Abstract
Immunoglobulin heavy-chain class-switch recombination (CSR) occurs between highly repetitive switch sequences located upstream of the constant region genes. However, the role of these sequences remains unclear. Mutant mice were generated in which most of the Iμ-Cμ intron was deleted, including all the repeats. Late B-cell development was characterized by a severe impairment, but not a complete block, in class switching to all isotypes despite normal germ line transcription. Sequence analysis of the Iμ-Cμ intron in in vitro activated–mutant splenocytes did not reveal any significant increase in activation-induced cytidine deaminase (AID)–induced somatic mutations. Analysis of switch junctions showed that, in the absence of any Sμ repeat, the Iμ exon was readily used as a substrate for CSR. In contrast to the sequence alterations downstream of the switch junctions, very few, if any, mutations were found upstream of the junction sites. Our data suggest that the core Eμ enhancer could be the boundary for CSR-associated somatic mutations. We propose that the core Eμ enhancer plays a central role in the temporal dissociation of somatic hypermutation from class switching.
Introduction
During B-cell development, the immunoglobulin (Ig) locus is the site of 2 types of rearrangements: V(D)J assembly that generates the variable (V) region exons at the heavy- and light-chain loci and class-switch recombination (CSR) at the heavy-chain (IgH) locus. Upon antigen challenge, mature B cells expressing IgM and/or IgD undergo diversification processes that affect both the V and the constant (C) genes. Point mutations and occasional insertions and deletions are introduced in the V regions during somatic hypermutation (SHM) and gene conversion eventually resulting in higheraffinity receptors. CSR specifically affects C genes through a deletional process whereby a downstream C-region gene is brought to proximity of a rearranged VDJ gene, allowing expression of one of the downstream isotypes (IgG, IgE, or IgA).1,2
CSR generally occurs between highly repetitive, G-rich switch (S) sequences located upstream of all the C genes except Cδ. S sequences differ both in size and in the nature of the repeats. In the mouse, Sμ, Sϵ, and Sα are composed of pentameric tandem repeats such as GGGGT, GGGCT, and GAGCT, while Sγ sequences, which also contain these elements, consist of repeats of a 49–base pair (bp) sequence.3 In addition, S sequences bind several protein complexes but the physiologic consequences of this binding are poorly understood.2
CSR involves DNA breaks within partner S sequences followed by repair and ligation through a nonhomologous end-joining (NHEJ) mechanism with looped-out deletion of the intervening DNA. The final steps of CSR involve components of the general DNA repair machinery as well as mismatch-repair mechanisms. In contrast, the early steps requiring recognition and cleavage of S DNAs are still unclear. Both double-strand breaks and staggered single-strand breaks have been involved in the early steps of CSR.4,5 Frequent mutations have also been found in the vicinity of the breakpoints in the absence of any obvious consensus sequence or homology at the junction of the recombined S sequences.2,3,6
CSR is preceded by germ line transcription of target S region, which is directed by the upstream I promoter. Activation and targeting of CSR is correlated with the ability of certain mitogens and cytokines to induce or suppress germ line transcription of specific C genes. Cis-regulatory elements located upstream of the I promoters and downstream of the IgH locus, accurate splicing of germ line transcripts, and the polarity of transcription are critical for the efficiency of CSR.2,6-8
Germ line transcription has been suggested to be necessary for the accessibility of S sequences to putative recombinases effecting cleavage.2,6,9 Several studies also showed that germ line transcripts remain on the template DNA leading to RNA-DNA hybrids10-13 and long R-loops, which may serve as substrates for putative recombinase(s).14,15
Despite extensive efforts, the molecular bases of CSR, SHM, and gene conversion are not fully understood. These processes clearly require the activation-induced cytidine deaminase (AID), a member of the RNA-editing deaminase family, specifically expressed in germinal center B cells.16-21
However, neither the target(s) of AID nor its potential cofactor(s) are precisely known and the exact role of AID is still controversial.9,22-25 It has been suggested to act on one (or more) mRNA encoding putative endonuclease(s) that cleaves DNA in the V-region genes and S sequences.16,26 In contrast, indirect genetic evidence strongly suggests that V-region genes and S sequences are themselves the substrates of AID.27,28 In addition, recent biochemical and genetic studies provide good evidence that a target for AID could be the exposed single-stranded DNA during the transcription process.29-33
An increasing body of evidence shows that AID is responsible for the DNA cleavage that initiates CSR as well as the intraswitch rearrangements.34-36 In addition, AID seems to be the only B-cell–specific factor required for CSR, as ectopic expression of this enzyme is sufficient to induce CSR in fibroblasts.37 However, other studies strongly suggest that, in addition to AID, class-specific factors regulate isotype switching.38,39
Whatever the role of AID, the S sequences are clearly (but not exclusively) the sites for cleavage. Sequencing of switch junctions unambiguously identified the tandem repeats as the main site of recombination, though in the case of Sμ, breakpoints were also found outside the tandem repeats.3,40 How the different S regions are recognized by the CSR machinery is unclear, although increasing evidence points to a role of higher-order structures in targeting CSR.6,9 Deletion of the core Sμ from the mouse genome reduced the efficiency of CSR but preserved recombination events within the remaining intron sequences.41 In contrast, deletion of most of the Iγ1-Cγ1 intron abolished switching to Cγ1.8
We thus sought to make a larger deletion of the Iμ-Cμ intron, including all the pentameric repeats and the sequences thought to play a regulatory role in μ gene expression.42-45 We show that, although severely impaired, CSR is not abolished, with a shift of switch junctions toward Iμ in the absence of proximal AID-induced mutations.
Materials and methods
Gene targeting
An Sμ targeting construct was generated using a plasmid containing an approximately 5 kilobase (kb) long 5′ arm (StuI-SpeI fragment in which StuI was replaced by NotI site) tailored with a 1.3-kb ClaI-SalI fragment encompassing a neor gene flanked by loxP sites and an approximately 5-kb 3′ arm XhoI fragment from phage MB8. An HSV tk gene was inserted in NotI site for negative selection. Embryonic stem (ES) cells (cell line CK35) were transfected by electroporation and selected using G418 (400 μg/mL) and ganciclovir (2 μM). Recombinant clones were identified by Southern blot analysis with an external probe (0.6 kb XhoI-XbaI fragment).
Two ES clones showing homologous recombination were injected into C57Bl/6 blastocysts; the male chimeras were then mated with C57Bl/6 females. Germ line transmission of the mutation was checked by Southern blot. Homozygous mutant mice N/N were mated with EIIa-cre transgenic mice (a kind gift of Dr Heiner Westphal, used under a noncommercial research license agreement from Dupont Pharma, Wilmington, DE). The progeny was checked by Southern blot for recombinase (Cre)–mediated deletion.
Northern blots
Total cellular RNA preparation and Northern blotting were carried out as described.46 Probes used for hybridization were as follows: for Cμ transcripts, a 1.2-kb XbaI-HindIII genomic fragment containing the murine Cμ1 to Cμ3 region; for γ transcripts, a 1.8-kb BamHI-SphI genomic fragment containing the Cγ3 region and cross-hybridizing with all γ constant transcripts; for mb-1 transcripts, a 0.5-kb PvuII mb-1 cDNA fragment.
RT-PCR analysis of germ line transcription
Total RNA was prepared by the Tripure technique (Roche GmbH, Mannheim, Germany). One microgram of RNA was then retrotranscribed by addition of reverse transcriptase (RT; Invitrogen, Gröningen, The Netherlands). The oligonucleotide sequences, the polymerase chain reaction (PCR) conditions, and the expected sizes of amplified products corresponding to spliced germ line transcripts were described.46
Spleen cell cultures
Splenocytes from 6- to 8-week-old mice were activated in vitro as described,46 except that we used cells at a density of 106 cells/mL. At days 0, 2, 4, and 5, aliquots of cells were removed in order to prepare RNA.
Fluorescence-activated cell sorter (FACS) analysis
Flow cytometry analysis of bone marrow cells. Bones from 6- to 8-week-old mice were flushed with 10% fetal calf serum (FCS)–containing RPMI 1640. After disaggregation and washing, cells were stained with spectral red–conjugated anti-B220 and phycoerythrin-conjugated anti–c-kit, anti-CD43, anti-CD25, anti-IgM, or anti-IgD (Southern Biotechnology, Birmingham, AL). Data were obtained on 1.5 × 104 viable cells by using a Coulter XL apparatus (Beckman Coulter, Fullerton, CA).
Flow cytometry analysis of spleen cells. At day 5 of stimulation, splenocytes (5 × 105 cells/assay) were stained with spectral red–conjugated anti-B220 and phycoerythrin-conjugated anti-IgM, anti-IgG3, or anti-IgG2b or fluorescein isothiocyanate (FITC)–conjugated anti-IgG1, anti-IgG2a, or anti-IgA (Southern Biotechnology). Data were obtained as described in the previous paragraph.
ELISA assays
Sera or supernatants from spleen cell cultures (harvested after 5 days of stimulation) were analyzed for the presence of the various Ig classes and subclasses by enzyme-linked immunosorbent assay (ELISA) as described.46
PCR and sequencing
For AID-induced mutations, in vitro stimulation of splenocytes, high– molecular weight DNA preparation, and PCR conditions were as described,36 except that we used 20 μg/mL lipopolysaccharide (LPS; Sigma, St Louis, MO) supplemented or not with 1 ng/mL of mouse recombinant interleukin 4 (IL4; PeproTech, Rocky Hill, NJ) and the cultures were stopped at day 5. PCR products were cloned in Topo I vector (Invitrogen), transfected in TG1 bacterial strain, and plated without preculture at 37°C to avoid generation of sister clones. Sequences were determined on both strands by using M13 universal primers and aligned by using the Clustalw program (Institut Pasteur, Paris, France).
Oligonucleotides
Amplification of germ line transcripts. Primers Imf, Cmr, Ig2af, Cg2ar, Ig2bf, Cg2br, Iaf, Car, Acti-4, and Acti-5 have been described.46 The primers were as follows: cEμf, 5′-GAGGATTCAGCCGAAACTGGAG-3′;5′NaeI, 5′-TCAGGTAAGAATGGCCTCTCC-3′.
Amplification of Iμ-Cμ region. The primers were as follows: 3′Eμf, 5′-CATTCTGGTCAAAACGGCTTCACAAATC-3′; 5′Sμb, 5′-GGTCTCTATTCTTTCTCAATTC-3′; 3′Sμf, 5′-GCCAGTGTCTCAGAGGGAAGCC-3′; H5′, 5′-GTAAGGAGGGACCCAGGCTAAG-3′; H3′, 5′-CAGTCCAGTGTAGGCAGTAGA-3′.
Amplification of JH4-Eμ enhancer region. The primers were as follows: VH7183, 5′-CAACCATTAGTAGTGGTGGTAG-3′; 5′Eμb, 5′-ATACACATACTTCTGTGTTCCTTTG-3′.
Amplification of switch breakpoints. Splenocytes from mutant homozygous (Δ/Δ) mice were activated with LPS for 5 days, the IgG3+ population was enriched by magnetic-activated cell separation (MACS; Miltenyi Biotec, Bergisch Gladbach, Germany), and high–molecular weight DNA was subjected to a PCR using cEμf primer located 15 nucleotides downstream of the octamer motif of Eμ enhancer and either γ3-2 or γ3-3 primers downstream of Sγ3.47 The smeary DNA with size ranging from 0.3 kb to 4.0 kb was eluted and cloned for sequencing. PCR products were cloned in Topo I vector as described in “PCR and sequencing.”
Results
Generation of mice carrying a large deletion of the Iμ-Cμ intron
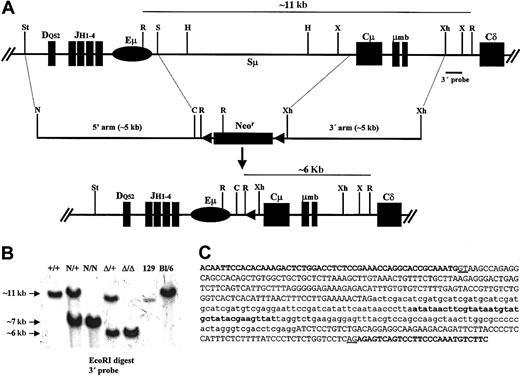
In order to delete a large portion of the Iμ-Cμ intron, a targeting vector was designed to remove all the scattered pentamers beyond the core Sμ (defined as a ∼2.9-kb HindIII fragment). Among 480 resistant clones, 2 clones showed a recombinant band with the expected size and both allowed germ line transmission. A total of approximately 4.6 kb were thus replaced by a neor cassette flanked by 2 loxP sites (Figure 1A-B). The neor cassette was removed by mating homozygous N/N mice with EIIa-Cre transgenic mice to yield heterozygous Δ/+ mice (Figure 1A-B). The removal of the neor cassette left an intron of 421 bp, including 174 bp of the loxP site and some cloning sites added in the course of making the targeting vector. From the germ line intron, 172 bp were left downstream of Iμ and 75 bp upstream of Cμ. No GGGGT, GGGCT, or GAGCT motifs were left. Conversely, no such motif was brought in by the loxP and the remaining cloning sites (Figure 1C).
Deletion of the Iμ-Cμ intron. (A) Structure of the targeted locus (not to scale). (Top) Wild-type allele showing an unrearranged IgH locus, the extent of the deletion, and the location of the 3′ probe (0.6-kb XhoI-XbaI fragment). (Middle) Structure of the targeting vector in which a neor cassette flanked by 2 loxP sites was used to replace the Iμ-Cμ intron. The proximal XhoI site was picked from the phage multiple cloning site; the endogenous StuI and SpeI sites were replaced by NotI and ClaI sites, respectively. (Bottom) The resulting locus after Cre-mediated deletion of neor cassette. C indicates ClaI; H, HindIII; N, NotI; R, EcoRI; S, SpeI; St, StuI; X, XbaI; and Xh, XhoI. Only the relevant restriction sites are shown. (B) Southern blot analysis of wild-type and mutant mice. High–molecular weight DNA was extracted from the indicated mice tails and digested with EcoRI. The 3′ probe distinguishes the germ line band (∼11 kb) from the neor recombinant band (∼7 kb) and the Δ recombinant band (∼6 kb) and enables detection of a polymorphism between 129 and C57Bl/6 strains. (C) Nucleotide sequence of the Iμ-Cμ intron after removal of the neor cassette. The distal part of Iμ exon and the proximal part of the Cμ1 exon are shown in bold capital letters. The remaining germ line intronic sequences are shown in normal capital letters; the canonical donor and acceptor splice sites are underlined. The exogenous sequences brought by the targeting vector are shown in lowercase letters with the loxP sequence in bold.
Deletion of the Iμ-Cμ intron. (A) Structure of the targeted locus (not to scale). (Top) Wild-type allele showing an unrearranged IgH locus, the extent of the deletion, and the location of the 3′ probe (0.6-kb XhoI-XbaI fragment). (Middle) Structure of the targeting vector in which a neor cassette flanked by 2 loxP sites was used to replace the Iμ-Cμ intron. The proximal XhoI site was picked from the phage multiple cloning site; the endogenous StuI and SpeI sites were replaced by NotI and ClaI sites, respectively. (Bottom) The resulting locus after Cre-mediated deletion of neor cassette. C indicates ClaI; H, HindIII; N, NotI; R, EcoRI; S, SpeI; St, StuI; X, XbaI; and Xh, XhoI. Only the relevant restriction sites are shown. (B) Southern blot analysis of wild-type and mutant mice. High–molecular weight DNA was extracted from the indicated mice tails and digested with EcoRI. The 3′ probe distinguishes the germ line band (∼11 kb) from the neor recombinant band (∼7 kb) and the Δ recombinant band (∼6 kb) and enables detection of a polymorphism between 129 and C57Bl/6 strains. (C) Nucleotide sequence of the Iμ-Cμ intron after removal of the neor cassette. The distal part of Iμ exon and the proximal part of the Cμ1 exon are shown in bold capital letters. The remaining germ line intronic sequences are shown in normal capital letters; the canonical donor and acceptor splice sites are underlined. The exogenous sequences brought by the targeting vector are shown in lowercase letters with the loxP sequence in bold.
Early B-cell development in Iμ-Cμ intron–deleted mice
The early B-cell compartment in mutant mice was analyzed by flow cytometry using a set of specific surface markers. Bone marrow cells were labeled with anti-B220 and anti–c-kit to check for pro-B cells. Normal pro-B-cell population was found in Δ/Δ mice (4.09%) compared with wild-type (wt) control (4.73%). In contrast, a 2- to 3-fold increase in the pre-B population was found in Δ/Δ mice (12.26% compared with 4.52% in wt mice) as detected by anti-B220 and anti-CD25 (see Supplemental Figure S1 on the Blood website; see the Data Set link at the top of the online article). The same increase was also detected by using anti-B220 and anti-CD43 (not shown). We then looked at IgM+ and IgD+ populations and found that surface IgM+B220+ and IgD+B220+ populations in Δ/Δ (6.43% and 4.42%, respectively) and wt mice (6.70% and 4.16%, respectively) were comparable (Figure S1).
These results indicate that deletion of the Iμ-Cμ intron leads to a slight increase in the pre–B-cell compartment but has no effect on the other developmental stages.
Serum antibodies and in vitro immunoglobulin production by mutant splenocytes
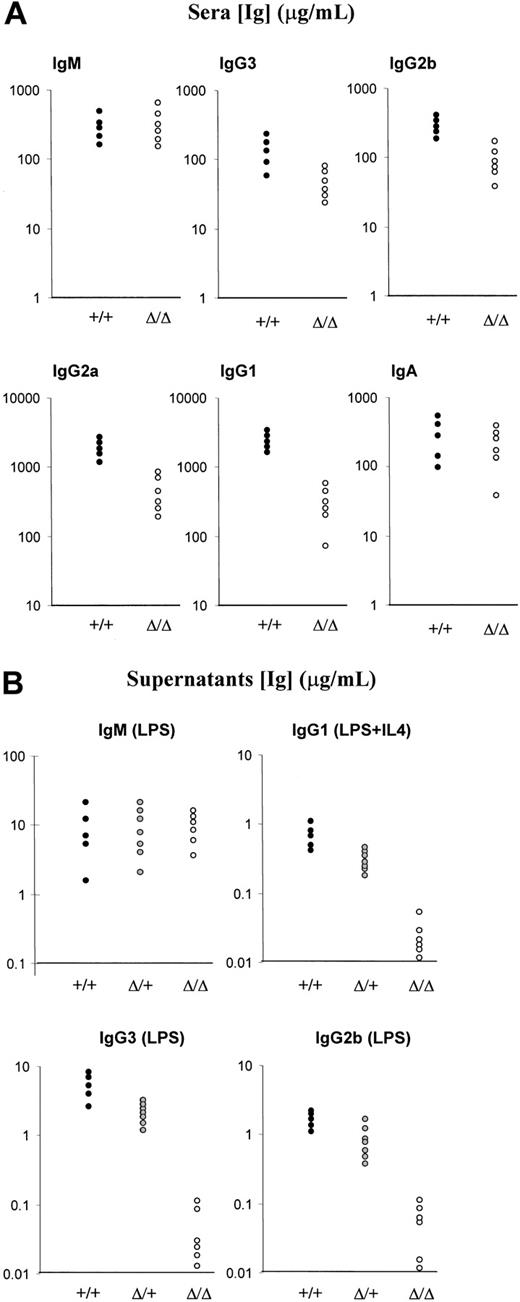
In order to analyze the sera, homozygous Δ/Δ mice were bred and the progeny bled at week 6. The serum isotype profile was analyzed by ELISA. The mutant mice displayed high titers of all isotypes tested. With regard to IgM, no difference could be detected between the mutant mice and the wt controls. As for IgG level, there was a slight decrease in IgG3 and IgG2b titers (2- to 5-fold reduction) and a relatively higher reduction in IgG1 and IgG2a (2- to 10-fold decrease). No obvious difference in IgA titers was found between the mutant and the wt mice (Figure 2A).
Analysis of Ig production in the sera and culture supernatants. (A) Ig production in unimmunized mice. Analysis of Ig secretion in 6-week-old mice was done by ELISA. Five wt mice and 6 Δ/Δ mice were analyzed. (B) ELISA analysis of Ig secretion after LPS or LPS+IL4 stimulation. Splenocytes from 5 wt (+/+), 7 heterozygous (Δ/+), and 6 homozygous (Δ/Δ) littermates were analyzed for LPS-induced IgM, IgG3, and IgG2b secretion and for LPS+IL4–induced IgG1 secretion.
Analysis of Ig production in the sera and culture supernatants. (A) Ig production in unimmunized mice. Analysis of Ig secretion in 6-week-old mice was done by ELISA. Five wt mice and 6 Δ/Δ mice were analyzed. (B) ELISA analysis of Ig secretion after LPS or LPS+IL4 stimulation. Splenocytes from 5 wt (+/+), 7 heterozygous (Δ/+), and 6 homozygous (Δ/Δ) littermates were analyzed for LPS-induced IgM, IgG3, and IgG2b secretion and for LPS+IL4–induced IgG1 secretion.
A totally different picture emerged from the analysis of Ig production in vitro. Splenocytes from littermates were stimulated with LPS supplemented or not with IL4 for 5 days, and supernatants were analyzed by ELISA. Whereas no difference was detected for IgM production between wt (+/+), heterozygous (Δ/+), and homozygous (Δ/Δ) mice, a severe impairment was found for IgG3, IgG2b, and IgG1 production. Ig levels from heterozygous splenocytes were roughly half those from wt splenocytes. In contrast, LPS-induced IgG3 and IgG2b and LPS+IL4–induced IgG1 production by homozygous mutant splenocytes was cut by about 10- to 100-fold compared with wt splenocytes (Figure 2B).
Altered cell surface immunoglobulin expression in Iμ-Cμ intron–deleted mice
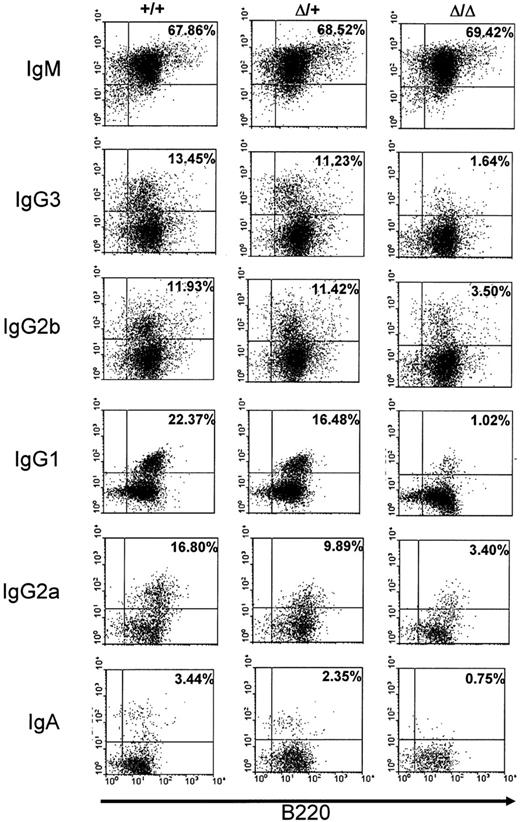
We also checked alteration of CSR in the mutant mice by studying cell surface expression of the different isotypes on activated splenic B cells. Splenocytes from wt, heterozygous, or homozygous mutant mice were activated with LPS alone to induce switching to IgG3 and IgG2b, with LPS+IL4 to induce switching to IgG1, with LPS+IFNγ to induce switching to IgG2a, and with LPS+TGFβ (transforming growth factor β) to induce switching to IgA. The percentage of surface Ig–expressing cells was quantified among total B220-positive cells. Whereas no clear-cut difference could be detected between wt and heterozygous splenocytes, a differential defect on cell surface expression was found on homozygous mutant splenocytes. The most severe defect was noted for surface expression of IgG1, IgG3, and IgA (roughly 5- to 20-fold decrease), whereas for IgG2b and IgG2a, a weaker alteration was found (2- to 5-fold decrease) in comparison with wt (+/+) splenocytes. No alteration was found for surface IgM expression in mutant splenocytes (Figure 3).
Cell surface immunoglobulin expression on stimulated splenocytes. Splenocytes from wt (+/+), heterozygous (Δ/+), or homozygous (Δ/Δ) mice were cultured for 5 days with LPS+IL4 (to induce IgG1), LPS (to induce IgG3 and IgG2b), LPS+IFNγ (to induce IgG2a), or LPS+TGFβ (to induce IgA) and stained with anti-B220 and anti-isotype antibodies. The data shown are representative of 4 independent experiments for IgM, IgG1, IgG3, and IgG2b and of 2 independent experiments for IgG2a and IgA. Controls included staining with anti-B220 and anti-IgG1 for LPS stimulation and with anti-B220 and anti-IgG3 or anti-IgG2b for LPS+IL4 (not shown). Percentages of surface Ig–expressing cells among total B220-positive cells are shown in subpanels.
Cell surface immunoglobulin expression on stimulated splenocytes. Splenocytes from wt (+/+), heterozygous (Δ/+), or homozygous (Δ/Δ) mice were cultured for 5 days with LPS+IL4 (to induce IgG1), LPS (to induce IgG3 and IgG2b), LPS+IFNγ (to induce IgG2a), or LPS+TGFβ (to induce IgA) and stained with anti-B220 and anti-isotype antibodies. The data shown are representative of 4 independent experiments for IgM, IgG1, IgG3, and IgG2b and of 2 independent experiments for IgG2a and IgA. Controls included staining with anti-B220 and anti-IgG1 for LPS stimulation and with anti-B220 and anti-IgG3 or anti-IgG2b for LPS+IL4 (not shown). Percentages of surface Ig–expressing cells among total B220-positive cells are shown in subpanels.
Altered expression of class-switched genes but normal germ line transcription in stimulated mutant splenocytes
We then asked if the pattern of class-switched gene expression would mirror that found for cell surface expression. To this end, total RNA from unstimulated, LPS-, or LPS+IL4–stimulated splenocytes was hybridized with a Cμ probe and with a Cγ3 probe that cross-hybridizes with all γ transcripts. Given that the RNA was prepared at day 5 after stimulation and that germ line transcript level is decreased by that time, the probe mainly hybridizes with class-switched RNA species. The γ gene expression is clearly depressed in homozygous mutant splenocytes whereas μ gene expression is not (Figure 4A).
Analysis of class-switched gene expression and germ line transcription. (A) Northern blot analysis of Ig gene transcription. Total RNA was isolated from wt (+/+) and mutant (Δ/Δ) unstimulated (UNS) splenocytes and from LPS- or LPS+IL4–stimulated splenocytes (day 5 after stimulation) and hybridized with a Cμ probe or with a Cγ3 probe that cross-hybridizes with all γ transcripts. The mb1 hybridization was used as a loading control. (B) Analysis of germ line transcription. RT-PCR was performed on wt (+/+) or mutant (Δ/Δ) germ line transcripts from LPS- or LPS+IFNγ–activated splenocyte's RNA (day 4) for Iμ-Cμ, Iγ2b-Cγ2b, or Iμ-Cγ2b or for Iμ-Cμ, Iγ2a-Cγ2a, or Iμ-Cγ2a germ line transcripts, respectively. Single-stranded cDNAs or dilutions thereof (1/5 and 1/25) were subjected to PCR using appropriate primers. PCR products of Iμ-Cγ2b and Iμ-Cγ2a hybrid transcripts were hybridized with the corresponding cDNAs (bottom panels).
Analysis of class-switched gene expression and germ line transcription. (A) Northern blot analysis of Ig gene transcription. Total RNA was isolated from wt (+/+) and mutant (Δ/Δ) unstimulated (UNS) splenocytes and from LPS- or LPS+IL4–stimulated splenocytes (day 5 after stimulation) and hybridized with a Cμ probe or with a Cγ3 probe that cross-hybridizes with all γ transcripts. The mb1 hybridization was used as a loading control. (B) Analysis of germ line transcription. RT-PCR was performed on wt (+/+) or mutant (Δ/Δ) germ line transcripts from LPS- or LPS+IFNγ–activated splenocyte's RNA (day 4) for Iμ-Cμ, Iγ2b-Cγ2b, or Iμ-Cγ2b or for Iμ-Cμ, Iγ2a-Cγ2a, or Iμ-Cγ2a germ line transcripts, respectively. Single-stranded cDNAs or dilutions thereof (1/5 and 1/25) were subjected to PCR using appropriate primers. PCR products of Iμ-Cγ2b and Iμ-Cγ2a hybrid transcripts were hybridized with the corresponding cDNAs (bottom panels).
We then sought to analyze germ line transcription, which is generally a prerequisite of CSR. Total RNA was prepared from appropriately stimulated splenocytes and was subjected to an RT-PCR in semiquantitative conditions, using a set of specific primers for germ line promoters and C exons. We found no obvious alteration in Iμ-Cμ and Iγ2b-Cγ2b transcripts in LPS-stimulated mutant splenocytes or in Iμ-Cμ and Iγ2a-Cγ2a transcripts in LPS+IFNγ–stimulated mutant splenocytes compared with wt controls (Figure 4B). In contrast, the hybrid transcripts in the form of Iμ-Cx (x denotes any constant gene after CSR) normally detected in B cells undergoing CSR48 were reduced in LPS- and LPS+IFNγ–stimulated mutant splenocytes (Iμ-Cγ2b and Iμ-Cγ2a, respectively; Figure 4B). We then subjected the PCR products of Iμ-Cγ2b and Iμ-Cγ2a hybrid transcripts to a Southern blot analysis in semiquantitative conditions using the corresponding cDNAs as probes. We found roughly a 5-fold decrease in Iμ-Cγ2b and a 10-fold decrease in Iμ-Cγ2a hybrid transcripts in mutant splenocytes compared with their wt counterparts (Figure 4B bottom panels), further confirming the differential switching to the different isotypes in the mutant mice.
Altogether, these results show that deletion of most of the Iμ-Cμ intron has no effect on germ line transcription but drastically affects class-switched gene expression and hybrid germ line transcription.
Lack of AID-induced somatic mutations in Iμ-Cμ intron–deleted splenocytes
Recent work34,36 has shown that AID introduces inducible DNA lesions in the Sμ region that are thought to initiate CSR. We sought to determine the mutation frequency in the Iμ-Cμ intron from homozygous mutant mice. We first amplified 868 bp between the 3′ matrix attachment region (MAR) and Cμ1 from unstimulated, LPS-, or LPS+IL4–activated Δ/Δ splenocytes. We found no significant increase in AID-induced mutations upon LPS stimulation or LPS+IL4 stimulation compared with unstimulated splenocytes (Table 1; Figure 5A). To check that these conditions induce CSR but not SHM, we also compared the sequences between JH4 and Eμ enhancer (614 bp) known to be the target of SHM in vivo, the upstream CDR3 sequences on the amplified regions being a straightforward marker for clonality. Although the mutation frequency was nearly twice that in the mutated Iμ-Cμ intron, there was no increase in mutation number in LPS- or LPS+IL4–stimulated splenocytes compared with the unstimulated ones (Table 1; Figure 5A).
Analysis of AID-induced mutations in Iμ-Cμ intron
. | Stimulation conditions . | . | . | ||
|---|---|---|---|---|---|
| Splenocytes . | UNS . | LPS . | LPS-IL4 . | ||
| Δ/Δ, no. (total no.) | |||||
| 3′Eμf/Cmr | 2/16 492 (19) | 3/17 360 (20) | 2/13 020 (15) | ||
| VH7183/5′Eμb | 5/5 526 (9) | 6/5 526 (9) | 4/3 684 (6) | ||
| +/+, no. (total no.) | |||||
| Imf/5′Sμb | 4/9 702 (18) | 7/9 163 (17) | — | ||
| 3′Sμf/Cmr | 0/7 980 (20) | 2/8 379 (21) | — | ||
| Δ/+, no. (total no.) | |||||
| Imf/Cmr | 2/13 320 (20) | 2/12 654 (19) | — | ||
| Imf/5′Sμb | 1/9 702 (18) | 3/10 241 (19) | — | ||
| 3′Sμf/Cmr | 0/8 379 (21) | 1/7 581 (19) | — | ||
| H5′/H3′ | 2/13 000 (20) | 11/14 3000 (22) | — | ||
. | Stimulation conditions . | . | . | ||
|---|---|---|---|---|---|
| Splenocytes . | UNS . | LPS . | LPS-IL4 . | ||
| Δ/Δ, no. (total no.) | |||||
| 3′Eμf/Cmr | 2/16 492 (19) | 3/17 360 (20) | 2/13 020 (15) | ||
| VH7183/5′Eμb | 5/5 526 (9) | 6/5 526 (9) | 4/3 684 (6) | ||
| +/+, no. (total no.) | |||||
| Imf/5′Sμb | 4/9 702 (18) | 7/9 163 (17) | — | ||
| 3′Sμf/Cmr | 0/7 980 (20) | 2/8 379 (21) | — | ||
| Δ/+, no. (total no.) | |||||
| Imf/Cmr | 2/13 320 (20) | 2/12 654 (19) | — | ||
| Imf/5′Sμb | 1/9 702 (18) | 3/10 241 (19) | — | ||
| 3′Sμf/Cmr | 0/8 379 (21) | 1/7 581 (19) | — | ||
| H5′/H3′ | 2/13 000 (20) | 11/14 3000 (22) | — | ||
The number of mutations per total number of bases sequenced is indicated for each category of splenocytes under the corresponding stimulation conditions. The total number of independent clones sequenced is indicated in the parentheses. The maps (not to scale) in Figure 5 indicate the relative position of the primers used and the sequences examined on the alleles of the homozygous mutant (Δ/Δ) (A), the wt (+/+) (B), and the heterozygous (Δ/+) (C) splenocytes.
— indicates not applicable.
Analysis of AID-induced mutations in Iμ-Cμ intron. The number of mutations per total number of bases sequenced is indicated for each category of splenocytes under the corresponding stimulation conditions. The total number of independent clones sequenced is indicated in Table 1. The maps (not to scale) indicate the relative position of the primers used and the sequences examined on the alleles of the homozygous mutant (Δ/Δ) (A), the wt (+/+) (B), and the heterozygous (Δ/+) (C) splenocytes.
Analysis of AID-induced mutations in Iμ-Cμ intron. The number of mutations per total number of bases sequenced is indicated for each category of splenocytes under the corresponding stimulation conditions. The total number of independent clones sequenced is indicated in Table 1. The maps (not to scale) indicate the relative position of the primers used and the sequences examined on the alleles of the homozygous mutant (Δ/Δ) (A), the wt (+/+) (B), and the heterozygous (Δ/+) (C) splenocytes.
One possibility to account for the lack of increased mutation frequency in the altered Iμ-Cμ intron could be that the remaining sequences are not prone to a high frequency of mutation even in the wt locus; we therefore sought to analyze splenocytes from wt (+/+) mice. This was achieved by sequencing the regions corresponding to those remaining in the mutated intron (3′ to Iμ and 5′ to Cμ1 exons); the length of the regions to be sequenced in the wt alleles was extended to include an equivalent number of nucleotides to that brought by the targeting vector (loxP site and cloning sites) in the mutant allele. A slight accumulation of mutations was found in the 5′ region (539 bp) as well as in the 3′ region (399 bp; 7 and 2 mutations, respectively) from LPS-activated splenocytes, although a higher “background” was found in the 5′ region from unstimulated splenocytes (4 mutations; Table 1 and Figure 5B). This could be due to some amplified sequences from preactivated splenocytes, from memory B cells, or from some rare class-switched splenocytes.
Altogether, these results suggest that the remaining sequences in the mutated Iμ-Cμ intron are poor targets of AID and that the deleted sequences offer better substrates for AID. We therefore asked if in heterozygous (Δ/+) splenocytes, AID will specifically target the Sμ sequences on the wt allele or whether it will have a bystander effect on the mutated allele. To this end, we sequenced the same regions as those sequenced in homozygous wt (+/+) and (Δ/Δ) mutated splenocytes. In addition, we sequenced as an internal control the proximal part of the core Sμ known to accumulate somatic mutations at a relatively high frequency in activated B cells.34,36 With regard to the regions in the vicinity of Iμ and Cμ1 on the wt allele and the Iμ-Cμ region on the mutated allele, the results show essentially the same pattern of low accumulation of mutations to those found in the corresponding regions from +/+ and Δ/Δ homozygous splenocytes (Table 1 and Figure 5). In contrast, the region including the 5′ part of the core Sμ (650 bp) on the wt allele clearly displays a higher number of mutations in LPS-activated splenocytes compared with the unstimulated ones (11 and 2 mutations, respectively; Table 1 and Figure 5C; Figure S2).
These data strongly suggest that AID specifically targets the Sμ sequences on the wt allele and has no obvious bystander effect on the mutated allele.
Iμ is readily used as a substrate for CSR in the absence of mutations upstream of switch junctions in Iμ-Cμ intron–deleted mice
Having shown that deletion of the major portion of the Iμ-Cμ intron leads to a severe impairment but not a complete abrogation of CSR and that this decrease is likely to be due to a decrease in CSR initiation as reflected by the lack of AID-induced mutations, we sought to analyze the junction sites and the flanking mutations on class-switched alleles. We sequenced 27 independent clones, the sequence of the recombined Sγ3 being the marker for clonality. The junctions fall into 2 groups: 18 breakpoints (∼67%) were found within Iμ and 9 breakpoints (∼33%) within the mutant Iμ-Cμ intron (Figure 6A group A and group B, respectively; Figure S3). Of the latter breakpoints, only 1 junction occurred within the loxP site (clone TS17), clearly indicating that the exogenous sequences have no effect on the ongoing CSR. Computer search for secondary structures showed that of the 27 switch junctions, 2 breakpoints occurred in single-stranded loops and 10 in necks49 (Figure S4).
CSR junctions in Iμ-Cμ intron–deleted mice and potential cryptic splice site in Iμ exon. (A) Localization of switch donor sites. The donor sites fall into 2 groups, group A (▴) comprises the switch donor sites within Iμ and group B (▵) those within the mutated Iμ-Cμ intron. The relative position of cEμf primer is indicated. cEμ indicates the core Eμ enhancer; H, HindIII; R, EcoRI; and X, XbaI. (B) Putative cryptic splice site in Iμ exon. (Top) Relative position of the primers used in RT-PCR. (Bottom) The first line shows the localization of the putative cryptic donor splice site (underlined) downstream of the octamer motif of the core Eμ enhancer. Three transcription initiation sites52 are indicated by asterisks. The second line shows the correctly spliced shortened Iμ (Iμ*) exon to Cμ exon. The third line shows the nucleotide sequence in the vicinity of Cμ acceptor splice site (underlined).
CSR junctions in Iμ-Cμ intron–deleted mice and potential cryptic splice site in Iμ exon. (A) Localization of switch donor sites. The donor sites fall into 2 groups, group A (▴) comprises the switch donor sites within Iμ and group B (▵) those within the mutated Iμ-Cμ intron. The relative position of cEμf primer is indicated. cEμ indicates the core Eμ enhancer; H, HindIII; R, EcoRI; and X, XbaI. (B) Putative cryptic splice site in Iμ exon. (Top) Relative position of the primers used in RT-PCR. (Bottom) The first line shows the localization of the putative cryptic donor splice site (underlined) downstream of the octamer motif of the core Eμ enhancer. Three transcription initiation sites52 are indicated by asterisks. The second line shows the correctly spliced shortened Iμ (Iμ*) exon to Cμ exon. The third line shows the nucleotide sequence in the vicinity of Cμ acceptor splice site (underlined).
Within the limits of our data set, neither a “hot spot” for CSR nor a clear-cut bias toward G-rich sequences in Iμ was found. No obvious microhomology was found within the 10 nucleotides flanking the switch junction. Interestingly, the most proximal switch donor site within Iμ was found in the 3′ MAR (clone TS23; Figure S3). When we looked at mutations upstream of switch junctions, very few mutations were found. In group A, only one mutation (clone TS19) and in group B, 3 mutations all clustered in a single clone (clone TS17) were found in the proximal Iμ sequences. In contrast, the usual alterations associated with CSR (point mutations, deletions, duplications) were observed downstream of Sγ3 acceptor sites (Figure S3 and data not shown). The very low frequency (or the lack) of mutations upstream of Iμ switch donor sites and the usual lesions in Sγ3 acceptor sites further confirm the results from in vitro–stimulated splenocytes: (1) the lack or the very low frequency of AID-induced mutations accounting for the decreased initiation of CSR, and (2) the exquisite specificity of AID.
Previous work strongly suggests that splicing of germ line transcripts is critical for CSR efficiency.50 In our case, all clones from group A have deleted the canonical Iμ splice donor site, which suggests that cryptic splice sites may have been activated. We adopted an RT-PCR approach on unstimulated and LPS-activated splenocyte's RNA. PCR and sequence analysis of spliced products mostly revealed normal use of the canonical splice sites (not shown), whereas a shorter and lesser abundant amplified product revealed an upstream cryptic splice site correctly spliced to the canonical Cμ1 acceptor site in LPS-activated splenocyte's RNA (Figure 6B). These results suggest that in addition to or in the absence of the canonical Iμ splice donor site, an upstream cryptic splice site on Iμ transcripts occasionally enables the splicing of Iμ-Cμ transcripts.
Discussion
We generated mice in which most of the Iμ-Cμ intron was deleted, leaving intact just the necessary sequences for correct splicing of μ transcripts. In addition to the core Sμ, the deletion removed all the remaining scattered pentamers involved in CSR41 as well as intronic sequences thought to play a regulatory role in Ig gene expression.42-45
We found no evidence for such a regulatory role in activated B cells. With regard to early B-cell development, no obvious abnormality could be detected in pro-B- and immature B-cell compartments. In contrast, we did find a 2-fold increase in the pre–B-cell compartment by FACS analysis of bone marrow, which could indicate a delayed or decreased pre–B-cell–receptor (pre-BCR) expression. The reasons for such an alteration are unclear but are unlikely to involve the absence of NFSμ-U1 and BSAP binding sites, since their deletion in the mouse genome had apparently no effect on pre-B cells.45
Late B-cell development was characterized by an impairment, but not a complete block, in CSR to all isotypes. In the serum, the decrease was in the range of 2- to 10-fold for IgG subclasses, whereas the IgA titer was unaffected, which is reminiscent of the core Sμ–deleted mice.41
In contrast, in vitro LPS- or LPS/IL4–activated Ig production was more severely decreased (10- to 100-fold reduction for IgG3, IgG2b, and IgG1), pointing to an autonomous defect in the ability to effect CSR by mutant splenocytes. Accordingly, whereas μ transcripts were equally abundant in Δ/Δ and +/+ mice, γ transcripts were clearly decreased in Δ/Δ mice.
Interestingly, germ line transcription was normal, be it from Iμ or from downstream I promoters reflecting their unaltered accessibility. In contrast, hybrid germ line transcripts in the form of Iμ-Cx (Cx being any C region gene apart from μ and δ)48 were reduced in activated mutant splenocytes. Although their function is unknown, they are a good marker for the efficiency of CSR and further show a CSR defect in mice devoid of the Iμ-Cμ intron.
In mice devoid of the core Sμ, CSR was decreased but not abrogated with an apparent bias toward the region downstream of the deletion.41 In our study, the bias was rather toward the region upstream of the deletion with a clear shift toward Iμ. The reason for this discrepancy may be due to the lack of any Sμ remnants in our mice.
Although PCR may induce some cloning bias, our sequences are likely representative of the spectrum of S junctions occurring in Δ/Δ mice, since they were derived from independent clones bearing fragments of different sizes, the different recombined Sγ3 sequences being a straightforward marker of clonality.
Based on the analysis of a large number of switch junctions from a cell line induced to switch to Cα, it has been suggested that the 3′ end of Iμ could be the upstream border for CSR, the rationale behind is that splicing (requiring the Iμ donor site) is critical for CSR.40 Our results clearly show that, in the absence of the intronic pentamers, Iμ can readily be used as a substrate for CSR. Our findings are supported by the results of the same group who found rare breakpoints involving Iμ as well as from earlier findings from myeloma cells.3,40
Requirement for a canonical splicing of Iμ may also be less stringent than expected. Thus, cryptic sites downstream of the Iμ exon were activated in cells with a mutation of the usual I donor site.51 In Iμ-Cμ intron–deleted mice, we identified by an RT-PCR approach a cryptic splice site just downstream 3 of the Iμ transcription initiation sites.52,53 This proximal site may be rarely used and it may have a physiologic meaning in that it lies upstream of all the S junctions we found in our Δ/Δ mice. Clearly, this topic requires further investigation.
Altogether, these data strongly suggest that the 3′ end of Iμ is not the 5′ border for CSR as suggested.40 Rather, we favor the view that it is the core Eμ enhancer itself that is the actual border (below).
That CSR may, in some extreme situations, be rescued by elements outside the core Sμ or even within Iμ suggests that CSR may be physiologically vital to the cells committed to this pathway. It may indicate that, upon appropriate stimulation and even in the absence of the specific motifs, the cleavage/repair machinery may somehow find its way to the cEμ-Cμ region. In that view, Sμ repeats would provide the optimal substrate for efficient recombination, while other components of the machinery would target CSR to that peculiar region. Whether this is due to the fact that this region is the initial and crucial switch donor site or whether it is a general feature of the sequences preceding C genes can now be grasped by comparing our Sμ deletion with a similar deletion of Sγ1.8 In the latter study, a nearly complete abrogation of CSR to Cγ1 was noted, whereas a less marked severity of CSR inhibition was observed in our case. Interestingly, the rare recombination events in Sγ1-deleted mice occurred in Iγ1 and not in downstream sequences of the Cγ1 locus8 similarly to the shift toward Iμ that we found.
This suggests that beyond structural (S sequences, I promoters, etc) and functional (germ line transcription, splicing, etc) similarities, there are likely other levels of regulation to account for such differential effects.
Analysis of switch junctions did not reveal any obvious microhomology indicating that the breaks were normally resolved by the NHEJ repair pathway. Strikingly, whereas the usual alterations (point mutations, deletions, duplications) were found downstream of the Sγ3 acceptor sites, indicating that recombination of Sγ3 was not the rate-limiting factor, very few (if any) mutations were found upstream of the switch junctions, questioning the contribution of an error-prone repair process in the resolution stage of CSR.
The very rare mutations upstream of the junction sites, the lack of obvious internal rearrangements in the mutant allele in preliminary experiments by Southern blot, together with the absence of a significant increase in AID-induced mutations in the Iμ-Cμ intron from mutant splenocytes or in the Iγ1-Cγ1 intron from mutant Sγ1-derived hybridomas8 have a direct bearing on the mechanism of CSR and are consistent with the model in which AID-induced mutations are the initiating event in CSR.
Computer search for secondary structures suggests that many stem-loop structures49 may form in the mutated Iμ-Cμ region. However, only about half the breakpoints were found within these structures indicating that the stem loops are not the main targets of CSR machinery.
Sequence analysis of the Iμ-Cμ intron in activated heterozygous splenocytes revealed an exquisite specificity of AID and a lack of a bystander effect on the mutated intron. This could be due to the removal, by our deletion, of DNA motifs recognized by AID or a putative cleaving enzyme or cofactor(s). Indeed, several proteins binding Sμ have been identified2 whose role is still unclear. Bearing in mind the biallelic nature of germ line transcription,8,54 this indicates that, although required, transcription per se is not sufficient for AID-induced mutations as suggested33 but needs the simultaneous formation of higher-order structures common to all S sequences, one likely candidate being the R-loop.14,15
We would like to propose the following model to account for the decreased initial cleaving activity involved in CSR in our mice. Upon appropriate stimulation, the cleaving enzyme(s) preferentially binds to the transcriptional machinery that initiates from germ line promoters similarly to what has been proposed for SHM.55,56 As transcription proceeds, the transcriptional complexes may be slowed down by the secondary (or higher) structures of S sequences generated by the first complexes, increasing the probability that the endonuclease(s) introduces mutations. In the absence of such structures, the complexes will proceed without pausing and the probability of introducing mutations will decrease. Conversely, in the absence of germ line promoter,57-59 the endonuclease(s) may still cleave the DNA, albeit at lower frequency.
In the case of μ gene, cEμ acts as both a germ line promoter and an enhancer of transcription initiating from the promoters of the variable regions (PVHs). An important implication of our model is that the endonuclease(s) will be targeted to the VH region (SHM) or to the Sμ region (CSR) depending on the commitment of cEμ. Therefore, cEμ may well be the key component of an activation-induced molecular switch that controls the temporal dissociation of SHM from CSR.
Prepublished online as Blood First Edition Paper, February 24, 2004; DOI 10.1182/blood-2003-10-3470.
Supported by grants from Association pour la Recherche sur le Cancer (grant no. 4403), Ligue Nationale Contre le Cancer, the Switch Network, and Conseil Régional du Limousin.
F.G. and Z.O. contributed equally to this work.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Marianne Brüggemann and Michael S. Neuberger for the kind gift of Cδ phages and to Chantal Kress for kindly providing ES cells. We would like to thank an unknown reviewer for very helpful comments.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal