| Hyperdiploid less than 50 chromosomes | 16 (5) | 11 (4) | 5 (8) | .20 |
| Diploid | 83 (26) | 72 (28) | 11 (18) | .14 |
| Pseudo | 35 (11) | 27 (11) | 8 (13) | .65 |
| Hypodiploid | 13 (4) | 11 (4) | 2 (3) | .99 |
| Structure abnormal | 80 (25) | 60 (23) | 20 (33) | .14 |
| 11q23 | 8 (3) | 8 (3) | 0 (0) | .36 |
| t(1;19) | 2 (1) | 2 (1) | 0 (0) | .99 |
| 6q− | 9 (3) | 7 (3) | 2 (3) | .69 |
| 9p− | 7 (2) | 3 (1) | 4 (7) | .03 |
| 12p | 10 (3) | 7 (3) | 3 (5) | .41 |
| Insufficient | 94 (30) | 74 (29) | 20 (33) | .54 |
| ND | 17 (5) | 15 (6) | 2 (3) | .54 |
. | Overall (%) . | Patients with 1 sequence (%) . | Patients with more than 1 sequence (%) . | P* . |
|---|---|---|---|---|
| No. | 317 | 256 (81) | 61 (19) | — |
| Median age, y (range) | 4.3 (0.01-17.9) | 4.3 (0.01-17.9) | 4.1 (0.9-16.5) | .56 |
| Sex | .89 | |||
| Male | 167 (53) | 134 (52) | 33 (54) | |
| Female | 150 (47) | 122 (48) | 28 (46) | |
| WBC, × 109/L | .38 | |||
| 0 to less than 20 | 211 (67) | 174 (68) | 37 (60) | |
| 20 to less than 50 | 54 (17) | 40 (16) | 14 (23) | |
| 50 to less than 100 | 28 (9) | 21 (8) | 7 (11) | |
| 100 or more | 23 (7) | 20 (8) | 3 (5) | |
| Relapse | .21 | |||
| No | 275 (87) | 225 (88) | 50 (82) | |
| Yes | 42 (13) | 31 (12) | 11 (18) | |
| Cytogenetic analysis | ||||
| Hyperdiploid more than 50 chromosomes | 53 (17) | 40 (16) | 13 (21) | .34 |
| Hyperdiploid less than 50 chromosomes | 16 (5) | 11 (4) | 5 (8) | .20 |
| Diploid | 83 (26) | 72 (28) | 11 (18) | .14 |
| Pseudo | 35 (11) | 27 (11) | 8 (13) | .65 |
| Hypodiploid | 13 (4) | 11 (4) | 2 (3) | .99 |
| Structure abnormal | 80 (25) | 60 (23) | 20 (33) | .14 |
| 11q23 | 8 (3) | 8 (3) | 0 (0) | .36 |
| t(1;19) | 2 (1) | 2 (1) | 0 (0) | .99 |
| 6q− | 9 (3) | 7 (3) | 2 (3) | .69 |
| 9p− | 7 (2) | 3 (1) | 4 (7) | .03 |
| 12p | 10 (3) | 7 (3) | 3 (5) | .41 |
| Insufficient | 94 (30) | 74 (29) | 20 (33) | .54 |
| ND | 17 (5) | 15 (6) | 2 (3) | .54 |
WBC indicates white blood cell count; ND, not done.
P value was obtained using the Wilcoxon rank-sum test for age. The Fisher exact test and the χ2 test were used for categoric variables.
In 52 (85%) of the 61 patients with more than one sequence at presentation, the IgH sequences were unrelated. Only 9 cases of the samples identified at presentation involved ongoing IgH rearrangement mechanisms, phenomena described previously to account for clonal evolution.41-44 Sequence analyses demonstrated 2 cases showing VH-VH replacement (cases 99 and 177), 6 cases showing VH to DHJH joining (cases 190, 246, 266, 269, 392, and 400), and 1 case showing an “open-and-shut” mechanism (Table 3).Although any of these sequences identified at presentation could be used for MRD detection, no relapses have occurred in any of these 9 children.
Sequence analysis showing ongoing rearrangement in 9 children with B-lineage ALL
Case no. . | VH− gene* . | CDR3 (n-DH-n) . | 5′-JH . | Molecular mechanisms . |
|---|---|---|---|---|
| 99 | VH1-69 | gatcggcggTTGTACTAATGGTGTATGCT (DH2-8) | AAC (JH5) | |
| VH3-52 | gaggct---------------------- (DH2-8) | ---(JH5) | VH-VH replacement | |
| 177 | VH3-11 | agagaccctgaacggTATTTTGACTGGTTATTA (DH3-9) | TTT (JH4) | |
| VH4-39 | cggatg------------------------------ (DH3-9) | ---(JH4) | VH-VH replacement | |
| 190 | VH3-13 | gatcgggagaAT (AGC) 3TGaGATATTGTAGTGGTGGTAGCTGCTAC (DH2-15) | TAC (JH6) | |
| VH1-3 | gatggccc------------------------ (DH2-15) | --- (JH6) | VH-DJH joining | |
| 246 | VH3-74 | gtcgattGGATATTGTAGTAGTACCAGCTGCTAT (DH2-2) | TAC (JH6) | |
| VH1-2 | tcacggacaacgcgacag----------------------- (DH2-2) | ---(JH6) | VH-DJH joining | |
| 266 | VH1-46 | gaaactctaggctaactcggta (n=22 bp) | TGA (JH4) | |
| VH5-51 | cgaaaaa---------------------- (n=29 bp) | --- (JH4) | VH-NJH joining | |
| 269 | VH3-48 | GATATTGTAGTAGTACCAGCTGCTA (DH2-2) | AAC (JH5) | |
| VH4-34 | ggcggacc------------------- (DH2-2) | --- (JH5) | VH-DJH joining | |
| 392 | VH3-23 | ggccgccctacgGTATTACTATGATAGTAGTGGTTATcgg (DH3-22) | CTA (JH4) | |
| VH4-30 | gcccct--------------------------- (DH3-22) | --- (JH4) | VH-DJH joining | |
| 400 | VH4-28 | aatttcttcCTATGATAGTAGTGGTTATTAaaaggggg (DH3-22) | TGG (JH5) | |
| VH3-13 | tggggggg-------------------------- (DH3-22) | --- (JH5) | VH-DJH joining | |
| 214 | VH3-30 | cTAGTAGTACCAGCTGCTATtttggtcggggtg (DH2-2) | ACT (JH6) | |
| VH3-30 | gagggtaggttt-------------------------------- (DH2-2) | --- (JH6) | “Open-and-shut” |
Case no. . | VH− gene* . | CDR3 (n-DH-n) . | 5′-JH . | Molecular mechanisms . |
|---|---|---|---|---|
| 99 | VH1-69 | gatcggcggTTGTACTAATGGTGTATGCT (DH2-8) | AAC (JH5) | |
| VH3-52 | gaggct---------------------- (DH2-8) | ---(JH5) | VH-VH replacement | |
| 177 | VH3-11 | agagaccctgaacggTATTTTGACTGGTTATTA (DH3-9) | TTT (JH4) | |
| VH4-39 | cggatg------------------------------ (DH3-9) | ---(JH4) | VH-VH replacement | |
| 190 | VH3-13 | gatcgggagaAT (AGC) 3TGaGATATTGTAGTGGTGGTAGCTGCTAC (DH2-15) | TAC (JH6) | |
| VH1-3 | gatggccc------------------------ (DH2-15) | --- (JH6) | VH-DJH joining | |
| 246 | VH3-74 | gtcgattGGATATTGTAGTAGTACCAGCTGCTAT (DH2-2) | TAC (JH6) | |
| VH1-2 | tcacggacaacgcgacag----------------------- (DH2-2) | ---(JH6) | VH-DJH joining | |
| 266 | VH1-46 | gaaactctaggctaactcggta (n=22 bp) | TGA (JH4) | |
| VH5-51 | cgaaaaa---------------------- (n=29 bp) | --- (JH4) | VH-NJH joining | |
| 269 | VH3-48 | GATATTGTAGTAGTACCAGCTGCTA (DH2-2) | AAC (JH5) | |
| VH4-34 | ggcggacc------------------- (DH2-2) | --- (JH5) | VH-DJH joining | |
| 392 | VH3-23 | ggccgccctacgGTATTACTATGATAGTAGTGGTTATcgg (DH3-22) | CTA (JH4) | |
| VH4-30 | gcccct--------------------------- (DH3-22) | --- (JH4) | VH-DJH joining | |
| 400 | VH4-28 | aatttcttcCTATGATAGTAGTGGTTATTAaaaggggg (DH3-22) | TGG (JH5) | |
| VH3-13 | tggggggg-------------------------- (DH3-22) | --- (JH5) | VH-DJH joining | |
| 214 | VH3-30 | cTAGTAGTACCAGCTGCTATtttggtcggggtg (DH2-2) | ACT (JH6) | |
| VH3-30 | gagggtaggttt-------------------------------- (DH2-2) | --- (JH6) | “Open-and-shut” |
For case 99 and case 177, DHJH regions were conserved but VH segment was replaced by the other VH segment, demonstrating a VH-VH replacement mechanism. For case 190, 1 sequence used VH3-13, 2 DH (DH6-13 and DH2-15) and JH6 segment, but in the other sequence VH1-3 is attached to the identical DH2-15-JH6 region. For case 246, 2 sequences shared the identical DH2-2-JH6 region but were attached to 2 different VH segments (VH3-34 and VH1-2). In case 266, 22 nucleotides in the N region and the JH4 segment were preserved but attached to different VH segments (VH1-46 and VH5-51). For cases 269, 392, and 400, an identical DHJH complex was conserved but attached to 2 different VH segments. Sequence analysis in these 6 cases demonstrates a VH-DHJH joining mechanism. Case 214 presented with 2 sequences that used the same V, D, and J segment but introduced 12 nucleotides in V-D joining with 1 C nucleotide deletion, indicating that an “open-and-shut” mechanism was involved in this case.
CDR indicates complementarity-determining region; bp, base pairs.
Identity to VH germ line genes was 99% to 100%.
Functional rearrangements in B-cell childhood ALL.
Among the 381 sequences, 302 (79%) were joined in a potentially nonproductive rearrangement, including out-of-frame joinings or in-frame joinings containing a stop codon. Only 21% of the IgH rearrangements could potentially result in production of heavy chain protein, and these cases demonstrated an association with privileged usage of VH segments with 36% of these sequences utilizing VH4 (Table 4).
Association between potential productive rearrangement and VH segment utilization
Gene family . | Potential productive (%) . | Potential Nonproductive (%) . | Percentage of productive within VH gene family . |
|---|---|---|---|
| VH1 | 4 (5) | 58 (19) | 6 (4 of 62) |
| VH2 | 5 (6) | 17 (6) | 23 (5 of 22) |
| VH3 | 40 (51) | 152 (50) | 21 (40 of 192) |
| VH4 | 22 (28) | 38 (13) | 37 (22 of 60) |
| VH5 | 3 (4) | 7 (2) | 30 (3 of 10) |
| VH6 | 5 (6) | 30 (10) | 14 (5 of 35) |
| Total | 79 (21) | 302 (79) |
Gene family . | Potential productive (%) . | Potential Nonproductive (%) . | Percentage of productive within VH gene family . |
|---|---|---|---|
| VH1 | 4 (5) | 58 (19) | 6 (4 of 62) |
| VH2 | 5 (6) | 17 (6) | 23 (5 of 22) |
| VH3 | 40 (51) | 152 (50) | 21 (40 of 192) |
| VH4 | 22 (28) | 38 (13) | 37 (22 of 60) |
| VH5 | 3 (4) | 7 (2) | 30 (3 of 10) |
| VH6 | 5 (6) | 30 (10) | 14 (5 of 35) |
| Total | 79 (21) | 302 (79) |
Association between VH family usage and potential productive rearrangements (P = .002 by the χ2 test, and P = .001 by the Fisher exact test).
In-frame rearrangements only were detected in 59 patients, out-of-frame rearrangements only in 239, and in 19 patients both in-frame and out-of-frame rearrangements were detected in the same patient (Table 5). We noted an association between the presence of more than one sequence and whether the rearrangements were in frame or out of frame (Table 5).
Association between potential productive rearrangement and clonality in childhood ALL
Clonality . | Potential productive (%) . | Potential nonproductive (%) . | Mixed with productive and nonproductive . |
|---|---|---|---|
| No. of cases | 59 | 239 | 19 |
| One IgH sequence | 57 (97) | 199 (83) | — |
| More than 1 IgH sequence | 2 (3) | 40 (17) | 19 |
Clonality . | Potential productive (%) . | Potential nonproductive (%) . | Mixed with productive and nonproductive . |
|---|---|---|---|
| No. of cases | 59 | 239 | 19 |
| One IgH sequence | 57 (97) | 199 (83) | — |
| More than 1 IgH sequence | 2 (3) | 40 (17) | 19 |
P = .006 by the Fisher exact test. — indicates association between detection of more than 1 IgH sequence and potential nonproductive rearrangement.
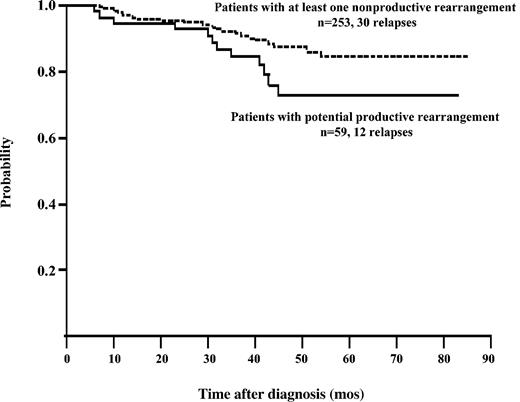
Only 3% of patients with only in-frame rearrangements had more than one sequence detested, compared with 17% of the cases with only out-of-frame rearrangements (P = .006 by the Fisher exact test). A trend toward a higher probability of risk of relapse was observed in patients with only productive rearrangements (P = .08 by the log-rank test), compared with children in whom at least one nonproductive rearrangement was identified at presentation (Figure 5).
Kaplan-Meier analysis. Analysis shows a trend toward a higher probability of risk of relapse in patients with only potential productive rearrangements compared with children in whom at least one nonproductive rearrangement was identified at presentation (P = .08 by the log-rank test, 2-sided). Five cases lost to follow-up for relapse were excluded from this analysis.
Kaplan-Meier analysis. Analysis shows a trend toward a higher probability of risk of relapse in patients with only potential productive rearrangements compared with children in whom at least one nonproductive rearrangement was identified at presentation (P = .08 by the log-rank test, 2-sided). Five cases lost to follow-up for relapse were excluded from this analysis.
Discussion
We assessed the VHDHJH gene usage profiles at presentation from 381 IgH sequences identified from 317 cases of B-lineage childhood ALL. This is the largest reported series of IgH sequences in childhood ALL and incorporates knowledge from the now complete map of the human IgH locus.
When VH gene usage is analyzed by position on the chromosome rather than simply by VH gene family, we observed a preferential usage of the DHJH-proximal VH segments including not only VH6-1 segment, the closest segment to the DHJH locus, but also other VH family segments near the DHJH locus, including VH1-2, VH1-3, VH2-5, VH3-7, VH3-9, VH3-11, VH3-13, and VH3-15 segments. The finding of position-dependent VH gene utilization supports the hypothesis that chromosomal order might in part regulate the VH sequences rearrangement.45 VH3 represents the largest gene family and was the most utilized family in children with B-lineage ALL, in keeping with the size of this VH family in the germ line.9,10 We observed a privileged usage of the VH6 segment, previously identified as a component of the quite restricted human fetal antibody repertoire.16,20 A privileged usage of VH6 has also been reported in immature B cells.30 Our observation is in line with previous reports from precursor B-ALL suggesting that ALL transformation arises at the early stage of B-cell development.30,32,34
The DH2 and DH7 segments were overrepresented and DH4 and DH5 segments underrepresented in childhood B-ALL compared with PBLs.11 A privileged usage of DH7 segments has been observed in both murine and human fetal cells (liver, spleen, and marrow B cells).16,19,46-48 However, in the present study we observed a lower frequency (4%) of utilization of DH7 in cases of ALL compared with those reported for use in nonmalignant B cells in fetal tissues by Sanz6 (14%) and by others (50%).16,48 We also observed that JH-distal DH segments were overrepresented. Therefore, it is unlikely that this can be explained by proximal locus regulation of the recombination machinery at the DHJH joining stage. In murine immature B-cell lines, a secondary DHJH rearrangement has been suggested by the finding that an initial DHJH rearrangement can be deleted and replaced by a more 5′ DH gene.49 Recent studies have shown that the specific recombination signal sequences (RSSs) and coding ends may play a role in the preferential joining of specific DH to JH genes,50-52 and the more closely an RSS resembles the consensus (CACAGTG-spacer-ACAAAAACC), the more often it is recombined in extrachromosomal substrates.53 Therefore, molecular mechanisms rather than location appear to govern the selection of DH gene segment during early B-cell development.
A characteristic JH gene usage pattern (JH4 > JH6 > JH5 > > JH3/2/1) has been reported for fetal, neonatal, childhood, and adult peripheral B-lymphocyte CDR3 regions.12,46,54 The usage of the JH genes in this study is in line with this order. It is likely that molecular mechanisms also govern the JH gene usage, with JH4 RSS more closely reassembling the consensus sequences followed by the RSSs of JH6, JH5, JH3, JH2, and JH1.12,54
In normal B cells, previous studies have demonstrated shorter CDR3 in preterm infants than in term infants and adults.55 In immature B cells, a recent study demonstrated that lack of expression of TdT was associated with the absence of shorter N sequences, which contributed to shorter CDR3 regions, especially at DHJH joining.56 TdT interacts with the DNA-dependent protein kinase (DNA-PK), particularly with the Ku proteins, suggesting that TdT expression may modulate gene segment recombination at the ligation step.57 Most cases of ALL presenting within the first 3 years of age have been suggested to arise from in utero transformation events by the finding of a high frequency (87.5%) of absence of N sequences at DHJH joining, similar to that found in human fetal life.31,33 This high frequency of absence of N sequences at DHJH joining was not observed in our study (data not shown). However, we observed that a short CDR3 was correlated with utilization of VH6 and DH7-27. This observation might be explained by developmental regulation of the recombination machinery with favored selection of JH-proximal VH and DH gene segments and by influence of TdT expression at an earlier stage of B-cell development.17,57
The overall incidence of cases with more than one IgH sequence in the present study was 19%. Although this is lower than in some reported studies (30% to 60%) using PCR fingerprinting or cloning strategies that may be better suited to find a minor subclone(s),44,58,59 our findings are in keeping with previous studies using Southern blot analysis.60-62 Some studies have shown an association between oligoclonality at presentation and poor prognosis in ALL patients.63,64 We did not observe an association between biclonality or oligoclonality and relapse or with any other clinical characteristic except for the presence of chromosome 9p deletion. The correlation between chromosome 9p deletion and more than one IgH sequence might be partly explained by the recent finding that illegitimate VDJ recombinations involved in chromosome 9p21 deletion in lymphoid leukemia are targeted at RSS-like sequences widely distributed in 9p21.65 Detailed sequence analysis showed that among the cases with more than one sequence, most sequences were unrelated with ongoing rearrangements found in only 15% of cases.
Similar to the findings previously reported for immature B cells,66 and in keeping with a previous study in ALL,33 only 21% of VH-JH rearrangements detected were potentially productive. Unlike normal B cells, this does not lead to loss of leukemic clone, demonstrating that ALL does not require signals mediated by Ig signaling for survival. Of note, a trend toward a higher probability of risk of relapse was found in patients with only productive rearrangements compared with children in whom at least one nonproductive rearrangement was identified at presentation. The reason potentially productive rearrangement would be relevant to clinical outcome is unclear. We did note an association between detection of more than one sequence and functional IgH rearrangement. Whether this relates to the stage of differentiation of the B cell at which malignant transformation occurs, or to lack of allelic exclusion by nonfunctional VDJ recombination, is currently unknown. In productive rearrangements we observed a privileged usage of VH4 segments. Of note, VH4-34 was found to be the third most overused VH segment (22 of 381, 5.8%) and the most frequently overrepresented VH4 family segment (22 of 60, 36.7%). The VH4-34 gene segment is located 500 kb upstream of the D gene locus and is markedly overrepresented in the normal B-cell repertoire.67 A biased usage of the VH4-34 segment in rearranged VH4 family genes has been reported in immature B cells.17 Increased usage of this gene segment also occurred in pathologic states including autoimmune disease and B-cell malignancies associated with autoimmune phenomena.68,69 The relevance of these findings to ALL is not clear, because ALL is not thought to be an antigen-driven process, although it is possible that this may be important in the subset of cases with potentially productive Ig gene rearrangements.
In summary, this study describes the VHDHJH gene utilization profile from 317 children with B-lineage ALL. A biased usage of VH6, and a shift from JH4 to JH6, was observed, similar to the patterns of immature B cells, in keeping with the widely held views that leukemic transformation occurs at the early stage of B-cell differentiation. The preferential usage of DHJH-proximal VH gene segments suggests that V to DJ recombination is somewhat dependent on physical location at the VH locus. However, DH and JH segment utilization is position independent and more likely governed by molecular mechanisms. Moreover, patients with only productive rearrangement demonstrated a trend toward an increased risk of relapse, suggesting that molecular pathophysiology might be relevant to clinical outcome.
Appendix
The members of the DFCI/ALL consortium are: Dana-Farber Cancer Institute, Children's Hospital, Boston, MA; St Justine Children's Hospital, Montreal, QC, Canada; San Jorge Children's Hospital, San Truce, Puerto Rico; University of Rochester Medical Center, Rochester, NY; Mount Sinai Medical Center, New York, NY; McMaster University Medical Center, Hamilton, ON, Canada; Maine Children's Cancer Program, Scarborough, ME; Centre Hospitalier de l'Universite Laval du CHUQ, Ste Foy, QC, Canada; and Tulane University, New Orleans, LA
Prepublished online as Blood First Edition Paper, March 9, 2004; DOI 10.1182/blood-2003-11-3857.
Supported by grant P01 CA68484 from the National Cancer Institute, National Institutes of Health (NIH).
A complete list of the participating institutions of the DFCI/ALL Consortium are listed in the “Appendix.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Peter Varney for help in the preparation of the manuscript and all members of the DFCI ALL Consortium for providing samples for this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal