Abstract
Caspase-3 plays a central role in apoptosis. It is also activated in normal erythropoiesis, with its activity peaking early during development (erythroid colony-forming unit [CFU-E] stage). In the present study, we have reduced the expression and subsequent enzymatic activity of caspase-3 by transfection of small interfering RNA (siRNA) directed to caspase-3 in a differentiating human erythroid culture system. We find that siRNA treatment yields a 50% reduction in cells that undergo enucleation with no change in the fraction of cells that undergo apoptosis, measured throughout the culture. Furthermore, a substantial fraction of treated cells are unable to complete the transition from pronormoblasts to basophilic normoblasts. These results demonstrate that caspase-3 is required for efficient erythropoiesis in this model system. (Blood. 2004;103:4310-4316)
Introduction
Members of the caspase family of aspartate-specific proteases have been found to be essential in a variety of cells and tissues for differentiation and homeostasis, by virtue of their role in causing apoptotic cell death. Due to its potent effect on cell viability, caspase activity is tightly regulated. These proteins are expressed as inactive proenzymes that must be proteolytically cleaved into large and small fragments, which then associate as a tetramer to form a catalytically active enzyme.1 Caspase activation proceeds by a cascade mechanism. External stimuli, such as death receptor (Fas, tumor necrosis factor-α [TNF-α]) stimulation or cytokine withdrawal, and internal stimuli, such as genotoxic stress or mitochondrial permeabilization, activate a subset of the initiator caspases, 2, 8, 9, and 10. These cleave the effector caspases, 3, 6, and 7, which in turn cleave additional caspases and vital cellular targets.1 Caspase substrates include structural proteins (actin, lamin B, and gelsolin2,3 ), proteins required for DNA repair (poly(ADP-ribose) polymerase [PARP]4 and DNA-dependent protein kinase5 ), and proteins with specific apoptotic function (DNA fragmentation factor that releases caspase-activated DNase6,7 ). Although there are distinct differences,8 the striking similarities between programmed cell death and late stages of erythropoiesis have been noted9,10 and include nuclear and chromatin condensation; cleavage of nuclear proteins, such as acinus, lamin B, and PARP; and possibly, caspase activation. However, the significance of these similarities, especially with respect to caspase activation, is not fully established.
Erythropoiesis is a complex multistage process encompassing the differentiation of pluripotent hematopoietic progenitor cells to mature erythrocytes. The earliest cell committed to the erythroid lineage is the erythroid burst-forming unit (BFU-E). These then become erythroid colony-forming unit (CFU-E) cells, which are followed in turn by the morphologically distinguishable stages of pronormoblast and then basophilic, polychromatic, and orthochromatic normoblast. A key event of late-stage erythropoiesis is nuclear condensation followed by extrusion of the nucleus to produce enucleated reticulocytes and finally mature erythrocytes.
Erythropoietin (EPO) positively regulates erythropoiesis by stimulating proliferation and differentiation of erythroid progenitors, at least in part by up-regulating the erythroid-specific transcription factor, GATA-1.11 In addition to activating a program of erythroid gene expression,12 GATA-1 up-regulates the expression of the antiapoptotic protein Bcl-XL, which is necessary for erythroblast survival.11,13,14 Withdrawal of EPO or stimulation of a death receptor such as Fas leads to activation of a subset of caspases, including caspase-3, -7, and -8, which then cleave GATA-1 and trigger apoptosis.15-18 However, a number of caspases are expressed and activated in normal, productive erythroid development.14,19,20 Consistent with this, several caspase substrates are cleaved in differentiating erythroid cells; this includes lamin B,20 a protein associated with the nuclear membrane, and acinus,19 whose activation by cleavage is required for chromatin condensation.2 Furthermore, it has been demonstrated that pancaspase inhibitors block erythroid development at an early stage of maturation.19,20 The demonstration of procaspase cleavage and the presence of caspase activity toward caspase-specific peptide substrates indicate a possible involvement of caspase-2, -3, -6, -7, and -9 in erythroid differentiation.19,20 Paradoxically, in normal erythroid development caspase activation does not lead to cleavage of GATA-1, although it is known to be a substrate of most caspases.17 Many additional hallmarks of apoptosis are also absent in these cells, including phosphatidylserine externalization and DNA cleavage. Although nuclear condensation is observed, it is followed by extrusion of the nucleus and not cell death. The precise requirement for any individual caspase is difficult to determine from these studies, because the irreversible inhibitors that were used do not have absolute substrate specificities.
The participation of caspase-3 in erythropoiesis has been called into question10,21,22 by the findings of 2 separate studies in which a single caspase-3 knock-out was examined in 2 different genetic backgrounds. One study22 found that caspase-3-deficient mice are viable at birth although they are born at a frequency lower than expected by mendelian genetics, are smaller than wild type, and die within 1 to 3 weeks.22 The major defects in these mice are profound neural abnormalities due to decreased apoptosis in the developing brain.22 However, the precise effects of caspase-3 deficiency on the rate or efficiency of erythropoiesis in these mice have, to our knowledge, never been studied. The second caspase-3-deficient mouse study21 showed the mouse had the ability to reach adulthood. However, the precise effects in erythropoiesis may be masked. This possibility was demonstrated by the finding that caspase-8 can carry out some of the normal functions of caspase-3 in a caspase-3-deficient system. In neither study was the process of erythropoiesis explicitly investigated.
Here, we address the requirement for caspase-3 in erythropoiesis in an ex vivo model system of human erythroid cell differentiation in liquid culture. We demonstrate that this is a valid model of erythropoiesis because virtually all the cells in culture are of the erythroid lineage and a large fraction (> 30%) progress to become enucleated reticulocytes. An additional population of more than 30% of the culture reaches the stage of orthochromatic normoblast. We find that primary erythroblasts in our model are susceptible to targeting by RNA interference. We have used small interfering RNA (siRNA) to caspase-3 to show that ablation of caspase-3 activity blocks erythropoiesis in a large fraction of the erythroblasts in culture.
Materials and methods
Plasmids and siRNA
The plasmid, pEGFP, was from Clontech (Palo Alto, CA). All siRNAs were obtained from the Functional Proteomics Project at Memorial Sloan-Kettering Cancer Center (MSKCC) and included green fluorescent protein (GFP) siRNA, CAAGCUGACCCUGAAGUUCTT; and siRNA-1, UGGAUUAUCCUGAGAUGGGTT (nucleotides [nt's] 351 to 369), and siRNA-2, AGUGAAGCAAAUCAGAAACTT (nt's 2148 to 2166), for caspase-3. In experiments regarding GFP expression, the unrelated (control) siRNA was a nonfunctional siRNA to a caspase-3 sequence, CAUUCAUAGAGGGGUGGAGTT (nt's 1722 to 1740). The control siRNA used for caspase-3 experiments was the GFP siRNA.
Isolation and culture of human CD34+ cells
Human peripheral blood, obtained from Drs R. Comenzo and S. Sharma (Stem Cell Laboratory, MSKCC) with physician consent, was excess to patient requirements. The mobilized blood from patients and healthy donors was collected 5 to 6 days after injection with granulocyte colony-stimulating factor (G-CSF) (6 μg/kg) and stored at -80°C. Thawed blood was diluted with a half volume of sterile 10% dextran 40 (Sigma, St Louis, MO) and a half volume of 5% human albumin (American Red Cross). Cells were washed extensively with Hanks buffered saline solution and mononuclear cells isolated using a Ficoll-Paque PLUS (Amersham Pharmacia Biotech, Piscataway, NJ) step gradient. The lymphocyte layer was washed 3 times with phosphate-buffered saline (PBS) supplemented with 2 mM EDTA (ethylenediaminetetraacetic acid) and 0.5% bovine serum albumin (BSA). CD34+ cells were isolated by positive selection using anti-CD34-tagged magnetic beads and a VarioMacs magnet (Miltenyi Biotec, Auburn, CA) according to the manufacturer's protocol. The recovered cells were typically more than 95% CD34+.
CD34+ cells were grown in serum-free liquid culture using QBSF-60 media (Quality Biological, Gaithersburg, MD) supplemented with 2 U/mL penicillin/streptomycin and a cocktail of cytokines (R&D Systems) as follows: days 0 to 3, 10 ng/mL interleukin-3 (IL-3) and 10 ng/mL stem cell factor (SCF); days 3 to 7, 10 ng/mL IL-3, 10 ng/mL SCF, and 2 U/mL EPO; days 7 to 9, 10 ng/mL SCF and 2 U/mL EPO; and after day 9, 2 U/mL EPO. After day 9, media were renewed every 3 to 4 days and cell density was maintained below 1 × 106/mL. At day 16, cells were transferred to a dish coated with human fibronectin (Sigma).
Assay of cell proliferation, morphology, and fluorescent protein expression
Cell number was assessed by counting cells after trypan blue dye exclusion staining. For morphology, cytospin preparations were viewed with Wright-Giemsa stain. The composition of the cell population was determined by assessing and counting cells in 5 different fields of view, for a total of 500 to 600 cells. Micrographs were collected to view the GFP fluorescence using cells that were cytocentrifuged onto glass slides 3 days after transfection. Images were collected on a Leica LSM 5010 laser scanning confocal microscope (Leica, Bannockburn, IL) and at the MSKCC cytology core facility on a Zeiss Axionert 200M using Metamorph software (Zeiss, Jena, Germany).
FACS analysis
Approximately 1 × 106 cells were collected, washed with PBS containing 1% BSA and 0.1% sodium azide, and incubated with monoclonal antibodies against CD34, CD36, CD39, CD71, band 3, glycophorin A (GPA; Pharmingen, San Diego, CA), or rabbit polyclonal Lutheran glycoprotein (Lut-G) (a kind gift from the International Blood Group, National Blood Service, United Kingdom) or with no antibody. Cells were then washed and stained with the appropriate fluorescein isothiocyanate-conjugated secondary antibody (Pharmingen). Cells transfected with a GFP-encoding plasmid were analyzed at day 3 without antibody treatment. Fluorescence-activated cell sorter (FACS) analysis was performed by the flow cytometry core facility, MSKCC, on a Facscalibur using CellQuest software (Becton Dickinson, Franklin Lakes, NJ).
Transfection
CD34+ cells were electroporated using a Nucleofector and a Human CD34 Cell Nucleofector Kit (Amaxa Biosystems, Gaithersburg, MD) according to the manufacturer's protocol. For each electroporation, 1 × 106 CD34+ cells in 100 μL were treated with 2 μL of 50 μM siRNA and/or 300 nM DNA.
Evaluation of caspase-3 activity
Samples of 1 × 106 cells were resuspended in cell lysis buffer and lysed by freeze-thaw. The cell lysate was centrifuged at 200g for 5 minutes and the supernatant assayed for caspase-3 activity using the ENZCHEK caspase-3 assay kit with a DEVD-AMC substrate (Molecular Probes, Eugene, OR) according to the manufacturer's instructions.
Measurement of apoptosis
Samples of 1 × 106 cells were washed once in cold PBS and processed using the TACS Annexin 5 Kit (TREVIGEN, Gaithersburg, MD) as per the manufacturer's instructions. The positive control was generated using CD34 cells cultured in the model system for 7 to 9 days and then washed and cultured in media without cytokines for 6 hours prior to assay.
Western blot
Aliquots of 1 × 106 cells were frozen in liquid nitrogen and stored at -80°C prior to analysis. Cells were lysed in 50 mM Tris (tris(hydroxymethyl)aminomethane)-Cl (pH 8.0), 140 mM NaCl, 0.5% Nonidet P-40, and protease inhibitors with EDTA (Roche, Indianapolis, IN) and centrifuged at 1000g for 5 minutes at 4°C. One fifth of each sample was analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted to nitrocellulose. Membranes were incubated with one of the following primary antibodies: antihuman caspase-3 mouse monoclonal, antihuman acinus goat polyclonal immunoglobulin G (IgG), antihuman lamin B rabbit polyclonal IgG (Santa Cruz Biotechnology, Santa Cruz, CA), or mouse monoclonal to human glyceraldehyde phosphate dehydrogenase (GAPDH; Chemicon International, Temecula, CA) followed by horseradish peroxidase-conjugated secondary antibodies and developed with enhanced chemiluminescence (ECL) reagents (Amersham).
Results
Differentiation in culture of CD34 progenitors
It was first necessary to establish a model system of erythroid differentiation that would be amenable to manipulation. For this, we isolated human CD34+ progenitor cells and grew them in serum-free liquid culture with a regimen of cytokines (see “Materials and methods”) that promoted erythropoiesis. As shown in Figure 1A, these culture conditions supported logarithmic cell growth until about days 10 to 12, resulting in an approximately 100-fold increase in cell number. Progression of the cultured cells through the stages of erythropoiesis was assessed by microscopic examination (Figures 1B and 4A). At day 7 of culture, cells were primarily pronormoblasts, an early intermediate in erythroid differentiation. By days 10 to 13, basophilic normoblasts were predominant; the nucleus had clearly moved off center and become more granular. In addition, the first enucleated cells were detected at this time. With time, the numbers of basophilic normoblasts decreased as they were replaced by polychromatic normoblasts to be succeeded, at later stages of culture, by orthochromatic normoblasts and reticulocytes (Figure 1B, days 17 and 18). The defining event of terminal erythroid differentiation is the enucleation process (Figure 1B, day 17). The percentage of cells that enucleated to become reticulocytes is shown graphically in Figure 1C. No enucleation was detected prior to day 10, after which enucleation rose to more than 30% of the total culture by days 18 to 20. This corresponds to more than 1.5 × 107 enucleated reticulocytes in a total culture of 5 × 107 cells.
A model system for in vitro erythropoiesis of normal human CD34+ progenitor cells. (A) Highly purified CD34+ cells were placed in culture as described in “Materials and methods” and assessed over time for total cell number. (B) Cells stained with Wright-Giemsa showing pronormoblasts (day 7), basophilic normoblasts (day 13), orthochromatic normoblasts, one of which is in the process of enucleating (day 17), and enucleated reticulocytes and orthochromatic normoblasts (day 18). Scale bar is 5 μm. (C) The percentage of enucleated reticulocytes in culture at the indicated times. (D) FACS analysis showing the percentage of cells expressing CD34, CD36, or glycophorin A cell surface markers. Error bars indicate SD.
A model system for in vitro erythropoiesis of normal human CD34+ progenitor cells. (A) Highly purified CD34+ cells were placed in culture as described in “Materials and methods” and assessed over time for total cell number. (B) Cells stained with Wright-Giemsa showing pronormoblasts (day 7), basophilic normoblasts (day 13), orthochromatic normoblasts, one of which is in the process of enucleating (day 17), and enucleated reticulocytes and orthochromatic normoblasts (day 18). Scale bar is 5 μm. (C) The percentage of enucleated reticulocytes in culture at the indicated times. (D) FACS analysis showing the percentage of cells expressing CD34, CD36, or glycophorin A cell surface markers. Error bars indicate SD.
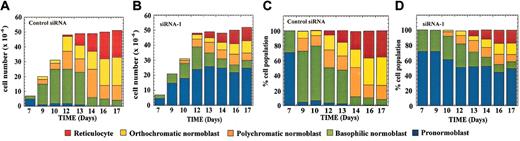
Caspase-3 siRNA alters erythroblast development. CD34+ progenitor cells were electroporated with control siRNA (A,C) or with siRNA-1 (B,D) at day 0 of culture. Cell numbers for each erythroblast population are shown in panels A and B, whereas percentage cell number for each culture is shown in panels C and D.
Caspase-3 siRNA alters erythroblast development. CD34+ progenitor cells were electroporated with control siRNA (A,C) or with siRNA-1 (B,D) at day 0 of culture. Cell numbers for each erythroblast population are shown in panels A and B, whereas percentage cell number for each culture is shown in panels C and D.
Analysis of cell surface markers confirmed that the cells in culture were progressing through the stages of normal erythroid development. At day 0, virtually all the cells were CD34+ (Figure 1D). After day 4, the percent of CD34+ cells declined until, by the point that cell proliferation had ceased (about days 14 to 15), less than 5% of the culture was CD34+. This is expected because CD34 is a marker of early progenitor cells and, in the erythroid lineage, is not expressed after the BFU-E stage.23 In contrast, the CD36 marker is first expressed on CFU-Es and maintained through the appearance of basophilic normoblasts and then declines on polychromatic normoblasts. As seen in Figure 1D, CD36+ cells increased in number rapidly after day 0 to reach 80% of the culture at day 7 and decreased in number thereafter. Glycophorin A (CD235a) is a marker of more mature erythroid cells, because it first appears at the basophilic normoblast stage of development after which its expression increases throughout the rest of erythroid differentiation up to and including erythrocytes. In our culture, GPA expression increased almost linearly with time until day 15, when about 90% of the cells are GPA-positive. In addition, a nonerythroid marker normally expressed on lymphoblasts, CD39, was not present on the cultured cells (data not shown). These data demonstrate that virtually all of the cells in culture were erythroblasts with most reaching the late stages of erythropoiesis and more than 30% undergoing enucleation to become reticulocytes. The ability of these cells to progress so far in erythroid development demonstrates that this culture system provides a valid model of erythropoiesis.
RNA interference functions in erythroid progenitor cells
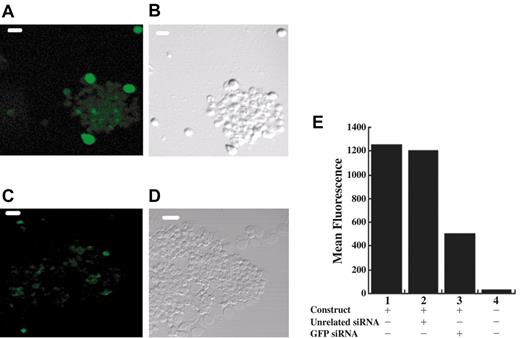
To determine if RNA interference can be used in the erythroblasts produced in our cell culture, CD34+ cells were transfected at day 0 with a GFP-expressing plasmid (pEGFP) alone or in combination with either an siRNA to GFP or a control siRNA of unrelated sequence. Examination by fluorescence microscopy after 3 days in culture allowed us to estimate that about 60% of cells cotransfected with pEGFP and a control siRNA expressed low to moderate GFP with a minor population expressing high levels of GFP (Figure 2A and data not shown). In contrast, cotransfection of pEGFP with siRNA specific to GFP resulted in a smaller population of cells (about 30%-40%) expressing low levels of GFP and no highly expressing cells (Figure 2C and data not shown). To quantitate the total reduction in GFP expression, which includes both the percentage of GFP-expressing cells and the intensity of green fluorescence per cell, we performed a FACS analysis, shown in Figure 2E. Cells transfected with pEGFP alone exhibited high levels of green fluorescence relative to background. Notably, treatment with a specific GFP siRNA decreased mean fluorescence by about 60%, whereas cotransfection of pEGFP with a control siRNA of unrelated sequence had no significant effect. These results indicate that RNA interference is operative in this primary culture of differentiating cells and that we are obtaining 50% to 60% transfection efficiency.
RNA interference functions in CD34+ cells. Cultured erythroblasts were transfected with a plasmid expressing GFP and either a control RNA (A-B) or an siRNA to GFP (C-D). Laser scanning confocal micrographs of cells showing fluorescent (A,C) or phase contrast (B,D) images of the same cells; scale bar is 40 μm. (E) FACS analysis at day 3 to measure the mean fluorescence of GFP expression in the GFP-positive populations from cells transfected at day 0 with pEGFP (lane 1), pEGFP plus an unrelated siRNA (lane 2), or pEGFP plus GFP-specific siRNA (lane 3) or of the total population of mock transfected cells (lane 4).
RNA interference functions in CD34+ cells. Cultured erythroblasts were transfected with a plasmid expressing GFP and either a control RNA (A-B) or an siRNA to GFP (C-D). Laser scanning confocal micrographs of cells showing fluorescent (A,C) or phase contrast (B,D) images of the same cells; scale bar is 40 μm. (E) FACS analysis at day 3 to measure the mean fluorescence of GFP expression in the GFP-positive populations from cells transfected at day 0 with pEGFP (lane 1), pEGFP plus an unrelated siRNA (lane 2), or pEGFP plus GFP-specific siRNA (lane 3) or of the total population of mock transfected cells (lane 4).
Caspase-3 is required for erythroid differentiation
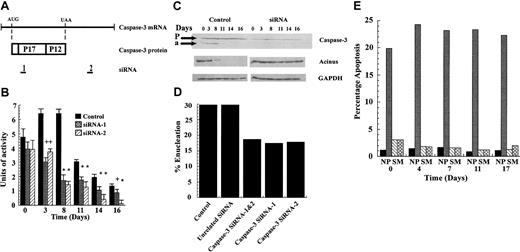
To test the role of caspase-3 in erythroid differentiation, we used siRNAs to 2 unique regions of caspase-3 mRNA, as diagrammed in Figure 3A (sequences in “Materials and methods”). siRNA-1 corresponds to sequences encoding the N-terminal region of the p17 polypeptide (nt's 351 to 369) and siRNA-2 to the 3′ untranslated region (3′UTR) (nt's 2148 to 2166). CD34+ cells at day 0 were transfected with siRNA-1, siRNA-2, or a control siRNA of unrelated sequence and then cultured as usual. The control culture displayed caspase-3 activity that peaked between day 3 and 8, declined rapidly between day 8 and 11, and declined slowly thereafter (Figure 3B). Cells treated with either siRNA-1 or siRNA-2 were indistinguishable from control at day 0. However, caspase activity was reduced by 40% at day 3 and by 70% at day 8, relative to control cells (Figure 3B). Remarkably, siRNA-2 was still effective at reducing caspase-3 activity by 60% to 90% out to day 16, despite a 100-fold expansion in total cell numbers between day 0 and 16 (Figure 1A).
siRNA to caspase-3 is effective in reducing caspase-3 activity and in blocking erythroid differentiation. (A) The relation of the 2 caspase-3 siRNAs to the sequence of the mRNA and protein-coding region of caspase-3. (B) CD34+ progenitor cells were electroporated with siRNA-1 or -2 or a control siRNA at day 0 and then assayed over time for caspase-3 enzymatic activity. Differences between control and siRNA-treated cultures were evaluated by t test. *P < .01; +P < .05. Error bars indicate SD. (C) Cell lysates were immunoblotted for protein levels of caspase-3, acinus and, as a control, GAPDH using control and caspase-3 siRNA-1-treated cells. Arrows indicate bands corresponding to procaspase-3 (P) and the cleaved 17-kDa active form of caspase-3 (a). (D) Percentage of cells that were enucleated on day 17. (E) Percentage of apoptotic cells in the culture at various times (N indicates negative control; P, positive control, S, caspase-3 siRNA-1 treated; M, mock transfected control).
siRNA to caspase-3 is effective in reducing caspase-3 activity and in blocking erythroid differentiation. (A) The relation of the 2 caspase-3 siRNAs to the sequence of the mRNA and protein-coding region of caspase-3. (B) CD34+ progenitor cells were electroporated with siRNA-1 or -2 or a control siRNA at day 0 and then assayed over time for caspase-3 enzymatic activity. Differences between control and siRNA-treated cultures were evaluated by t test. *P < .01; +P < .05. Error bars indicate SD. (C) Cell lysates were immunoblotted for protein levels of caspase-3, acinus and, as a control, GAPDH using control and caspase-3 siRNA-1-treated cells. Arrows indicate bands corresponding to procaspase-3 (P) and the cleaved 17-kDa active form of caspase-3 (a). (D) Percentage of cells that were enucleated on day 17. (E) Percentage of apoptotic cells in the culture at various times (N indicates negative control; P, positive control, S, caspase-3 siRNA-1 treated; M, mock transfected control).
Immunoblot analysis for caspase-3 protein in control cells detected stable precursor expression throughout the culture. Levels of active cleaved caspase peaked between day 3 and day 8, after which protein levels dropped below the limit of detection. In siRNA-treated cells, procaspase-3 was diminished but still detectable throughout the time course (Figure 3C), but no active form of the caspase was detected. To verify the levels of caspase-3 activity in vivo, cell lysates were immunoblotted for a known substrate of caspase-3, acinus, a nuclear protein that becomes involved in condensation of nuclear chromatin after cleavage.2,19 Figure 3C displays a representative experiment using siRNA-1, but similar results were obtained with siRNA-2 (data not shown). In control cells, the 100-kDa band representing uncleaved acinus, clearly visible at day 0, is diminished after 3 days and nearly gone after 8 days. In contrast, in cells treated with siRNA-1, the band for acinus is undiminished at day 3 and at best reduced by 2-fold at later times of culture. Notably, the antibody used in this experiment did not detect the 20-kDa cleavage product of acinus. Also, the almost total processing of acinus in the control cells after day 8 is in line with the observation that, although only 30% of cells enucleate, nearly all pass through the pronormoblast stage (Figure 4A), the stage with peak caspase-3 activity. Similar results were obtained for lamin B (data not shown). An immunoblot of GAPDH is shown as a control. Taken together, these data demonstrate that siRNA to caspase-3 has diminished but not fully eradicated expression and enzymatic activity of caspase-3 in these developing erythroblasts.
Our initial readout for the ability of treated cultures to mature was to score the percentage of cells that become enucleated reticulocytes. As seen in Figure 3D, both siRNAs, used singly or in concert, resulted in a 50% decrease in the percent enucleation.
This raised the question of whether the observed decrease in enucleating reticulocytes was due to apoptosis of specific subpopulations of cells, a process in which the participation of caspase-3 has been widely documented. To answer this question, cells were again cultured with or without siRNA to caspase-3 and assayed at various time points for the presence of phosphatidylserine on the outer surface of the plasma membrane, an accepted marker for apoptosis in erythroid cells.19 Figure 3E demonstrates that there was no significant difference in apoptosis between the mock siRNA-treated (M) or the caspase-3 siRNA-treated (S) cells, especially at early time points. Both siRNA-treated cultures displayed a slight elevation in apoptosis relative to control cells at day 0, probably due to the nucleoporation treatment.
A more detailed examination of cell morphology at different times of culture revealed that a significant fraction of the cells that received caspase-3 siRNA were arrested at the pronormoblast stage. At day 7, cell composition was identical between cells transfected with control siRNA or siRNA-1 (compare Figure 4A,C to Figure 4B,D). With time, the number of pronormoblasts in the control culture (Figure 4A) dropped to near zero, whereas in the siRNA-treated cells (Figure 4B) it rose 5-fold. Thus, in the control, virtually all of the pronormoblasts were able to progress to basophilic normoblasts while in the siRNA-treated culture a fraction of the pronormoblasts were blocked in development. This resulted in 50% of the siRNA-treated culture being pronormoblasts by day 17 (Figure 4D). For later erythroblast populations, changes in cell numbers followed a similar pattern in control and siRNA-treated cultures, although with lower numbers of siRNA-treated cells. In both cases, numbers of basophilic and polychromatic normoblasts rose, then fell, peaking at days 10 to 12 and at days 13 to 14, respectively. Numbers of orthochromatic normoblasts and reticulocytes rose steadily after about day 10. Final numbers of these 2 populations in the siRNA-treated cultures were approximately half that in the control culture. Remarkably, total cell number for control and siRNA-treated cultures was essentially identical at each time point, despite the large differences between individual cell populations (Figure 4). In particular, growth of both cultures plateaued after day 12. The pronormoblast population in the siRNA-treated culture expanded rapidly from day 7 to 12 and plateaued thereafter.
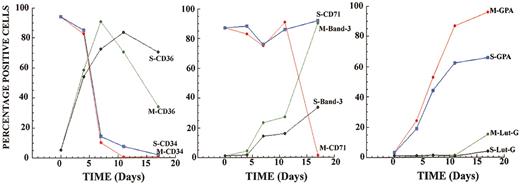
In support of the identification of cell populations reported in Figure 4, FACS analyses were performed throughout the culture period after treatment of cells with either caspase-3 siRNA-1 (S) or control siRNA (M) (Figure 5). The markers were selected to represent changes in cell surface expression that occur early (CD34, CD36), intermediate (band 3, GPA), and late (band 3, CD71, and Lut-G) during erythroid maturation.24,25 The results clearly demonstrate that before days 5 to 7 of culture, only minimal differences were seen between caspase-3 and control siRNA-treated cultures. Thus, both cultures displayed a rapid decline in CD34 expression, a major increase in CD36 expression, and stable, high levels of CD71 (also known as transferrin receptor) expression. The markers expressed, especially the very high levels of CD71, indicate that most cells in both cultures are indeed erythroid progenitors.9,26 After about day 5, expression levels of the different markers in the 2 cultures began to diverge. By day 17 of culture, cells treated with caspase-3 siRNA retained a higher level of CD36 (70% versus 30% in control cultures) but lagged behind control cultures in expression of band 3 (38% versus > 90%) and GPA (65% versus > 90%). Lutheran glycoprotein, which is transiently expressed on orthochromatic normoblasts shortly before enucleation, was expressed on 15% of the control culture on day 17 but on only 5% of the caspase-3 siRNA-treated culture. Finally, CD71, which is reported to disappear from the cell surface at very late stages of erythropoiesis, was no longer detectable in control cultures but was maintained at high levels after treatment with capsase-3 siRNA. For every marker examined, the expression pattern for the cells treated with caspase-3 siRNA points to an arrest in the maturation of these cells. Taken together, these results support the analysis shown in Figure 4 and are entirely consistent with a block in development at the end of the pronormoblast stage.
Caspase-3 siRNA disrupts the expression of erythroid cell surface markers. Cells were taken at various time points during the experiment described for Figure 4, and FACS analysis was performed. The 2 cell populations were defined: S for caspase-3 siRNA treated and M for control cells. The percentage of positive cells for each time point is shown.
Caspase-3 siRNA disrupts the expression of erythroid cell surface markers. Cells were taken at various time points during the experiment described for Figure 4, and FACS analysis was performed. The 2 cell populations were defined: S for caspase-3 siRNA treated and M for control cells. The percentage of positive cells for each time point is shown.
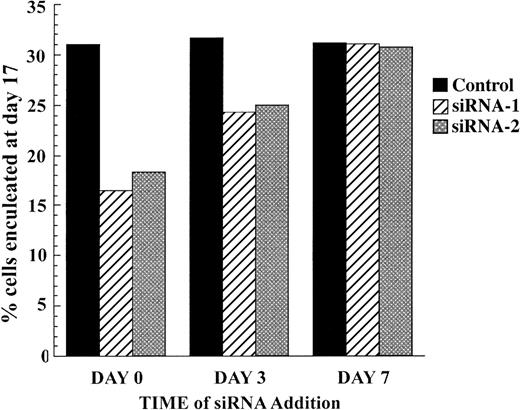
A block in differentiation at the pronormoblast stage suggests that the requirement for caspase-3 activity is early in erythropoiesis, when caspase-3 expression peaks. To test this, we introduced siRNA to caspase-3 at later times in the culture of the erythroblasts to determine whether this would be able to decrease the percentage of cells that become enucleated. We first confirmed that pEGFP and siRNA to GFP could be successfully transfected at later time points (data not shown). Figure 6 shows that introduction of siRNA to caspase-3 at day 7 had no effect on the percentage of cells that have enucleated by day 17. The maximal effect was seen with cells treated with siRNA at day 0. As in the experiment shown in Figure 3, this produced a 50% drop in percent enucleation compared with control cells. Introduction of siRNA at day 3 had an intermediate effect, decreasing enucleation from 32% for control cells to 24% and 25% for cells treated with siRNA-1 and siRNA-2, respectively.
Caspase-3 acts early in erythropoiesis. CD34+ progenitor cells were electroporated at day 0, day 3, or day 7 with control siRNA, siRNA-1, or siRNA-2 as indicated. The graph shows the percentage of cells that were enucleated on day 17.
Caspase-3 acts early in erythropoiesis. CD34+ progenitor cells were electroporated at day 0, day 3, or day 7 with control siRNA, siRNA-1, or siRNA-2 as indicated. The graph shows the percentage of cells that were enucleated on day 17.
Discussion
Feedback regulation of erythropoiesis relies on a balance between EPO promoting cell survival and Fas/Fas ligand (FasL) inducing cell death. When red blood cells are in excess, EPO is low and immature erythroblasts, which express Fas, are especially sensitive to killing by mature erythroblasts expressing FasL. When red blood cell production is again required, EPO levels increase, progressively inhibiting Fas-induced apoptosis, and erythropoiesis is able to proceed.15-18 In this scheme, caspase activation by Fas/FasL leads to apoptosis. However, accumulating evidence implicates caspases in a nonapoptotic role in erythropoiesis. It has been shown that caspase inhibitors arrest erythroid development in human cells19 and in mouse fetal liver.20 The lack of absolute specificity of these inhibitors, which bind irreversibly to the enzyme's active site, does not permit a firm conclusion to be drawn as to which caspases are involved.27 In other studies, analysis of Raf-1-deficient mouse embryos suggested that the severe anemia they exhibit is a result of premature caspase activation leading to inappropriate erythropoiesis with depletion of hematopoietic progenitors.20 Conversely, Raf-1 overexpression in an erythroblast cell line led to decreased lamin B cleavage, indicative of decreased caspase activity, and delayed erythroid differentiation. Our studies were initiated to explore the specific contribution of caspase-3 to erythropoiesis. We determine that caspase-3 activity that appears in developing erythroid cells does not trigger detectable apoptosis. Further, we find that inhibition of caspase-3 expression and activity by treatment of cultures with caspase-3-specific siRNA significantly reduces the ability of erythroblasts to progress through erythropoiesis. The developmental arrest produced by caspase-3 inhibition appears to occur at the transition between pronormoblasts and basophilic normoblasts.
These experiments rely on a model system for erythropoiesis in which early CD34+ erythroid progenitors isolated from human peripheral blood are cultured in a defined serum-free medium (see “Materials and methods”). Under our culture conditions, virtually all of the cells become erythroblasts, as shown by cell morphology (Figure 1B) and cell surface markers (Figures 1D and 5). By days 17 to 20 of culture, more than 30% of the cells have enucleated to become reticulocytes with another 30% left as orthochromatic normoblasts (Figures 3D, 4A, and 5). Further time in culture does not result in a significant increase in the percentage of enucleated cells (data not shown). Thus, only half of the cells that become orthochromatic normoblasts are able to progress to the next stage to become reticulocytes. This may, at least in part, be due to the absence of the normal supporting cell populations, such as macrophages.28,29 However, the earlier stages of erythropoiesis are transited much more efficiently. Essentially all of the CD34+ BFU-Es that constitute the erythroid-committed cells at day 0 develop through the pronormoblast stage to become basophilic normoblasts (Figures 4A and 5). Due to this, by day 14 of culture, the pronormoblast population is reduced to less than 0.5% of the culture. Caspase-3 activity peaks early in erythroblast development, between days 3 and 8, when most of the cells are expressing CD36 and correspond to CFU-Es or pronormoblasts (Figures 1D, 3B, and 5A). Consistent with this early peak in caspase activity, the caspase-3 substrate, acinus, is fully cleaved by day 11 of culture (Figure 3C).
Introduction of caspase-3 siRNA into these cells at day 0 has a profound effect on their capacity to complete erythropoiesis. Although siRNA theoretically acts to decrease levels of caspase-3 mRNA in the culture, the ultimate goal of the experiment was to test the effect of a reduction in caspase-3 enzymatic activity. Treatment with either of 2 unique siRNAs reduces caspase-3 activity about 50%, measured both in vitro in cell lysates and ex vivo. Given that we observe a transfection efficiency for GFP of 50% to 60% in these cells, it is likely that nearly every cell that actually receives caspase-3 siRNA loses caspase-3 activity. Similarly, the percent of orthochromatic normoblasts and reticulocytes that develop in siRNA-treated cultures is reduced by about 50% compared with a control culture (Figures 3D, 4, and 5), indicating that the reduction in caspase-3 activity blocks erythropoiesis. The accumulation of pronormoblasts in siRNA-treated cultures points to a developmental arrest at the pronormoblast stage. An early time point for the action of caspase-3 is also supported by our result that introduction of siRNA into the cultures at day 3 or day 7 does not block erythropoiesis to the same extent, if at all, compared with addition at day 0. Our results confirm and extend the observations of Zermati et al, who showed that caspase inhibitors blocked erythropoiesis prior to the basophilic normoblast stage.19
These observations support a scenario in which approximately half of the pronormoblast population at day 7 continues along the developmental pathway and half is blocked at the pronormoblast stage, although apparently able to go through several more cell divisions without further maturation. Alternatively, we cannot entirely exclude that some cells that receive caspase-3 siRNA are still able to develop normally, possibly due to some redundancy among the different caspases. Indeed, redundancy may be the key as to why caspase-3-deficient mice do not appear to have obvious erythropoietic defects. For example, in caspase-3-deficient mice, caspase-8 has been demonstrated to cleave the normal caspase-3 target protein PARP.28 It is further possible that such redundancy is triggered by interaction with other cell types that are not available in our ex vivo culture system. In either case, the profound disruption in erythropoiesis produced by reducing caspase-3 activity (Figures 4, 5) establishes that a nonapoptotic function of caspase-3 is necessary for optimal erythroid development. Similarly, it has been established that in mature T cells, caspase-8 is required for cell proliferation after activation.30 The next challenge will be to learn what features distinguish the action of caspases in these nonapoptotic roles from the better-understood apoptotic pathway.
Prepublished online as Blood First Edition Paper, February 19, 2004; DOI 10.1182/blood-2003-09-3362.
Supported by grants from the National Institutes of Health (NIH) (HL60889) (M.W) and by the DeWitt Wallace Fund and Memorial Sloan-Kettering Cancer Center (MSKCC) (M.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Thanks to Drs Malcolm Moore, Michel Sadelain, Thomas Mayer, and Paul Szabo for their help and advice during this work. Further thanks to Udo Többen for his assistance during manuscript preparation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal