Abstract
Heterozygous mutations of the gene encoding neutrophil elastase (ELA2) have been associated with cyclic neutropenia (CN) and severe congenital neutropenia (SCN). To date, 30 different mutations have been reported, but no correlation has been found with the degree of neutropenia. To address this issue, we analyzed the clinical, hematologic, and molecular characteristics of 81 unrelated patients with SCN (n = 54) or CN (n = 27). We identified mutations in 31 patients, two thirds of whom had sporadic forms. Familial cases were consistent with dominant inheritance. Seventeen novel mutations were identified, showing that the mutational spectrum encompasses not only the region encoding the mature enzyme but also the prodomains and promoter region. Genotype-phenotype analysis strongly suggested that ELA2 mutations correlate with more severe expression of neutropenia, specifically in patients diagnosed with SCN. This study underlines the importance of ELA2 molecular screening to identify patients who may be at particular risk of severe bacterial infections and/or acute myeloid leukemia/myelodysplasia. By phenotypic analysis of affected relatives and carriers of the same ELA2 mutations, we showed that the expression of neutropenia in CN and SCN may be either homogeneous or variable according to the type of mutations, suggesting different pathogenetic mechanisms.
Introduction
Severe chronic neutropenia is a heterogeneous group of rare disorders because of intrinsic defects of myeloid cell proliferation and maturation. Despite improved understanding of molecular and cellular factors involved in granulopoiesis, the pathogenetic mechanisms underlying these disorders are unknown. Nosological classification is thus based on clinical and hematologic characteristics of severe congenital neutropenia (SCN) and cyclic neutropenia (CN).1 Typically, SCN is defined by extremely low peripheral blood neutrophil counts (usually < 0.5 × 109/L), maturation arrest of bone marrow myeloid precursors at the promyelocyte-myelocyte stage, and recurrent bacterial infections.2 Initially described as an autosomal recessive disease in a Swedish family, most reported cases of SCN have been sporadic.3,4 A few cases consistent with autosomal dominant inheritance have been identified.5 CN is characterized by recurrent severe neutropenia (< 0.5 × 109/L) with a 21-day cycle.6 Patients are susceptible to opportunistic infections during neutropenic periods. Both sporadic and autosomal dominant inherited cases of CN have been reported.7,8 Classical forms of SCN and CN are readily distinguishable, but a continuum of phenotypes hinders clinical diagnosis in some cases. The severity of neutropenia can vary in SCN, and patients may have different clinical histories and/or hematologic profiles. Periodic oscillations have also been observed in patients with SCN.9 Likewise, the length of neutropenic periods can be variable in CN.
Heterozygous mutations of the ELA2 gene coding for neutrophil elastase (NE) have been associated with both CN and SCN.8,10 As NE protein is synthesized in promyelocytes, and SCN is characterized by bone marrow maturation arrest at this stage of differentiation, accelerated intramedullary apoptosis of promyelocytes bearing mutant NE may be involved in SCN.11,12 Molecular screening of the ELA2 gene in CN and SCN has identified about 30 different mutations.8,10,13-16 However, in the absence of detailed phenotypic studies of large cohorts of patients with severe neutropenia, no relationship has yet been shown between ELA2 mutations and clinical expression.
Here we describe the clinical, hematologic, and molecular characteristics of 81 unrelated patients with severe congenital or cyclic neutropenia. We also discuss the variable expression of neutropenia by analyzing 13 pairs of first-degree affected relatives and carriers of the same ELA2 mutations.
Patients and methods
Patients
Eighty-one patients diagnosed with severe neutropenia (SCN or CN) in French hematology departments and included in the French Neutropenia Register were screened for mutations in the ELA2 gene. Severe neutropenia was defined by an absolute neutrophil count (ANC) of less than 0.5 × 109/L on at least 3 separate occasions during the 3 months before enrollment.1 Patients were subdivided into 2 groups around the median of all ANC values recorded during the disease course, during periods without granulocyte colony-stimulating factor (G-CSF) therapy. The first subgroup consisted of 54 patients with a median ANC value of less than 0.5 × 109/L and constituted the SCN group. The second group consisted of 27 patients with a median ANC value of less than 1 × 109/L, because of oscillating neutropenia, and constituted the CN group. Patients with any of the following diagnoses were excluded from the study: autoimmune neutropenia, neutropenia associated with B- or T-lymphocyte deficiency, drug-induced neutropenia, glycogen storage disease type Ib, or Shwachman-Diamond syndrome.
Clinical investigations
The clinical histories and hematologic data collected during the disease course were recorded in the French Neutropenia Register. Demographic data, including age at diagnosis of neutropenia, severe bacterial and fungal infections, gingivitis, and hematologic complications were analyzed. A family tree with the relevant clinical history was drawn in each case. G-CSF treatment was analyzed, taking into account the dose of G-CSF per injection (microgram per kilogram) and the frequency of injections over 10-day periods.
Absolute counts of neutrophils, monocytes, platelets, and red cells were obtained from routine blood samples. Bone marrow was systematically studied in patients with SCN. The diagnoses were confirmed by systematic review of all clinical and hematologic data by 2 independent experts.
Mutation screening of the ELA2 gene
A blood sample was collected for genetic screening in every case. Written informed consent was obtained in every case, in keeping with French legislation. Genomic DNA was extracted from peripheral blood using a salting-out procedure.17 The minimal promoter and the 5 exons of the ELA2 gene were amplified by polymerase chain reaction (PCR) and purified with Millipore (San-Quentin en Yvelines, France) PCR purification kits. The purified PCR products were sequenced by using Big Dye Terminator Chemistry (Applied Biosystems, Courtaboeuf, France) on an Applied Biosystems 3100 capillary sequencer. Detailed PCR protocols and primer sequences are available on request. Direct diagnosis in relatives was performed by sequencing the corresponding PCR product.
Structural analysis of ELA2 missense mutations
Bioinformatics analyses of ELA2 missense mutations were based on the crystal structures of human NE protein reported by Navia et al18 (Protein Data Bank entry, 1HNE) and Cregge et al19 (Protein Data Bank entry, 1B0F). Structures were analyzed by using the DeepView-SwissPdbViewer program.20 The thermodynamic stability of mutants was estimated by using Popmusic21 and Fold-X.22 Multiple alignments of NE protein sequences were done by using Clustal W1.8.23
Statistical analysis
Demographic, clinical, and hematologic data and G-CSF schedules were recorded in the Access database (Microsoft, Redman, WA). Stata version 7 (Stata, College Station, TX) software was used for all statistical analyses. Unless otherwise stated, median values of hematologic and clinical characteristics were used for analysis, because of the non-gaussian data distribution. Categorical data were compared with Fisher exact test and quantitative data with the Mann-Whitney test. All tests were 2-tailed. A P value of less than .05 was considered to indicate statistical significance, unless otherwise stated.
Results
Molecular analysis of the ELA2 gene
Human NE, like other serine proteases, is synthesized as a precursor protein composed of 2 amino- and carboxy-terminal domains flanking the mature NE enzyme.24 We analyzed the sequence of the promoter region and of all the exons with their intron-exon boundaries, including the sequence of the mature enzyme and its prodomains, in the 81 unrelated patients. A heterozygous mutation was found in 31 patients. When a mutation was identified in a proband, first-degree relatives were offered specific screening, regardless of their clinical status. Among the 43 relatives thus tested, 11 were found to carry an ELA2 mutation, and all had either SCN or recurrent severe neutropenia (CN). None of the 32 first-degree relatives with normal clinical findings had detectable mutations. Cosegregation in the 11 relatives with ELA2 mutations was consistent with dominant inheritance, as all were either children or parents of the affected probands. Six probands had de novo mutations, as their parents did not carry the corresponding mutation; parental relationships were confirmed by testing a panel of 15 microsatellite markers (data not shown).
Twenty-two of the 31 identified ELA2 mutations were different (Table 1). Seventeen had not previously been reported, bringing to 47 the number of distinct mutations so far identified in the ELA2 gene (Table 1). All but 3 of the 22 distinct mutations observed here affected a residue of the mature NE protein. One was located at the methionine start codon of the amino-terminal domain, leading to a valine residue (I1 -29A>G) that should prevent the translation of the precursor NE protein; one was localized in the carboxyterminal domain (P233L), and the third was a nucleotide change (1246C>A) identified in a GC box of the promoter region, 18 base pairs upstream of the transcription start site. Of the 19 mutations located in the region encompassing exons 2 to 5, which encodes the mature NE protein, 15 were missense mutations resulting in amino acid substitutions. Two mutations identified in exon 5, a nonsense mutation (C194ter) and a one–base pair deletion (D201fsX210), led to a truncated protein. Finally, 2 mutations were located in the donor splice site of intron 4, at positions +1 and +5 (IVS4 +1 and +5G>A). These splicing defects result in the activation of a cryptic donor splice site 30 nucleotides upstream, leading to an in-frame deletion of the last 10 residues of exon 4.8
Characteristics of mutations identified in the ELA2 gene
No. . | Location . | Nucleotide change* . | Protein level† . | Mutation type . | Phenotype‡ . | Reference . |
|---|---|---|---|---|---|---|
| 1§ | Promoter | 1246C>A | — | Expression abnormality ? | 1 | This report |
| 2§ | Exon 1 | 1287A>G | I1-29A>G | Expression abnormality ? | 1 | This report |
| 3§ | Exon 2 | 1843C>T | P13L | Missense | 1 | This report |
| 4§ | Exon 2 | 1847C>A | F14L | Missense | 2 | This report |
| 5§ | Exon 2 | 1855C>T | S17F | Missense | 2 | This report |
| 6§ | Exon 2 | 1858T>C | L18P | Missense | 1 | This report |
| 7§ | Exon 2 | 1876A>T | H24L | Missense | 1 | This report |
| 8 | Exon 2 | 1882G>A | C26Y | Missense | 1 | Ancliff et al, 200113 |
| 9 | Exon 2 | 1887G>A | A28T | Missense | 1 | Dale et al, 200010 |
| 10 | Exon 2 | 1897T>C | I31T | Missense | 1 | Dale et al, 200010 |
| 11 | Exon 2 | 1900C>T | A32V | Missense | 2 | Horwitz et al, 19998 ; Ancliff et al, 200113 ; Kawaguchi et al, 200316 |
| 12 | Exon 2 | 1929T>C | C42R | Missense | 1 | Ancliff et al, 200213 |
| 13 | Exon 2 | 1929T>A | C42S | Missense | 1 | Dale et al, 200010 |
| 14§ | Exon 3 | 2190G>C | R52P | Missense | 1 | This report |
| 15§ | Exon 3 | 2192G>A | V53M | Missense | 2 | This report |
| 16§ | Exon 3 | 2199T>C | L55P | Missense | 1 | This report |
| 17 | Exon 3 | 2202G>A | G56E | Missense | 1 | Ancliff et al, 200113 |
| 18 | Exon 3 | 2249G>A | V72M | Missense | 1 | Dale et al, 200010 |
| 19§ | Exon 3 | 2310T>C | L92P | Missense | 1 | This report |
| 20 | Intron 3 | IVS3 - 8C>A | InsPQ94 | Splicing defect | 1 | Dale et al, 200010 ; Ancliff et al, 200113 |
| 21 | Exon 4 | 4495C>T | S97L | Missense | 1, 2 | Dale et al, 200010 ; Ancliff et al, 200113 ; Kawaguchi et al, 200316 ; this report |
| 22§ | Exon 4 | 4497G>C | A98P | Missense | 1 | This report |
| 23 | Exon 4 | 4534C>T | P110L | Missense | 1, 2 | Dale et al, 200010 ; this report |
| 24 | Exon 4 | 4569T>A | C122S | Missense | 1 | Ancliff et al, 200113 |
| 25 | Exon 4 | 4570G>A | C122Y | Missense | 1 | Germeshausen et al, 200114 |
| 26§ | Exon 4 | 4573T>C | L123P | Missense | 1 | This report |
| 27 | Exon 4 | 4638-4661del | V145-C152del | In-frame deletion | 1 | Dale et al, 200010 |
| 28 | Exon 4 | 4675-4715del | V157-F170del | In-frame deletion | 2 | Horwitz et al, 19998 |
| 29 | Exon 4 | 4708-4709ins5T | V168fsX184 | Nonsense | 1 | Dale et al, 200010 |
| 30 | Intron 4 | IVS4 + 1G>A | V161-F170del | In-frame deletion | 2, 1 | Horwitz et al, 19998 ; Dale et al, 200010 ; Ancliff et al, 200113 ; this report |
| 31 | Intron 4 | IVS4 + 3G>A | — | Splicing defect | 2 | Horwitz et al, 19998 |
| 32 | Intron 4 | IVS4 + 5G>A | — | Splicing defect | 2, 1 | Dale et al, 200010 ; Horwitz et al, 19998 ; this report |
| 33 | Exon 5 | 4898C>G | P176R | Missense | 1 | Ancliff et al, 200113 |
| 34 | Exon 5 | 4902T>C | L177F | Missense | 2 | Horwitz et al, 19998 |
| 35 | Exon 5 | 4913G>T | G181V | Missense | 1 | Dale et al, 200010 |
| 36 | Exon 5 | 4924G>A | G185R | Missense | 1 | Dale et al, 200010 ; this report |
| 37§ | Exon 5 | 4939G>A | V190I | Missense | 2 | This report |
| 38 | Exon 5 | 4943G>A | R191Q | Missense | 2 | Horwitz et al, 19998 |
| 39 | Exon 5 | 4945G>T | G192ter | Nonsense | 1 | Dale et al, 200010 |
| 40§ | Exon 5 | 4953C>A | C194ter | Nonsense | 1 | This report |
| 41 | Exon 5 | 4958C>A | S196ter | Nonsense | 1 | Dale et al, 200010 |
| 42 | Exon 5 | 4968C>A | Y199ter | Nonsense | 1 | Dale et al, 200010 |
| 43 | Exon 5 | 4971delC | P200fsX210 | Nonsense | 1 | Dale et al, 200010 |
| 44§ | Exon 5 | 4972delG | D201fsX210 | Nonsense | 1 | This report |
| 45 | Exon 5 | 4986delC | P205fsX210 | Nonsense | 1 | Dale et al, 200010 |
| 46 | Exon 5 | 5054C>T | P228L | Rare polymorphism | 1 | Ancliff et al, 200113 |
| 47§ | Exon 5 | 5069C>T | P233L | Missense or rare polymorphism ? | 2 | This report |
No. . | Location . | Nucleotide change* . | Protein level† . | Mutation type . | Phenotype‡ . | Reference . |
|---|---|---|---|---|---|---|
| 1§ | Promoter | 1246C>A | — | Expression abnormality ? | 1 | This report |
| 2§ | Exon 1 | 1287A>G | I1-29A>G | Expression abnormality ? | 1 | This report |
| 3§ | Exon 2 | 1843C>T | P13L | Missense | 1 | This report |
| 4§ | Exon 2 | 1847C>A | F14L | Missense | 2 | This report |
| 5§ | Exon 2 | 1855C>T | S17F | Missense | 2 | This report |
| 6§ | Exon 2 | 1858T>C | L18P | Missense | 1 | This report |
| 7§ | Exon 2 | 1876A>T | H24L | Missense | 1 | This report |
| 8 | Exon 2 | 1882G>A | C26Y | Missense | 1 | Ancliff et al, 200113 |
| 9 | Exon 2 | 1887G>A | A28T | Missense | 1 | Dale et al, 200010 |
| 10 | Exon 2 | 1897T>C | I31T | Missense | 1 | Dale et al, 200010 |
| 11 | Exon 2 | 1900C>T | A32V | Missense | 2 | Horwitz et al, 19998 ; Ancliff et al, 200113 ; Kawaguchi et al, 200316 |
| 12 | Exon 2 | 1929T>C | C42R | Missense | 1 | Ancliff et al, 200213 |
| 13 | Exon 2 | 1929T>A | C42S | Missense | 1 | Dale et al, 200010 |
| 14§ | Exon 3 | 2190G>C | R52P | Missense | 1 | This report |
| 15§ | Exon 3 | 2192G>A | V53M | Missense | 2 | This report |
| 16§ | Exon 3 | 2199T>C | L55P | Missense | 1 | This report |
| 17 | Exon 3 | 2202G>A | G56E | Missense | 1 | Ancliff et al, 200113 |
| 18 | Exon 3 | 2249G>A | V72M | Missense | 1 | Dale et al, 200010 |
| 19§ | Exon 3 | 2310T>C | L92P | Missense | 1 | This report |
| 20 | Intron 3 | IVS3 - 8C>A | InsPQ94 | Splicing defect | 1 | Dale et al, 200010 ; Ancliff et al, 200113 |
| 21 | Exon 4 | 4495C>T | S97L | Missense | 1, 2 | Dale et al, 200010 ; Ancliff et al, 200113 ; Kawaguchi et al, 200316 ; this report |
| 22§ | Exon 4 | 4497G>C | A98P | Missense | 1 | This report |
| 23 | Exon 4 | 4534C>T | P110L | Missense | 1, 2 | Dale et al, 200010 ; this report |
| 24 | Exon 4 | 4569T>A | C122S | Missense | 1 | Ancliff et al, 200113 |
| 25 | Exon 4 | 4570G>A | C122Y | Missense | 1 | Germeshausen et al, 200114 |
| 26§ | Exon 4 | 4573T>C | L123P | Missense | 1 | This report |
| 27 | Exon 4 | 4638-4661del | V145-C152del | In-frame deletion | 1 | Dale et al, 200010 |
| 28 | Exon 4 | 4675-4715del | V157-F170del | In-frame deletion | 2 | Horwitz et al, 19998 |
| 29 | Exon 4 | 4708-4709ins5T | V168fsX184 | Nonsense | 1 | Dale et al, 200010 |
| 30 | Intron 4 | IVS4 + 1G>A | V161-F170del | In-frame deletion | 2, 1 | Horwitz et al, 19998 ; Dale et al, 200010 ; Ancliff et al, 200113 ; this report |
| 31 | Intron 4 | IVS4 + 3G>A | — | Splicing defect | 2 | Horwitz et al, 19998 |
| 32 | Intron 4 | IVS4 + 5G>A | — | Splicing defect | 2, 1 | Dale et al, 200010 ; Horwitz et al, 19998 ; this report |
| 33 | Exon 5 | 4898C>G | P176R | Missense | 1 | Ancliff et al, 200113 |
| 34 | Exon 5 | 4902T>C | L177F | Missense | 2 | Horwitz et al, 19998 |
| 35 | Exon 5 | 4913G>T | G181V | Missense | 1 | Dale et al, 200010 |
| 36 | Exon 5 | 4924G>A | G185R | Missense | 1 | Dale et al, 200010 ; this report |
| 37§ | Exon 5 | 4939G>A | V190I | Missense | 2 | This report |
| 38 | Exon 5 | 4943G>A | R191Q | Missense | 2 | Horwitz et al, 19998 |
| 39 | Exon 5 | 4945G>T | G192ter | Nonsense | 1 | Dale et al, 200010 |
| 40§ | Exon 5 | 4953C>A | C194ter | Nonsense | 1 | This report |
| 41 | Exon 5 | 4958C>A | S196ter | Nonsense | 1 | Dale et al, 200010 |
| 42 | Exon 5 | 4968C>A | Y199ter | Nonsense | 1 | Dale et al, 200010 |
| 43 | Exon 5 | 4971delC | P200fsX210 | Nonsense | 1 | Dale et al, 200010 |
| 44§ | Exon 5 | 4972delG | D201fsX210 | Nonsense | 1 | This report |
| 45 | Exon 5 | 4986delC | P205fsX210 | Nonsense | 1 | Dale et al, 200010 |
| 46 | Exon 5 | 5054C>T | P228L | Rare polymorphism | 1 | Ancliff et al, 200113 |
| 47§ | Exon 5 | 5069C>T | P233L | Missense or rare polymorphism ? | 2 | This report |
— indicates not applicable; ? indicates unknown mutation effect.
Nucleotide nomenclature is given according to genomic sequence GenBank Y00477
Protein nomenclature is given considering the Ile of the mature NE as the first amino acid; mature enzyme is composed of 218 amino acids; amino- and carboxy-terminal domains are peptides of 29 and 20 residues, respectively
Phenotype 1 refers to SCN and 2 refers to CN, main phenotype associated to the mutation is numbered first
Novel mutation
Bioinformatics analysis based on multiple alignment of NE sequences of different species, secondary structure, and protein stability predictions, as well as ligand interaction analysis of modeled mutant NE proteins compared with human NE, was performed on the 15 observed missense mutations (Table 2). Most (13 of 15) involved amino acids defining the chymotrypsin-like family of serine proteases, which are conserved in the NE sequences of mouse, rat, and pig. Eleven of the 15 missense mutations modified the charge and/or polarity and/or stereochemical constraints. The study of the thermodynamic stability parameters of modeled mutant proteins suggested that 12 of the 15 amino acid substitutions located in β-sheet motifs would decrease protein stability (more specifically, missense mutations introducing or removing a proline residue) and consequently impair substrate interactions. Furthermore, 6 mutations were predicted to interact with residues defined as key positions for ligand interaction in the crystal structure of human NE. The likely consequences on NE structure or function remained ambiguous for only one mutation, namely amino acid substitution S97L.
Bioinformatics analysis of ELA2 missense mutations
. | . | . | Predicted effects*on human ELA2 protein . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| No. . | Mutation* . | Conserved residue† . | Polarity and/or charge modifications . | Stereochemical constraints . | Secondary structure . | Ligand interaction . | Phenotype‡ . | |||
| 1 | P13L | Yes | - | + | + | - | 1 | |||
| 2 | F14L | No | - | - | + | + | 2 | |||
| 3 | S17F | Yes | + | - | + | - | 2 | |||
| 4 | L18P | Yes | - | + | + | - | 1 | |||
| 5 | H24L | Yes | + | - | + | + | 1 | |||
| 6 | R52P | Yes | + | + | + | + | 1 | |||
| 7 | V53M | Yes | - | - | + | + | 2 | |||
| 8 | L55P | Yes | - | + | + | - | 1 | |||
| 9 | L92P | Yes | - | + | + | - | 1 | |||
| 10 | S97L | No | - | - | - | - | 1, 2 | |||
| 11 | A98P | Yes | - | + | - | - | 1 | |||
| 12 | P110L | Yes | - | + | - | - | 1, 2 | |||
| 13 | L123P | Yes | - | + | + | - | 1 | |||
| 14 | G185R | Yes | + | - | + | + | 1 | |||
| 15 | V1901 | Yes | - | - | + | + | 2 | |||
. | . | . | Predicted effects*on human ELA2 protein . | . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| No. . | Mutation* . | Conserved residue† . | Polarity and/or charge modifications . | Stereochemical constraints . | Secondary structure . | Ligand interaction . | Phenotype‡ . | |||
| 1 | P13L | Yes | - | + | + | - | 1 | |||
| 2 | F14L | No | - | - | + | + | 2 | |||
| 3 | S17F | Yes | + | - | + | - | 2 | |||
| 4 | L18P | Yes | - | + | + | - | 1 | |||
| 5 | H24L | Yes | + | - | + | + | 1 | |||
| 6 | R52P | Yes | + | + | + | + | 1 | |||
| 7 | V53M | Yes | - | - | + | + | 2 | |||
| 8 | L55P | Yes | - | + | + | - | 1 | |||
| 9 | L92P | Yes | - | + | + | - | 1 | |||
| 10 | S97L | No | - | - | - | - | 1, 2 | |||
| 11 | A98P | Yes | - | + | - | - | 1 | |||
| 12 | P110L | Yes | - | + | - | - | 1, 2 | |||
| 13 | L123P | Yes | - | + | + | - | 1 | |||
| 14 | G185R | Yes | + | - | + | + | 1 | |||
| 15 | V1901 | Yes | - | - | + | + | 2 | |||
- indicates no predicted effect; +, predicted effect.
Bioinformatics analysis is based on the crystal structure of human ELA2 protein (Protein Data Bank ID, 1HNE)
Conserved residues in NE sequences of pig, rat, and mouse
Phenotype 1 refers to patients with SCN and 2 refers to patients with CN
Relationships between the ELA2 genotype and clinical and hematologic characteristics
For this analysis, patients were divided into 2 groups on the basis of all ANC values collected during the disease course, in periods without G-CSF treatment. Neither the clinical history nor bone marrow findings were taken into account. The first group consisted of 54 patients with a median ANC value below 0.5 × 109/L (SCN group). The second group consisted of 27 patients with a median ANC value below 1 × 109/L, due to oscillations between < 0.5 to 1.5 × 109/L (CN group). Variable periods of severe neutropenia (< 0.5 × 109/L) were observed in the CN group. We determined the frequency of ELA2 mutations in both groups and analyzed the presence (NM genotype) or absence (NN genotype) of ELA2 mutations according to the clinical history and hematologic data. ELA2 mutations were found in 31 (38%) of the 81 patients studied, 19 (35%) of the 54 patients with SCN and 12 (44%) of the 27 patients with CN.
Severity and complications of neutropenia according to the ELA2 genotype. Four parameters were analyzed to assess the severity of neutropenia and its complications: (1) age at diagnosis of neutropenia, (2) blood and bone marrow data, (3) G-CSF treatment, and (4) complications (infections, gingivitis, and leukemic transformation).
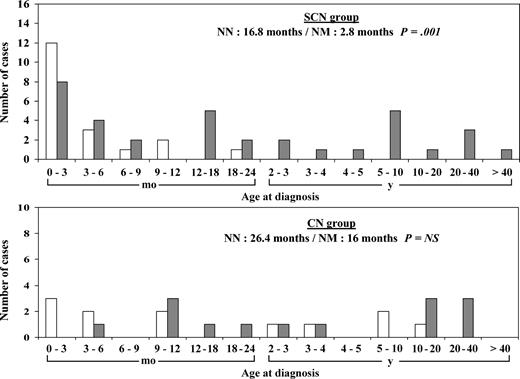
In the SCN group, the presence of an ELA2 mutation correlated with a younger median age at diagnosis (2.8 months [0.2 year], versus 16.8 months [1.4 years], P = .001) (Figure 1). Ninety-five percent (18 of 19) of the patients with NM genotype were diagnosed before age 18 months, compared with 57% (20 of 35) of the patient with NN genotype. No such difference was observed in the CN group, in which neutropenia was diagnosed at age older than 18 months in about 50% of cases irrespective of the ELA2 genotype (Figure 1).
Age distribution at diagnosis of neutropenia. Age at diagnosis is given in months for patients younger than 2 years and in years above this cutoff. ▦ indicate patients with no ELA2 mutation (NN genotype); and □, patients with an ELA2 mutation (NM genotype).
Age distribution at diagnosis of neutropenia. Age at diagnosis is given in months for patients younger than 2 years and in years above this cutoff. ▦ indicate patients with no ELA2 mutation (NN genotype); and □, patients with an ELA2 mutation (NM genotype).
The hematologic profile was analyzed on the basis of median values for an average of 17 blood samples per patient collected during the disease course, in periods without G-CSF (Table 3). In the SCN group, neutrophil counts were significantly lower in patients with NM genotype than in patients with NN genotype (0.13 × 109/L and 0.38 × 109/L, respectively; P = .002). Patients with ELA2 mutations in both subgroups had significantly elevated circulating monocyte counts (1.33 × 109/L versus 0.64 × 109/L in the SCN group; P = .01, and 0.69 × 109/L versus 0.36 × 109/L in the CN group; P = .007). Analysis of bone marrow smears showed typical maturation arrest at the promyelocyte-myelocyte stage in all patients with the NM genotype of the SCN group, with a significant decrease in the percentage of myeloid precursors and neutrophils (Table 3). Two thirds (14 of 21) of the patients with the NN genotype had a similar myeloid differentiation profile. Among patients with CN, the median ANC value did not differ between the NM and NN genotypes. Retrospective analysis of all blood samples showed that the 12 CN patients with ELA2 mutations had more regular periods of severe neutropenia than their mutation-free counterparts.
Demographic and hematologic profile by ELA2 genotype
. | SCN group . | . | . | CN group . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | NN . | NM . | P . | NN . | NM . | P . | ||||
| Peripheral blood | ||||||||||
| No. patients | 35 | 19 | — | 15 | 12 | — | ||||
| Mean no. of analyzed blood samples | 16 | 20 | — | 13 | 21 | — | ||||
| Neutrophils, × 109/L* | 0.38 | 0.13 | .002 | 0.84 | 0.64 | NS | ||||
| Monocytes, × 109/L* | 0.64 | 1.33 | .01 | 0.36 | 0.69 | .007 | ||||
| Bone marrow cytology, normal range* | NA† | NA | — | |||||||
| No. patients. | 19 | 19 | — | |||||||
| Myeloblasts, 0.3-2.4, % | 2.7 | 3.5 | NS | |||||||
| Promyelocytes-myelocytes, 12-25, % | 13.2 | 4.9 | .002 | |||||||
| Metamyelocytes-neutrophils, 33-48, % | 13.3 | 3.1 | .002 | |||||||
. | SCN group . | . | . | CN group . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | NN . | NM . | P . | NN . | NM . | P . | ||||
| Peripheral blood | ||||||||||
| No. patients | 35 | 19 | — | 15 | 12 | — | ||||
| Mean no. of analyzed blood samples | 16 | 20 | — | 13 | 21 | — | ||||
| Neutrophils, × 109/L* | 0.38 | 0.13 | .002 | 0.84 | 0.64 | NS | ||||
| Monocytes, × 109/L* | 0.64 | 1.33 | .01 | 0.36 | 0.69 | .007 | ||||
| Bone marrow cytology, normal range* | NA† | NA | — | |||||||
| No. patients. | 19 | 19 | — | |||||||
| Myeloblasts, 0.3-2.4, % | 2.7 | 3.5 | NS | |||||||
| Promyelocytes-myelocytes, 12-25, % | 13.2 | 4.9 | .002 | |||||||
| Metamyelocytes-neutrophils, 33-48, % | 13.3 | 3.1 | .002 | |||||||
NS indicates not significant; NA, not analyzed; NN genotype, patient with no ELA2 mutation; NM genotype, patient with a heterozygous ELA2 mutation; and —, not applicable.
Measurements are expressed as the median of counts
No analysis of bone marrow data in patients with CN because of variable periods of neutropenia
G-CSF was prescribed to 48 (59%) of the 81 patients overall, and to 33 (61%) of the 54 patients with SCN (Table 4) and 15 (55%) of the 27 patients with CN. In the SCN group, the presence of an ELA2 mutation correlated with the need for G-CSF therapy, as 95% of patients with the NM genotype and 43% of the patients with NN genotype received G-CSF (P = .0001). Furthermore, patients with NM genotype required a higher frequency of G-CSF injections (P = .007). These relationships with the ELA2 genotype were less striking in the CN group (75% of patients with NM genotype and 40% of patients with NN genotype received G-CSF). As expected, the median dose of G-CSF per injection was higher in the SCN group (9.5 μg/kg) than in the CN group (4.8 μg/kg), regardless of the ELA2 genotype.
Characteristics of G-CSF therapy by ELA2 genotype
. | SCN group . | . | . | CN group . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | NN . | NM . | P . | NN . | NM . | P . | ||||
| No. patients | 35 | 19 | — | 15 | 12 | — | ||||
| Prevalence of patients undergoing G-CSF therapy, n (%) | 15 (43) | 18 (95) | .0001 | 6 (40) | 9 (75) | NS | ||||
| Median G-CSF dose per injection, μg/kg (minimum-maximum) | 9.2 (2-32) | 9.9 (2.8-36) | NS | 4.2 (1.6-5) | 5.1 (2-9.9) | NS | ||||
| Frequency of G-CSF injections* | 2.1 | 5.3 | .007 | 5 | 2.5 | NS | ||||
. | SCN group . | . | . | CN group . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | NN . | NM . | P . | NN . | NM . | P . | ||||
| No. patients | 35 | 19 | — | 15 | 12 | — | ||||
| Prevalence of patients undergoing G-CSF therapy, n (%) | 15 (43) | 18 (95) | .0001 | 6 (40) | 9 (75) | NS | ||||
| Median G-CSF dose per injection, μg/kg (minimum-maximum) | 9.2 (2-32) | 9.9 (2.8-36) | NS | 4.2 (1.6-5) | 5.1 (2-9.9) | NS | ||||
| Frequency of G-CSF injections* | 2.1 | 5.3 | .007 | 5 | 2.5 | NS | ||||
NS indicates not significant; NN genotype, patient with no ELA2 mutation; NM genotype, patient with a heterozygous ELA2 mutation; and —, not applicable.
Frequency of G-CSF injections is given for a 10-day period
Complications were studied during periods without G-CSF therapy (Table 5). In the SCN group, the prevalence of bacterial infections (mainly cellulitis and pneumonia) was significantly higher in patients with NM genotype than in patients with NN genotype (P = .003). Recurrent infections were significantly more frequent in patients with ELA2 mutations (P = .008). Infections were rare in patients with CN, but they were nonetheless more frequent in patients with mutations (P = .006). Gingivitis, a chronic complication of neutropenia, was observed in all patients. However, in the SCN group, recurrent gingivitis was significantly more frequent in patients with mutations (P = .03). Four patients developed hematologic malignancies (myelodysplasia in 2 cases, acute myeloid leukemia and acute lymphoblastic leukemia in 1 case each). All these patients had SCN, ELA2 mutations, and G-CSF therapy. The cumulative incidence of leukemic transformation was thus 21% in patients with SCN associated with ELA2 mutations, after a median follow-up of 8 to 9 years. Interestingly, the G-CSF dose per injection in these 4 patients was markedly higher than in the other patients of this group with the NM genotype (16.8 μg/kg versus 6.9 μg/kg, P = .04), whereas the frequency of G-CSF injections was similar.
Prevalence of severe infections, gingivitis, and leukemic transformation by ELA2 genotype
. | SCN group . | . | . | CN group . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | NN . | NM . | P . | NN . | NM . | P . | ||||
| No. patients | 35 | 19 | — | 15 | 12 | — | ||||
| Period of follow-up without G-CSF* | 9.4 | 4.1 | — | 7.4 | 8.2 | — | ||||
| Prevalence of infections, n | ||||||||||
| All infections (%) | 15 (42) | 16 (84) | .003 | 3 (20) | 9 (75) | .006 | ||||
| Omphalitis | 3 | 4 | NS | 0 | 1 | NS | ||||
| Cellulitis | 7 | 11 | .006 | 1 | 4 | NS | ||||
| Perianal abscess | 5 | 5 | NS | 0 | 2 | NS | ||||
| Pneumonia | 6 | 11 | .003 | 2 | 2 | NS | ||||
| Sepsis | 1 | 3 | NS | 1 | 2 | NS | ||||
| Aspergillosis | 3 | 2 | NS | 3 | 1 | NS | ||||
| Prevalence of gingivitis, n (%) | 17 (49) | 11 (58) | NS | 5 (33) | 5 (42) | NS | ||||
| Prevalence of leukemic transformation, n (%) | 0 | 4 (21) | .04 | 0 | 0 | — | ||||
. | SCN group . | . | . | CN group . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | NN . | NM . | P . | NN . | NM . | P . | ||||
| No. patients | 35 | 19 | — | 15 | 12 | — | ||||
| Period of follow-up without G-CSF* | 9.4 | 4.1 | — | 7.4 | 8.2 | — | ||||
| Prevalence of infections, n | ||||||||||
| All infections (%) | 15 (42) | 16 (84) | .003 | 3 (20) | 9 (75) | .006 | ||||
| Omphalitis | 3 | 4 | NS | 0 | 1 | NS | ||||
| Cellulitis | 7 | 11 | .006 | 1 | 4 | NS | ||||
| Perianal abscess | 5 | 5 | NS | 0 | 2 | NS | ||||
| Pneumonia | 6 | 11 | .003 | 2 | 2 | NS | ||||
| Sepsis | 1 | 3 | NS | 1 | 2 | NS | ||||
| Aspergillosis | 3 | 2 | NS | 3 | 1 | NS | ||||
| Prevalence of gingivitis, n (%) | 17 (49) | 11 (58) | NS | 5 (33) | 5 (42) | NS | ||||
| Prevalence of leukemic transformation, n (%) | 0 | 4 (21) | .04 | 0 | 0 | — | ||||
NS indicates not significant; NN genotype, patient with no ELA2 mutation; NM genotype, patient with a heterozygous ELA2 mutation; and —, not applicable.
Expressed as the median time in years
Expression of neutropenia in carriers of ELA2 mutations. Finally, we studied the hematologic profile and clinical expression of neutropenia in affected relatives and also in unrelated patients carrying the same ELA2 mutations. We first analyzed the severity of neutropenia in 9 families with at least 2 affected members. Thus, 13 pairs of affected first-degree relatives (10 parent-child and 3 sibling pairs) were examined. On the basis of ANC levels, as described in “Patients,” both members of all but one of the pairs were classified in the same group of neutropenia (ie, SCN [3 pairs] or CN [9 pairs]). The discordant pair corresponded to a parent and a child with median ANC values of 1.4 × 109/L and 0.1 × 109/L, respectively; the child had more severe neutropenia, more frequent infections, and higher G-CSF requirements. Despite the similar neutrophil counts in the other 12 pairs, a marked difference in G-CSF requirements was observed, especially in familial cases of CN. Four of the 9 CN pairs had markedly different G-CSF requirements (eg, one patient needed regular injections, whereas the affected relative never received G-CSF). Interestingly, the ELA2 mutations found in these 4 families (S97L in 2 cases, IVS4 + 1G>A and IVS4 + 5G>A) have already been linked to CN or SCN.8,10,13
To determine the contribution of ELA2 mutations to the variability of neutropenia, we analyzed the clinical and hematologic profile of unrelated patients carrying the same mutations. Two mutations (G185R and S97L) could be studied, respectively, in 3 and 4 unrelated patients. The G185R mutation was always associated with severe neutropenia. All 3 carriers were aged younger than 3 months at diagnosis, had typical bone marrow maturation blockade at the promyelocytic stage, and had a median ANC value below 0.1 × 109/L. Two of them required high-dose G-CSF therapy (above 15 mg/kg) and developed hematologic malignancies, whereas the third patient was refractory to 100 μg/kg/d G-CSF. The S97L mutation was identified in 4 patients, 2 with CN (ANC between 0 and 2.7 × 109/L) and 2 with SCN (ANC always below 0.5 × 109/L). The 4 patients required similar doses of G-CSF per injection (5-10 μg/kg) but different frequencies of injections. G-CSF requirements were higher in the 2 patients with SCN.
Discussion
In the largest such series reported to date, we describe the spectrum of ELA2 gene mutations and the clinical and hematologic characteristics of 81 unrelated patients with severe neutropenia.
Thirty-one patients had a total of 22 different ELA2 gene mutations, 17 of which were previously unknown. Our results show that the mutational spectrum encompasses not only the coding region of the mature enzyme but also the prodomains and promoter region. The NE prodomains are eliminated by proteolysis during processing of the mature enzyme.25 Surprisingly, we identified a variant sequence affecting the translation start site of the amino-terminal domain. As the amino-terminal peptide is required for enzymatic activation of human NE, and as the downstream initiation codon is located in exon 2 of the sequence encoding the mature enzyme, an upstream translation start site may be used. We also found an amino acid substitution affecting a proline residue (P233L) in the carboxy-terminal domain. Its functional consequences are unknown, but it is noteworthy that all reported nonsense mutations are located in the last exon, giving rise to truncated proteins lacking the carboxy-terminal domain. Recently, Benson et al26 suggested, by analogy with canine cyclic neutropenia, that the C-terminal domain of NE may interact with an adaptator protein complex 3 (AP3) involved in protein trafficking. An abnormality of the C-terminal domain because of mutations might, therefore, impair the intracellular trafficking of NE. However, Ancliff et al13 reported the mutation P228L located in the same region and suggested that it might be a rare polymorphism, being detected in one of 110 control subjects. It has also been reported that the carboxy-terminal prodomain is not essential for the proteolytic activity, folding, activation, and granular targeting of NE.27 Finally, in the promoter region, we identified a de novo mutation (1246C>A) in a GC-box binding sequence that can bind to transcription factors of the Sp1 family. GC-box mutations have previously been linked to decreased promoter activity.28,29
Mutations located in sequences encoding the mature enzyme mainly consisted of missense substitutions (79%); changes resulting in a truncated protein and splicing defects each represented 10.5% of mutations. Converging evidence supports the pathogenicity of missense mutations. First, structural analysis predicted changes in the NE protein structure, resulting in reduced protein stability and/or impaired function, leading to abnormal protein-ligand interactions. Second, none of the amino acid changes were detected in 120 unrelated subjects who had neutropenia without ELA2 mutations, showing that these changes were not rare polymorphisms. Finally, cosegregation of neutropenia and ELA2 mutations was observed in 9 families, 7 of which carried missense mutations.
We found that ELA2 mutations correlated with a more severe expression of neutropenia. In the group of patients with SCN, ELA2 mutations were associated with younger age at diagnosis (< 2 years) and recurrent severe bacterial infections (mainly cellulitis and pneumonia). As a result, these patients received more intensive G-CSF therapy. Four (7.4%) of the 54 patients with SCN developed myelodysplasia syndrome/acute myeloid leukemia (MDS/AML), and this proportion rose to 21% (4 of 19) among carriers of ELA2 mutations. The estimated incidence of leukemic transformation in the international SCN register is 12%.2 All 4 patients received high doses of G-CSF. Thus, the presence of an ELA2 mutation, together with the use of high G-CSF doses, might be risk factors for the development of MDS/AML. Bone marrow and blood samples confirmed a more severe impairment of granulopoiesis in patients with mutated ELA2, with blockade of granulocytic maturation at the promyelocytic stage and a relatively high monocyte count. No mutations were identified in patients with severe neutropenia and no maturation arrest. However, only 57% of patients with granulopoietic maturation arrest carried ELA2 mutations. This proportion is lower than that reported by Dale et al10 (88%), suggesting a greater genetic heterogeneity of SCN. In our series, most SCN patients with ELA2 mutations had sporadic forms. Autosomal dominant inheritance was observed in 3 families, but the study of affected members of these families showed no evidence of incomplete penetrance or mosaicism, as previously reported.14,15
In the group of patients with CN, neither age at diagnosis nor the circulating neutrophil count discriminated between carriers and noncarriers of ELA2 mutations, although carriers tended to have more bacterial infections and, therefore, higher G-CSF requirements. Familial cases were only observed among patients with ELA2 mutations. Patients with more regular oscillations, probably had the same phenotype as the pedigrees initially used for positional cloning of ELA2.8 By contrast with the initial report, they only represented 44% of patients with CN in our series, suggesting that CN is more heterogeneous than previously thought. All other patients with CN had sporadic forms, with more variable neutrophil oscillations. However, the only characteristic clearly differentiating them from patients with the classical CN phenotype was the absence of ELA2 mutations. Our results confirm the diagnostic value of ELA2 molecular screening in patients with CN, especially as it is extremely difficult to demonstrate the length of neutropenic periods in early childhood. Furthermore, identification of patients with CN lacking ELA2 mutations is of interest in the search for other possible genetic factors.
Variations in the expression of neutropenia were not clearly explained by the location or nature of the mutations.30 However, examination of the known mutational spectrum of ELA2 reveals some interesting features. In patients with SCN, mutations are spread over the entire sequence, leading to molecular alterations of the mature NE, the prodomains, and the promoter region. Most are private mutations. Mutations resulting in truncated proteins are only observed in patients with SCN. By contrast, mutations associated with CN are preferentially located in exons 2, 3, and 4 and are restricted to amino acid substitutions and splice defects in the splice donor site of intron 4, previously described as a mutational hotspot.8 Overall analysis of the 31 mutations we identified failed to show, as previously suggested by Dale et al,10 that CN mutations preferentially affect the active site, whereas SCN mutations cluster on the opposite face of the molecule. Different molecular alterations likely have different functional effects on NE protein, which may contribute to the clinical phenotype. For example, mutation G185R, which interacts directly with the catalytic site of the protein and is predicted to impair NE function, was associated with a severe phenotype in 3 unrelated patients in our series. By contrast, mutation S97L, corresponding to a change in a polar residue located on the surface of the protein, has less clear-cut predicted effects on NE. Interestingly, this variant has been associated with different phenotypes in literature and in affected relatives in our series. These results suggest that the same phenotype can be generated by several different pathogenetic mechanisms. On the one hand, molecular alterations of NE resulting in a truncated protein or affecting the catalytic site could be directly involved in the impairment of granulopoiesis and, consequently, in the observed neutropenia. In this case, the expression of neutropenia may be quite homogeneous. The pathogenicity of ELA2 mutations might be due to accelerated apoptosis of myeloid precursors.11,12 On the other hand, the variable clinical expression of neutropenia could be explained by the combination of more than one genetic factor, including NE. Recently, a zinc finger transcription factor, Gfi1, controlling the expression of granulocyte-specific genes, including ELA2, was linked to neutropenia in mice and humans.31,32 The researchers showed that Gfi1 represses ELA2 and linked these 2 genes in a transcriptional network of factors involved in myeloid differentiation.33 The genetic dissection of such networks could lead to the identification of novel candidate genes that may interact with NE and play the role of modifier genes modulating disease expression.
Prepublished online as Blood First Edition Paper, February 12, 2004; DOI 10.1182/blood-2003-10-3518.
Supported by INSERM-AFM (research grant 4MR11F), the Société d'Hématologie et Immunologie Pédiatrique, Amgen SA, and Chugai Aventis.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all the physicians and patients who participated in the study. We also thank David Young for editing the text.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal